Abstract
Cell Adhesion Molecules (CAMs) play a crucial role in regulating immune responses and repairing damage caused by hypoxia. However, the relationship between the expression characteristics of CAMs in yak lung tissues and their adaptation to the plateau environment remains unclear. To address this question, we compared lung tissues from yaks and cattle at the same altitude. After digesting the lung tissues with trypsin or Type I collagenase for varying durations, we observed that fewer cells were isolated from yak tissues compared to cattle. RNA sequencing (RNA-seq) analysis revealed that the Differentially Expressed Genes (DEGs) in lung tissues of yaks and cattle were significantly enriched in cell adhesion-related pathways. Quantitative real-time PCR (qRT-PCR) further identified changes in the expression levels of five distinct types of CAMs. Among these, the cadherin family (CDH1, CDH2, CDH11, PCDH12, CD34) exhibited significantly higher expression in yaks than in cattle. These cadherins play a critical role in regulating lung inflammation and maintaining the alveolar-capillary barrier, thereby ensuring the structural stability of the lungs. Immunohistochemical staining demonstrated that the expression patterns of cell adhesion-related proteins (CDH1, CDH11, ITGB6, SELP, CD44) were largely consistent with the qRT-PCR results. In conclusion, compared to cattle, the enhanced cell adhesion capacity of yak lung tissues contributes to their superior adaptation to the harsh plateau environment.
Similar content being viewed by others
Introduction
The Qinghai-Tibet Plateau is a unique geographical unit that serves as a vital ecological security barrier for China and Asia1. This plateau is characterized by low partial pressure of oxygen, low ambient temperatures, and intense ultraviolet radiation, which pose serious challenges to the survival of both humans and animals inhabiting the area2. The indigenous animals of the plateau have developed excellent adaptations to the high-altitude environment through a long evolutionary and adaptive process3. For instance, the plateau pika (Ochotona curzoniae), a small mammal endemic to the Tibetan Plateau, enhances anaerobic glycolysis in somatic cells by upregulating the expression of LDH-C4 under hypoxic conditions, thereby reducing its reliance on oxygen and improving its ability to thrive in hypoxic environments4. Similarly, the Tibetan antelope (Pantholops hodgsonii) significantly enhances oxygen acquisition and storage under hypoxic conditions by increasing the expression of myoglobin in cardiac and skeletal muscles5. Among these indigenous species, the yak (Bos grunniens), as the only bovine species in the genus Bos that breeds in the harsh climatic conditions of the Qinghai-Tibet Plateau, has long been settled and widely distributed there6. Over long periods, yaks have evolved specialized mechanisms to adapt to the hypoxic and cold environments of the plateau through prolonged natural selection, ensuring their survival and reproduction in these extreme conditions7. As a result, the yak has become an ideal model species for researchers studying the environmental adaptations of plateau indigenous animals.
The lung is a critical organ for gas exchange between mammals and the external environment, and lung tissue is among the first to exhibit compensatory responses under hypoxic conditions on the plateau. Yak lung tissue has evolved a series of adaptive changes to cope with the plateau environment, including shorter and wider bronchi, well-developed respiratory muscles, larger alveolar surface areas, thicker alveolar septa, and thinner air-blood barriers8,9. Studies have shown that many physiological functions of lung tissue are linked to the expression of CAMs. CAMs are cell-surface glycoproteins that mediate physical interactions between neighboring cells and between cells and the extracellular matrix (ECM)10, thereby maintaining the structural stability of tissues. Based on their structural differences, CAMs can be classified into five categories: the cadherin family, the immunoglobulin superfamily, the selectin family, the integrin family, and other adhesion molecules11,12. Research has revealed a connection between CAMs and adaptation to hypoxia. HIFs significantly enhance the adhesion of cancer cells and vascular endothelial cells through pathways mediated by selectins and integrins13. Hypoxia influences integrin-mediated adhesion of mast cells to fibronectin, thereby alleviating inflammation14. Additionally, hypoxia upregulates the expression of VCAM-1, enhancing the adhesion of sickle red blood cells to aortic endothelial cells and contributing to vascular injury repair15. The upregulation of adhesion molecules under hypoxic conditions helps cells maintain a favorable environment for recovery at injury sites16. CEACAM-1, a member of the immunoglobulin superfamily, is considered as an angiogenic factor17. In a study using mouse and rat models to simulate chronic hypoxic conditions, hypoxia-preconditioned animals exhibited increased expression of CEACAM-1, suggests that CAMs, such as CEACAM-1, promote angiogenesis under hypoxic conditions, thereby helping animals avoid ischemic injuries and protecting tissues18.
