Abstract
The incidence of HPV-related oral squamous cell carcinomas (OSCC)and oropharyngeal squamous cell carcinomas (OPSCC) has increased significantly in recent years, but the role of HPV and P16 in OSCC and OPSCC remains controversial. Here, we evaluate the prevalence and prognostic significance of HPV-DNA, HPV E6E7 and P16 protein expression in OSCC and OPSCC patients. Additionally, we explore the correlation between P16 protein expression and HPV infection in these cases. The results show that the HPV DNA infection rate was significantly higher in OPSCC at 16.7% compared to 3.6% in OSCC (P = 0.002), HPV DNA positive cases were more prevalent in poorly- differentiated cases (P = 0.009). HPV E6E7 positive cases (10.4%) were only detected in OPSCC. P16 (+++) was observed in 6 of 48 OPSCC cases (12.5%) and in 1 of 140 OSCC cases (0.7%). P16 (+++) was significantly higher in OPSCC than in OSCC (P = 0.003). However, in OPSCC, Using P16 (+++) as a marker for HPV DNA infection yielded a sensitivity of 62.5%, Kappa coefficient between HPV DNA and P16 (+++) was 0.67 (P <0.0001). HPV DNA infection and P16 (+++) were not linked to prognosis (DFS: P = 0.35, P = 0.51; OS: P = 0.99, P = 0.96), Moreover, In OSCC, Age, T, N, and clinical stages were correlated with prognosis. Our results indicate that P16 (+++) can act as a biomarker for HPV- related high-risk tumors in OPSCC, yet not in OSCC. Besides, Older patients, less favorable T, N, and clinical stages having a worse prognosis in OSCC. In those with OPSCC, only the younger age of patients was associated with a better prognosis.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) mainly denotes cancers arising in oral tissues, such as the tongue, upper and lower gums, oral floor, palate, and buccal mucosa. Oropharyngeal squamous cell carcinoma (OPSCC) principally refers to malignancies emerging in the soft palate, palatine tonsils, tongue base, regions around the epiglottis, and pharyngeal wall. Both are integral parts of Head- and -Neck squamous cell carcinoma (HNSCC). As the sixth most common cancer in the world, HNSCC accounts for nearly 700,000 new diagnoses and 350,000 deaths annually1. The multidisciplinary treatment approach for HNSCC includes surgery, anticancer drug therapy, and radiotherapy. However, metastasis to the regional lymph nodes reduces the 5-year survival rate by 50% compared to that of patients with early-stage disease2. Known risk factors for HNSCC include tobacco and alcohol use, betel nut chewing, and autoimmune disorders3,4.
In addition to these well-established risk factors, recent advances in next-generation sequencing (NGS) have significantly accelerated the identification of cancer driver genes, leading to a deeper understanding of OSCC and OPSCC biology5. OSCC and OPSCC was among the first cancers to be extensively studied using NGS with a relatively large cohort of patient tumors6. Research shows that overexpression of epidermal growth factor receptor (EGFR) has been observed in 25–77% of colon cancers and in more than 90% of HNSCC cases, establishing EGFR as a valuable biomarker for HNSCC7. Apart from potential markers related to genes, another significant trend in OSCC and OPSCC is the increasing incidence of HPV-related cases, particularly in Europe. Studies estimate that HPV is present in 24.2% of HNSCC cases overall, and in up to 67.9% of OPSCC8. However, the reported rates of HPV infection in OSCC and OPSCC are highly variable, making it a topic of ongoing global debate. In China, few studies have examined HPV-related OSCC and OPSCC, and the prevalence of HPV positivity varies widely across different regions. A study from Hong Kong reported that 20.8% of OPSCC and 29.0% of tonsillar SCC cases were HPV-related9. Conversely, a retrospective study from Harbin found only 5.31% of OPSCC cases to be HPV-positive10. Additionally, the relationship between HPV status and survival outcomes in OSCC and OPSCC patients remains unclear.