In summary, there appears to be a correlation between CAMs and the adaptation of animals to the plateau environment. However, studies on the expression characteristics of CAMs in yak lung tissues and their relationship to adaptation to the plateau environment remain limited. Therefore, we selected lung tissues from yaks and cattle at the same altitude for this study. By comparing and analyzing the differences in cell adhesion capacity and CAMs expression in the lung tissues of yaks and cattle, we explored the significance of CAMs-specific expression in yak lung tissues for hypoxic adaptation. This study provides a theoretical foundation for further research on the mechanisms underlying yak adaptation to plateau environment.
Materials and methods
Experimental animals and treatments
In this study, three healthy adult male plateau-type yaks (XH-Y) and three healthy adult male cattle (XH-C), totaling six animals, were selected from Xunhua County, Qinghai Province (altitude 2,600 m). At the same altitude, yaks are a bovine species endemic to the Tibetan Plateau and serve as the experimental group, while cattle are a common bovine species and serve as the control group. The selected animals were all approximately 3–5 years old, as individuals in this age range have reached physiological maturity, with fully developed lung tissues and a high degree of adaptation to the plateau environment, ensuring an accurate representation of lung tissue characteristics. Yaks and cattle are killed by bloodletting in local slaughterhouse. Fresh tissues from the left lung diaphragmatic lobe of yaks and cattle were collected within 30 min of death. One portion of the tissues was placed in cryotubes after removing the fascia and stored in liquid nitrogen and brought back for spare, while the other portion was cut into small pieces of approximately 1 cm3 and fixed in 4% paraformaldehyde solution. All lung tissue samples were collected locally in Xunhua County, Qinghai Province, China, and not purchased. Animal experiments in this study were approved by the Animal Welfare and Ethics Committee of Qinghai University, and conducted in full compliance with the Guide for the Care and Use of Laboratory Animals, and adhered to the ARRIVE Guidelines. All methods were performed in accordance with the relevant guidelines and regulations.
Cell digestion experiment
Yak lung tissue was placed in a sterile Petri dish containing PBS with double antibodies, and the tissue was minced into pieces approximately 1 mm3 in size after removing the fascia. Subsequently, 2–3 mL of the minced tissue mixture was transferred to a 15 mL centrifuge tube, and 4–5 times its volume of either 0.25% trypsin or 2 mg/mL Type I collagenase was added for digestion at 37 °C. These two enzyme concentrations represent the minimum concentrations required to effectively dissociate the tissue. During digestion, shake every 10 min to observe the state of digestion. At 1 h, 2 h, and 3 h of tissue digestion, the filtrate was collected, and the digestion was terminated by adding an equal volume of culture solution to the cell filtrate. Subsequently, the mixture was centrifuged at 1500 r for 5 min, the supernatant was discarded, and 3 mL of erythrocyte lysate was added and mixed thoroughly. After standing for 10 min, the mixture was centrifuged again under the same conditions, and the resulting white precipitate represented the target cell mass. The supernatant was discarded, and 3 mL of culture solution was added, then the pipette was used to gently blow and mix the solution to ensure the cells were dispersed as single cells. Finally, 3 mL of the cell suspension was transferred into cell culture flasks. The cells were observed and photographed under an inverted fluorescence microscope for counting. After completion, the cell culture flasks were placed in a constant-temperature incubator at 37 ℃ with 5% CO2 for further cultivation.
The cell digestion method employed in this study utilizes the enzymatic properties to disrupt cellular connections in lung tissue. Collagenase and trypsin are two enzymes with distinct mechanisms of action. Collagenase effectively hydrolyzes proteins, polysaccharides, and lipids within the ECM of connective and epithelial tissues, making it more suitable for tissue digestion19,20. Trypsin degrades cell surface proteins, leading to cell lysis and the release of intracellular DNA21. The lung tissues of yaks and cattle differ significantly in structure and composition. Yak lung tissue is rich in collagen fibers, with a more stable and elastic basement membrane structure, whereas cattle lung tissue is softer and structurally simpler22. Therefore, the use of Type I collagenase to digest yak lung tissues can effectively dissociate the tissues, while the use of trypsin to digest cattle lung tissues may yield higher cell viability and recovery rates. However, differences in tissue structure lead to variations in digestion times between the two species.
RNA-seq and data analysis
Lung tissue samples collected from yaks and cattle were sent to Guangzhou Kidio Biotechnology Co. for RNA-seq. The study employed the Illumina paired-end sequencing method, which offers several advantages, including improved accuracy in data mapping, enhanced genome assembly quality, and increased capability for detecting structural variants23. RNA-seq primarily involves several key steps: extracting mRNA, reverse transcription, constructing sequencing libraries on the machine, synthesizing cDNA’s 1st and 2nd strand, building the cDNA libraries, and sequencing on the machine. For detailed methodology, please refer to the eukaryotic transcriptome experimental methods published on the Guangzhou Kidio Biotechnology Co., Ltd. Website (https://www.genedenovo.com/). We finally obtained the cDNA libraries and sequencing results of yak and cattle lung tissues. After sequencing, data analysis involves quality control and filtering of raw data, alignment to reference genomes, quantification, differential expression analysis, and gene annotation, among other steps. The reference genome version was BosGruv2.0 (http://ftp.ensembl.org/pub/release-110/fasta/bos_mutus/), and we used |Fold Change| ≥ 2 and FDR < 0.05 as the screening criteria for DEGs. The screened DEGs were functionally analyzed using the GO database (http://www.geneontology.org/) and the KEGG Pathway24 database (https://www.genome.jp/kegg/) to uncover their potential biological functions and pathways.
Measurement of ion concentrations related to cell adhesion
The freeze-drying method preserves samples by freezing water-containing cells or tissues into a solid state at low temperatures and removing water through vacuum sublimation, thereby maximizing the retention of cellular structure and active components25. Approximately 10 g of lung tissue from yaks and cattle was freeze-dried using this method to obtain freeze-dried lung tissue. The freeze-dried tissue samples were then pulverized using a grinder. Accurately weigh 0.1 g of the lung tissue dry powder sample using a precision balance, and place it in a dedicated microwave digestion tank. First, 7 mL of HNO₃ was added to the digestion vessel, followed by 2 mL of H2O2. The combination of microwave heating and strong acids rapidly and thoroughly decomposes the biological tissue, releasing the metal ions within. Leave it overnight for adequate pre-dissolution, followed by microwave dissolution. Following digestion, allow it to cool and then place it in an acid drive unit with the temperature set at 170 ℃ for approximately 130 min to remove excess acid. Once the acid drive is complete, rinse the microwave digestion tank and lid with ultrapure water, and make up to 50 mL in a volumetric flask for spare. The sample solution was diluted 1,000-fold, and the calcium and magnesium ion concentrations were determined using inductively coupled plasma mass spectrometer (ICP-MS).