This is increasingly sparking exploration into the role of HPV in the pathogenesis of OSCC and OPSCC. Within the realm of cervical cancer research, the integration of HPV genome into the host cell genome emerges as a linchpin event. This genomic merger precipitates the constitutive activation of the viral oncoproteins E6 and E7, orchestrating a cascade of oncogenic events11. In particular, for the high-risk HPV16 and HPV18 strains, their E6 proteins engage in a sophisticated molecular dialogue with E3 ubiquitin-protein ligases. This liaison culminates in the ubiquitination and subsequent proteolytic degradation of the tumor suppressor p53, effectively dismantling a crucial cellular safeguard. Concurrently, the E7 protein wields its phosphorylating prowess to neutralize the retinoblastoma protein (Rb), liberating an upsurge in free eukaryotic transcription factor E2F (E2F). The resultant perturbation in cellular homeostasis begets an elevation in cyclin-dependent kinase inhibitor P1612,13, cumulatively propelling the initiation and advancement of cervical cancer.
P16 is a tumor suppressor protein that inhibits the progression of the cell cycle by suppressing cyclin-dependent kinases. P16 protein expression is commonly used as a surrogate marker for HPV infection in cervical cancer14. In cervical cancer, human papillomavirus (HPV) infection can trigger the expression of P1612. Notably, when evaluating P16 immunohistochemical staining, some studies have used a cut off value of 50%15, while others contend that P16 must be expressed at 70% in both the nucleus and the cytoplasm to serve as an HPV marker16. According to the 2018 College of American Pathologists Guidelines, HPV is a major cause of oropharyngeal squamous cell carcinomas, and HPV status (or P16 as a surrogate marker) is a key prognostic factor that influences clinical management17. However, conflicting data on the role of P16 expression in HPV-associated OSCC and OPSCC and its impact on clinical outcomes has left its significance uncertain. Some studies, such as A. K. Bryant et al.‘s work, suggest that P16 positivity is associated with improved prognosis18,19. while others, including J. Blahak’s findings, indicate no correlation between P16 positivity and overall survival (OS) in HNSCC patients20.
In this study, we investigated the frequency of HPV DNA infection, HPV E6E7 status and P16 protein expression in OSCC and OPSCC to assess whether P16 can serve as a predictive biomarker for HPV infection. Additionally, through a four-year follow-up, we evaluated the potential role of HPV DNA infection and P16 expression in OSCC and OPSCC prognosis.
Methods
Cohort establishment
188 patients with histologically confirmed OSCC and OPSCC from the First and the Seventh Medical Center of PLA General Hospital between 2019 and 2020 were enrolled in this study. Additionally, 30 benign biopsy samples from the head and neck area were randomly selected as controls. Samples were collected from patients with available formalin-fixed paraffin-embedded (FFPE) blocks of cancer specimens obtained post-surgery. Each tissue sample was histopathologically validated to confirm the cancer diagnosis (Supplementary Table 1).
Immunohistochemistry (IHC)
IHC was performed on available FFPE samples from patients with OSCC and OPSCC to detect P16 expression. Four-micrometer thick tissue microarray sections were deparaffinized and stained using a fully automated immunohistochemical stainer (Benchmark Ultra, Roche, USA). The deparaffinized sections underwent antigen unmasking using Cell Conditioning Solution (Roche, USA) at 100 °C for 64 min, followed by quenching of exogenous peroxidases with 3% H₂O₂ at 36 °C for 4 min. Sections were then incubated with primary P16INK4a monoclonal mouse antibody (dilution 1:200, MXB Biotechnologies, China) at 37 °C for 30 min. A universal secondary antibody reagent was applied, and staining was visualized using DAB. Cervical cancer tissue with known P16 expression served as a positive control. Staining intensity was recorded based on the proportion of positive carcinoma cells: negative (0), weak (1+: 10–25%), moderate (2+: 25–70%), or strong (3+: more than 70%). Weak, moderate, and strong staining were classified as P16-positive expression.