Ca²⁺ and Mg²⁺ are key regulators of cell adhesion26. Ca²⁺ regulates cell adhesion by mediating calcium transients, which promote the recruitment of connexins and the reorganization of the cytoskeleton27. On the other hand, Mg²⁺ modulates cell adhesion by binding to integrins and inducing conformational changes that alter their binding affinity to ligands, thereby influencing cell adhesion28. Therefore, we examined changes in Ca²⁺ and Mg²⁺ concentrations to evaluate the functional state of cell adhesion.
qRT-PCR
Retrieve the CDS sequences of cell adhesion-related genes, such as CDH1, CDH2, and CDH11, for yaks and cattle from the NCBI database. Specific degenerate primers for these cell adhesion-related genes in yaks and cattle were designed using Oligo 7.0 software, and sent to Sangon Biotech (Shanghai) Co., Ltd. for synthesis. The primer sequences are listed in Table 1. qRT-PCR is a widely used technique for the quantitative analysis of gene expression, and the specificity of primers is critical to ensure accurate detection of target gene29. Our sources of primers include those reported in the literature and those designed in-house. All primers were validated before use, and the fluorescence quantitative PCR curves were single-peaked and stable with good experimental results. The primers were specific and effective.
Approximately 0.1 g of lung tissue from yaks and cattle was weighed, and total RNA was extracted from these tissues following the instructions provided with the Total RNA Extraction Kit. The RNA concentration and OD values were measured using an ultramicro spectrophotometer (NanoDrop 2000). In this method, the instrument blank was zeroed, and 2.5 µL of the RNA solution was placed on the detection pedestal. The sample arm was lowered, and the absorbance values were measured using the accompanying software. The calculation formulas of RNA concentration and OD values are shown in Eqs. (1) and (2).
Where C is the sample concentration (µg/mL), A is the absorbance value, ε is the wavelength-dependent molar extinction coefficient (ng cm/mL), and b is the optical path length (cm).
Where OD represents the purity, A260 is the absorbance value at 260 nm with a 10 mm optical path length, and A280 is the absorbance value at 280 nm with a 10 mm optical path length. Additionally, \(\:\frac{{A}_{260}}{{A}_{280}}\) > 1.8.
Subsequently, 1.2 µg of RNA was used for reverse transcription following the instructions provided with the Reverse Transcription Kit. The reverse transcription program was as follows: 94 ℃ for 15 min, 42 ℃ for 3 min, and 4 ℃ ∞. The resulting cDNA was stored in the refrigerator at -20 ℃ for spare. The qRT-PCR amplification system (total volume of 20.0 µL) consisted of: 2.0 µL of cDNA, 0.5 µL each of the upstream and downstream primers for the target gene and the internal reference gene, 10.0 µL of TB Green Premix Ex Taq II enzyme, and 7 µL of ddH2O, prepared as a standard reaction system. GAPDH was used as the internal reference, and three replicates were set up for both the target and internal reference genes. The qRT-PCR program followed a three-step protocol: 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s.
Immunohistochemical staining
Immunohistochemical staining of lung tissues was primarily performed using the Streptavidin-Biotin-Peroxidase Complex (SABC) technique, with diaminobenzidine (DAB, purchased from BOSTER) used for visualization. The staining procedure was performed according to the kit instructions and included the following steps: deparaffinization of paraffin sections to water, antigen repair, blocking of endogenous peroxidase, serum blocking, addition of primary and secondary antibodies, DAB color development, restaining of cell nuclei, ethanol dehydration, and sealing of the sections with neutral balsam. Nuclei stained with hematoxylin appeared blue, and DAB-positive staining was visualized as brown. Weak or background-like brown staining was considered negative. Images were captured using a DP70 microscopic image acquisition system and analyzed under a light microscope. The mean optical density of positive areas was calculated using Image-Pro Plus 6.0 software, the calculation formula is shown in Eq. (3).