HPV DNA analysis
Blocks with at least 30% tumor content were selected for HPV molecular analysis. DNA was extracted from these tissues using the FFPE tissue extraction kit (AmoyDx, China). The concentration and purity of the DNA samples were assessed using a Nanodrop spectrophotometer (Thermo Fisher Scientific) and then stored at − 20 °C until use. HPV genotyping was performed with the HPV assay kit (Hybribio Corp, Guangdong), which detects 21 HPV genotypes: 15 high-risk types (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, and 68) and 6 low-risk types (6, 11, 42, 43, 44, and 81). Primers specific for the HPV L1 gene fragment were used for amplification, and the products were classified using a nylon membrane with 21 type-specific probes. The PCR amplification was performed in a 25 µl reaction volume using the TC-96/G/H (BIOER Technologies, China) with the following thermal cycling parameters: 1 cycle at 95 °C for 9 min, 40 cycles at 95 °C for 20 s, 55 °C for 30 s, and 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. Quality control for HPV genotyping included: (1) β-globin, a human housekeeping gene, as an endogenous internal control; and (2) positive and negative controls using cervical exfoliated cells.
HPV E6 and E7 mRNA analysis
Using the PinpoRNA HPV kit (Guangzhou Tuoshan Technology Co., Ltd., China), through the in-situ hybridization method, the transcripts RNA of E6 and E7 genes of 18 high-risk types of HPV were detected. These 18 types include HPV16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82. Subsequently, the binding signals were amplified by labeled long-chain single-stranded DNA probes with specific sequences. Finally, the enzyme linked to the amplified probe acts on the substrate, causing the color to be amplified and enhanced, thereby enabling the identification of specific RNA transcripts of HPV.
Statistical analysis
Statistical analyses were conducted using SPSS 19.0. Specificity, sensitivity, Kappa coefficient were calculated to assess the feasibility of P16 as a potential marker for HPV DNA and HPV E6E7 positivity. Correlations between HPV DNA status, P16 expression, and variables such as age, smoking, alcohol consumption, cancer site, tumor differentiation, T/N stage and Clinical staging were analyzed using the chi-square test. Survival analysis was performed using log-rank tests. DFS was defined as the time from diagnosis to recurrence or death, while OS was defined as the time from diagnosis to the date of death from any cause. P-values less than 0.05 were considered statistically significant.
Results
Clinical and pathological characteristics of patients
The cohort comprised 131 males (69.7%) and 57 females (30.3%), with a mean age of 62.81 years. The patient population included 48 cases of oropharyngeal carcinoma (25.5%), 46 cases of tongue carcinoma (24.5%), 21 cases of lip carcinoma (11.2%), 24 cases of buccal carcinoma (12.8%), 34 cases of gum carcinoma (18.1%), and 15 cases of oral floor carcinoma (8%). All cases were squamous cell carcinomas. We categorized patients into OPSCC and OSCC groups for further analysis. Smoking and drinking habits were noted, with 80 smokers and 81 drinkers (42.6% and 43.1%, respectively). The majority of patients had T1 or T2 tumors (37.8% and 36.1%), and nearly half of the tumors were highly differentiated (47.9%) (Supplementary Table 1).
HPV-DNA status and P16 expression
All 30 benign biopsy samples from the head and neck area were negative for both HPV DNA and P16 immunostaining. The results of HPV testing and P16 immunostaining for patients show that the overall HPV DNA infection rate in OPSCC and OSCC was 6.9% (HPV16+: 11, HPV31+: 1, HPV16/58+: 1). The infection rate was 16.7% for OPSCC and 3.6% for OSCC, with the difference being statistically significant (P = 0.002). In OPSCC, the P16 (+++) expression showing a positive rate of 12.5% compared to 0.7% in OSCC cases. The positive rate of P16 (+++) expression in OPSCC was significantly higher than in OSCC (P = 0.003). Furthermore, we found that the HPV DNA infection rate was 26.7% in low differentiated tissues, which was significantly higher than that 5.2% in high and moderate differentiated tissues (P = 0.009). There was no significant association between HPV infection and P16 protein expression with respect to sex (P = 0.37; P = 0.28), age (P = 0.96; P = 0.83), smoking habits (P = 0.76; P = 0.26), alcohol consumption (P = 0.42; P = 0.99), T/N and clinical staging (Supplementary Tables 2 and Table 3).
Association of P16 protein expression with HPV DNA and HPV E6E7
The correlation between HPV status and P16 protein expression was analyzed using Kappa analysis. Our data revealed that 38.5% (5/13) of HPV DNA-positive tumors did not express P16, while 24.6% (43/175) of HPV-negative tumors were P16-positive, HPV E6E7 positivity (5 out of 48, 10.4%) was only observed in OPSCC cases with P16 (+++) expression (Supplementary Table 4). If we define only P16 (+++) as positive P16 protein expression, in oral squamous cell carcinoma (OSCC), there were no cases of P16 (+++), and there was no correlation between P16 (+++) and HPV DNA status (P = 0.85). However, in oropharyngeal squamous cell carcinoma (OPSCC), The sensitivity of P16 (+++) relative to HPV DNA status was 62.5%, the specificity was 97.5%, and there was a significant correlation between P16 (+++) and HPV DNA status (P < 0.0001). In both OSCC and OPSCC patients, the sensitivity of P16 (+++) relative to HPV E6E7 status was 100%, the specificity was 98.9%, and there was a significant correlation between P16 (+++) and HPV E6E7 status (P < 0.0001) (Supplementary Table 5).
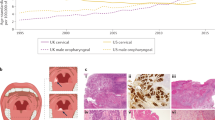
However, if we define P16 (+ - +++) as positive P16 protein expression, then in neither OSCC nor OPSCC was there a correlation between P16 (+ - +++) and HPV DNA status (P = 0.07; P = 0.06) (Supplementary Table 4). The images of Hematoxylin and eosin staining (HE), P16 immunohistochemistry, and the status of HPV E6E7 in different cases are shown in Fig. 1.
Representative images of Hematoxylin and eosin (H&E) staining, P16 immunohistochemistry and HPV E6E7 status in patients with OSCC and OPSCC of different sites. A–C HPV DNA negative, P16(+++) and HPV E6E7 negative in the gingiva area. D–F HPV DNA positive, P16(-) and HPV E6E7 negative in the lip area. G–I HPV DNA positive, P16(+++) and HPV E6E7 positive in the oropharynx area. J–L HPV DNA positive, P16(+) and HPV E6E7 negative in the tongue area.
Survival of patients with OSCC and OPSCC according to P16 expression, HPV DNA status, clinical and pathological characteristics
To further investigate the relationship between HPV DNA infection, P16 (+++) expression, and prognosis in OSCC and OPSCC patients, we conducted a follow-up study on 181 patients. The results indicated no significant differences in 4-year DFS or OS between patients with P16 (+++) and P16 (- ~ ++) expression tumors (DFS: hazard ratio [HR], 1.47, 95% CI: 0.37–5.90, P = 0.51; OS: HR, 0.96, 95% CI: 0.14–6.76, P = 0.96). No differences were observed in 4-year survival between patients with HPV-positive and HPV-negative tumors (DFS: HR, 0.67, 95% CI: 0.25–1.82, P = 0.35; OS: HR, 0.99, 95% CI: 0.23–4.21, P = 0.99) (Fig. 2). Our study concludes that HPV and P16 status do not correlate with the prognosis of OSCC and OPSCC patients.
Survival of patients with OSCC and OPSCC by HPV DNA, P16 expression. A–C Survival of patients with tumors positive and negative for HPV DNA status. Disease-free survival (A) and Overall survival (C). B–D Survival of patients with P16(+++) and P16(-~++) tumors by immunohistochemistry. Disease-free survival (B) and Overall survival (D).
Additionally, In OSCC, we observed that the patient’s age, T stage, N stage, and clinical stage were correlated with 4-year DFS and/or OS(DFS: age, P<0.0001, T stage, P = 0.002, N stage, P = 0.09, clinical stage, P = 0.0007;OS: age, P<0.0001, T stage, P = 0.0005, N stage, P = 0.02, clinical stage, P = 0.0007). In those with OPSCC, only patients’ age was associated with 4-year DFS (P = 0.01) (Fig. 3).