Where MOD is the mean optical density, OD represents the converted optical density value, calculated as \(\:OD={\text{log}}_{10}(\:\frac{I}{{I}_{0}}\:)\). Here, I is the light intensity of the sample (corresponding to the gray value), and I0 is the light intensity of the background (without the sample). Additionally, ODi denotes the optical density value of the i-th pixel in the region, and N is the total number of pixels in the region. These calculations are automatically performed by the Image-Pro Plus software.
The resulting data were analyzed for statistical significance using SPSS 25.0 software, with results expressed as mean ± standard deviation (X̅±SD). Data visualization was performed using Origin 2021 software (https://www.originlab.com/), with P < 0.05 as the criterion for the significance of differences.
Results
Results of cell digestion experiment
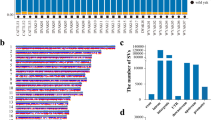
Lung tissues from yaks and cattle at the same altitude were digested using trypsin and Type I collagenase, respectively. Digestion was terminated at 1 h, 2 h, and 3 h. After gently blowing and mixing, cells of equal size were selected for photographing and counting. Cell counting was performed using Image-Pro Plus 6.0 software, and data were analyzed using SPSS 25.0 software, with results expressed as mean ± standard deviation (X̅±SD). Visualization of the results was performed using Origin 2021 software. The results of cell digestion and counting are shown in Fig. 1. Quantitative analysis revealed that the number of cells isolated from yak lung tissues at different time points and with different enzymes was significantly lower than that from cattle (P < 0.05). We hypothesize that this difference arises because the connections between cells in yak lung tissues are significantly tighter, making it more difficult to separate cells during enzymatic digestion. As a result, fewer cells were isolated from yak lung tissues compared to cattle. This finding could indicate that the cell adhesion capacity of yak lung tissues is stronger than that of cattle, indicating a correlation between the number of isolated cells and cell adhesion capacity. Enhanced cell adhesion may help yaks maintain the structural stability of lung tissues and thus adapt to the plateau environment.
Results of digesting yaks and cattle lung tissues at the same altitude using different enzymes. (a) and (b) show the results of cell digestion and counting by trypsin. (c) and (d) show the results of cell digestion and counting by Type I collagenase. Black arrows (→) refer to the isolated cells. An asterisk (*) denotes significant difference between the two groups, P < 0.05.
GO and KEGG enrichment analysis of DEGs
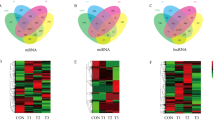
Following differential analysis of RNA-seq data from lung tissues of yaks and cattle at the same altitude, a total of 2,296 DEGs were identified in yaks compared to cattle, including 1,460 up-regulated genes and 836 down-regulated genes. Functional annotation and enrichment analysis of these DEGs were performed using the GO and KEGG Pathway databases. The results revealed that the DEGs were significantly enriched in GO terms such as biological adhesion (GO:0022610), cell adhesion (GO:0007155), and cell periphery (GO:0071944), as shown in Fig. 2a. Furthermore, the DEGs were enriched in KEGG pathways including Complement and coagulation cascades, Staphylococcus aureus infection, Hematopoietic cell lineage, ECM-receptor interaction, and Cytokine-cytokine receptor interaction. Notably, numerous DEGs were enriched in the calcium signaling pathway, which is associated with the cell adhesion process, as shown in Fig. 2b. The ECM-receptor interaction pathway is a key regulatory mechanism for cell adhesion. ECM components such as collagen and fibronectin regulate cell-matrix adhesion through interactions with cell surface receptors30. Genes such as COL1A1, COL1A2, FN1, and ITGB4 are enriched in this pathway. COL1A1 and COL1A2 encode type I collagen, while FN1 encodes fibronectin. Both proteins are essential components of the ECM, and their expression levels can influence cell adhesion capacity31,32. ITGB4 mediates cell-basement membrane adhesion by binding to laminin through its extracellular domain33. This enhanced adhesion capacity is advantageous for yaks’ adaptation to the plateau environment.