Discussion
This study examined the characteristics of OSCC and OPSCC patients tested for HPV DNA and P16 immunostaining. The HPV DNA infection rate and P16 (+++) expression in OPSCC patients were 16.7% and 12.5%, respectively, while the rates were lower in OSCC patients. These findings align with previous studies that reported HPV positivity rates between 14.5% and 48.7% for OPSCC and between 1.5% and 21.8% for OSCC21,22,23,24. The variation in HPV detection rates may be attributed to differences in sample selection and detection methods. Previous reports have also suggested that the most frequent virus type worldwide was HPV16 followed by HPV6, HPV18, HPV33 and other rare types21,25. Interestingly, we identified one case of HPV31 infection in an OPSCC patient, marking the first report of this infection type in China. Our results suggest that in the Chinese population, The positive rates of HPV DNA and p16(+++) are relatively high in OPSCC, yet this is not observed in OSCC. This might be attributed to regional differences or the fact that the oral cavity has a more open environment and is easier to clean compared to the oropharynx.
P16 expression is frequently used as a predictive biomarker for HPV-related tumors. The latest AJCC/UICC staging system for OPSCC uses P16 overexpression as a surrogate marker for HPV presence, distinguishing HPV-mediated (P16+) from HPV-unrelated (P16-) OPSCC26. In our study, when P16 (+ ~ +++) was defined as positive, P16 could not function as a marker for HPV DNA. Only the expression of P16 (+++) could serve as a marker for HPV DNA. To further explore the correlation between HPV status and P16 expression, we performed validation of HPV E6E7 in-situ hybridization on 188 tissue sections. This is because the progression from HPV infection to the integration of the HPV genome into host cells is a gradual process. In the initial stage of HPV infection, it may remain solely at the infective phase, whereas once the genome integrates into the host cells, it begins to exert an impact on tissue cells, thereby contributing to tumorigenesis. The detection of HPV E6E7 provides a more effective means of verifying the state of HPV virus integration into host cells.
We discovered that five cases tested positive for HPV E6E7, Meanwhile, these five cases were positive for HPV DNA and demonstrated P16 (+++) expression, A statistically significant difference was observed in the correlation between P16 (+++) expression and HPV DNA/HPV E6E7. For other cases with positive HPV DNA but P16 (- - ++), we postulate that this could be attributed to HPV remaining solely at the infective stage. Additionally, the poor correlation between P16 expression and HPV status may be due to genetic and epigenetic instability caused by HPV oncoproteins27, while other mechanisms unrelated to HPV may drive P16 expression in some cases28. Notably, HPV E6E7 positivity was detected solely in OPSCC. This might suggest to us that there is a subset of HPV - related tumors in OPSCC. Meanwhile, it seems that there might not be a strong causal link between HPV E6E7 and OSCC. I. H. Nauta et al. have also reported that HPV E6 mRNA was detected in only 2.2% of OSCC cases in the Netherlands29. Our findings suggest that although HPV infection may be a risk factor for OPSCC, it plays a minor role in OSCC.
The prognostic significance of HPV and P16 status in OSCC and OPSCC remains unclear. We assessed the DFS and OS of patients four years after surgery. Although studies have suggested that HPV positivity and P16 expression are associated with better survival30,31, our results showed no significant differences in DFS and OS between HPV DNA + and HPV DNA- or P16 (+++) and P16 (-~++) patients in OSCC and OPSCC. These findings are consistent with that HPV-positive of OSCC patients did not have significantly better survival outcomes compared to HPV-negative patients32. However, the study by Mehanna et al. compared a total of 7,654 patients from different cohorts in multiple countries, suggesting that P16+/HPV + oropharyngeal squamous cell carcinoma (OPSCC) may have a relatively favorable prognosis33. In our study, the small number of HPV DNA-positive (n = 13), including those with HPV E6E7-positive(n = 5) patients in our study may limit the generalizability of our results, and larger studies are needed to better understand the relationship between HPV E6E7 status and patient outcomes in OSCC and OPSCC.
The positive expression rate of P16 (+++) was 12.5% in OPSCC, 0.7% in OSCC. Consistent with our results, studies indicated that high expression of P16 is low in OPSCC and no significant association was noted with overall DFS in the expression of P16 in OSCC and OPSCC20,34. Additionally, In OSCC, we’ve uncovered that a younger age, as well as earlier T, N, and clinical stages, suggest a better prognosis. In those with OPSCC, only the younger age of patients was associated with a better prognosis.
Conclusion
In summary, our findings indicate that in the Chinese population, the positive rates of HPV DNA and p16(+++) are relatively high in OPSCC than in OSCC. The role of HPV as a driver of OSCC requires further investigation. Furthermore, P16 (+++) expression can be reliably used as a surrogate marker for HPV DNA and HPV E6E7 infection in OSCC and OPSCC, and neither HPV DNA nor P16 (+++) status is associated with improved survival outcomes, but there is a better prognosis for younger, earlier T, N, and clinical stages in OSCC patients.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- OSCC:
-
Oral squamous cell carcinoma
- OPSCC:
-
Oropharyngeal squamous cell carcinoma
- HNSCC:
-
Head- and -Neck squamous cell carcinoma
- NGS:
-
Next-generation sequencing
- Rb:
-
Retinoblastoma protein
- E2F:
-
Eukaryotic transcription factor
- FFPE:
-
Formalin-fixed paraffin-embedded
- HE:
-
Hematoxylin and eosin staining
- DFS:
-
Disease-free survival
- OS:
-
Overall survival
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Gogna, S., Kashyap, S. & Gupta, N. Neck Cancer Resection and Dissection, StatPearls, Treasure Island (FL), (2021).
Su, Y. Y. et al. Betel nut chewing history is an independent prognosticator for smoking patients with locally advanced stage IV head and neck squamous cell carcinoma receiving induction chemotherapy with docetaxel, cisplatin, and fluorouracil. World J. Surg. Oncol. 14, 86 (2016).
Pillai, A., Adilbay, D., Matsoukas, K., Ganly, I. & Patel, S. G. Autoimmune disease and oral squamous cell carcinoma: A systematic review. J. Oral Pathol. Med. 50, 855–863 (2021).
Xia, H. et al. MultiPrime: A reliable and efficient tool for targeted next-generation sequencing, Imeta, 2 e143. (2023).
Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157 (2011).
Nair, S., Bonner, J. A. & Bredel, M. EGFR mutations in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 23, 3818 (2022).
Ndiaye, C. et al. HPV DNA, E6/E7 mRNA, and P16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 15, 1319–1331 (2014).
Lam, E. W. et al. Prevalence, clinicopathological characteristics, and outcome of human Papillomavirus-Associated oropharyngeal cancer in Southern Chinese patients. Cancer Epidemiol. Biomarkers Prev. 25, 165–173 (2016).
Meng, H. X. et al. Association of P16 as Prognostic Factors for Oropharyngeal Cancer: Evaluation of P16 in 1470 Patients for a 16 Year Study in Northeast China, BioMed research international, (2018) 9594568. (2018).
Zhao, Q. et al. Identification of transcriptionally-active human papillomavirus integrants through nanopore sequencing reveals viable targets for gene therapy against cervical cancer. J. Med. Virol. 96, e29769 (2024).
Chung, C. H. & Gillison, M. L. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin. Cancer Res. 15, 6758–6762 (2009).
Stiasny, A. et al. Immunohistochemical evaluation of E6/E7 HPV oncoproteins staining in cervical cancer. Anticancer Res. 36, 3195–3198 (2016).
da Mata, S. et al. P16 and HPV genotype significance in HPV-Associated cervical Cancer-A large cohort of two tertiary referral centers. Int. J. Mol. Sci. 22, 2294 (2021).
Prigge, E. S., Arbyn, M., von Knebel Doeberitz, M. & Reuschenbach, M. Diagnostic accuracy of P16 (INK4a) immunohistochemistry in oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Int. J. Cancer. 140, 1186–1198 (2017).
Bishop, J. A., Lewis, J. S. Jr., Rocco, J. W. & Faquin, W. C. HPV-related squamous cell carcinoma of the head and neck: an update on testing in routine pathology practice. Semin Diagn. Pathol. 32, 344–351 (2015).
Lewis, J. S. Jr. et al. Human papillomavirus testing in head and neck carcinomas: guideline from the college of American pathologists. Arch. Pathol. Lab. Med. 142, 559–597 (2018).
Bryant, A. K. et al. Prognostic role of P16 in nonoropharyngeal head and neck cancer. J. Natl. Cancer Inst. 110, 1393–1399 (2018).
Girardi, F. M., Wagner, V. P., Martins, M. D., Abentroth, A. L. & Hauth, L. A. Prevalence of P16 expression in oropharyngeal squamous cell carcinoma in Southern Brazil. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 130, 681–691 (2020).
Blahak, J. et al. HPV, protein P16 and squamous cell carcinoma of the oral cavity. Biomed. Pap Med. Fac. Univ. Palacky Olomouc Czech Repub. 164, 292–299 (2020).
Castellsague, X. et al. I.C.O.I.H.i. head, G. neck cancer study, HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J. Natl. Cancer Inst. 108, djv403 (2016).
Nopmaneepaisarn, T. et al. Low prevalence of P16-positive HPV-related head-neck cancers in Thailand: tertiary referral center experience. BMC Cancer. 19, 1050 (2019).
Dona, M. G. et al. Evolving Profile of HPV-Driven Oropharyngeal Squamous Cell Carcinoma in a National Cancer Institute in Italy: A 10-Year Retrospective Study 8 (Microorganisms, 2020).
Mehanna, H. et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck. 35, 747–755 (2013).
Janecka-Widla, A. et al. Active HPV infection and its influence on survival in head and neck squamous-cell cancer. J. Cancer Res. Clin. Oncol. 146, 1677–1692 (2020).
American Cancer Society and American Joint Committee on Cancer Workshop on Molecular. Markers in the Classification and Staging of Cancer, Atlanta, Georgia, December 13–14, 1990 Vol. 69, 1519–1644 (Cancer, 1992).
Zafereo, M. E. et al. Squamous cell carcinoma of the oral cavity often overexpresses P16 but is rarely driven by human papillomavirus. Oral Oncol. 56, 47–53 (2016).
Halec, G. et al. H.P.V.T.T.S. Group, pathogenic role of the eight probably/possibly carcinogenic HPV types 26, 53, 66, 67, 68, 70, 73 and 82 in cervical cancer. J. Pathol. 234, 441–451 (2014).
Nauta, I. H. et al. The unveiled reality of human papillomavirus as risk factor for oral cavity squamous cell carcinoma. Int. J. Cancer. 149, 420–430 (2021).
Tian, S. et al. Survival outcomes by high-risk human papillomavirus status in nonoropharyngeal head and neck squamous cell carcinomas: A propensity-scored analysis of the National cancer data base. Cancer 125, 2782–2793 (2019).
Chung, C. H. et al. P16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J. Clin. Oncol. 32, 3930–3938 (2014).
Zhu, Y. et al. Prognostic implications of human papillomavirus type 16 status in non-oropharyngeal head and neck cancer: a propensity score matching analysis. Ann. Transl Med. 7, 759 (2019).
Mehanna, H. et al. H.-E. Group, prognostic implications of P16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol. 24, 239–251 (2023).
Hashmi, A. A. et al. P16 Immunohistochemical Expression in Head and Neck Squamous Cell Carcinoma Vol. 12, e8601 (Association With Prognostic Parameters, 2020).
Author information
Authors and Affiliations
Contributions
Conceptualization, AJ.L. and Y.L.; data acquisition and analysis, ZR.L. and K.Z.; Software, XY.L.; preparation of the manuscript: AJ.L., Y.L.,and ZR.W.; supervision, LW.XS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethical approval and consent to participate
The protocol of this research has been approved by the Ethics Committee of the First and the Seventh Medical Center of PLA General Hospital. The research complied with the Declaration of Helsinki. All patients have signed written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, Y., Li, Z., Zhang, K. et al. Correlation and prognosis analysis of human papillomavirus infection and P16 expression in oral and oropharyngeal squamous cell carcinomas. Sci Rep 15, 11270 (2025). https://doi.org/10.1038/s41598-025-95643-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95643-1
Keywords
This article is cited by
-
p16 Expression as an Independent Prognostic Marker in Oral and Oropharyngeal Squamous Cell Carcinoma: A Meta-Analysis
Indian Journal of Otolaryngology and Head & Neck Surgery (2026)
-
Saliva-based molecular diagnostics in oral squamous cell carcinoma (OSCC): a non-invasive frontier in oncology
European Archives of Oto-Rhino-Laryngology (2025)