GO and KEGG enrichment analyses in lung tissues of yaks and cattle at the same altitude showed that DEGs were mainly enriched in cell adhesion-related pathways. (a) GO enrichment analysis of DEGs, displaying the top 20 enriched GO terms with their significance levels. The vertical coordinate is − log10 (Qvalue), the horizontal coordinate is the z-score value (the difference between the number of up-regulated differential genes and the number of down-regulated differential genes as a proportion of the total number of differential genes), and the yellow line represents the threshold of Qvalue = 0.05. (b) KEGG pathway enrichment analysis of DEGs highlighting the top 20 significantly enriched pathways, KEGG images are licensed from Kanehisa Laboratories (www.kegg.jp/kegg/kegg1.html). The horizontal coordinate is the ratio of the number of differential genes annotated to the total number of differential genes, the vertical coordinate is the KEGG pathway. The shade of the color represents the significance levels, the darker the color the more significant it is.
Measurement of cell adhesion-related ion concentrations
Ca2+ and Mg2+ affect adhesion capacity by regulating integrin function, influencing signaling pathways, and organizing the cytoskeleton34,35. Within a certain concentration range, increased levels of both Ca2+ and Mg2+ enhance cell adhesion capacity, with their synergistic effect being particularly significant36. The ion concentrations in the samples were measured using ICP-MS after constant-volume dilution. The results showed that the Ca2+ concentration in yak lung tissues was 48.0343 µg, significantly higher than the 44.9467 µg observed in cattle at the same altitude (P < 0.05), whereas the Mg2+ concentration showed no significant change, as shown in Fig. 3. Ca²⁺ enhances the tightness of intercellular and cell-ECM interactions by binding to cadherins or activating integrin-mediated signaling pathways37,38. This enhanced cell adhesion is achieved through the regulation of CAMs function by elevated Ca²⁺ levels, which helps reduce tissue damage and promote tissue repair under hypoxic conditions, thereby facilitating yaks’ adaptation to the plateau environment.
Quantitative analysis of cell adhesion-related genes
Following RNA extraction from lung tissues of yaks and cattle at the same altitude, qRT-PCR analysis was performed. The expression profiles of five different types of CAMs are shown in Fig. 4. In yak lung tissues, the relative expression levels of genes in the cadherin family (CDH1, CDH2, CDH11, PCDH12, CD34), integrin family (ITGA3, ITGA5, ITGAV), immunoglobulin superfamily (VCAM1, SIGLEC1, CEACAM1), and other cell adhesion-related genes (CD44, ESAM, JAM3) were significantly higher than those in cattle (P < 0.05). In contrast, selectin family-related genes (SELL, SELPLG) were significantly lower in yak lung tissues compared to cattle (P < 0.05). Among them, CDH2 and SIGLEC1 exhibited the highest expression levels in yak lung tissues, approximately three times higher than those in cattle. While SELL showed the lowest expression level in yak lung tissues, with a value of 0.3800. Notably, the expression levels of ITGB6, ICAM3, ALCAM, and SELP showed no significant differences between yaks and cattle. Among these genes, ITGB6 mediates cell-cell and cell-ECM adhesion39, ICAM3 and ALCAM are involved in immune cell adhesion and trafficking40,41, and SELP plays a critical role in leukocyte-endothelial cell adhesion and the regulation of inflammatory responses42. The lack of significant changes in the expression of these genes suggests that they are stably expressed in lung tissues. This stability indicates their potential role in maintaining fundamental cell adhesion functions and ensuring the basic functional stability of lung tissues.
Relative mRNA expression of cell adhesion-related genes in lung tissues of yaks and cattle at the same altitude. (a) is cadherin family-related gene; (b) is integrin family-related gene; (c) is immunoglobulin superfamily-related gene; (d) is selectin family-related gene; (e) is other cell adhesion genes.
Protein expression analysis of cams
Following immunohistochemical staining of selected CAMs in lung tissues of yaks and cattle, we observed that the positive areas of CAMs were localized on the cell membrane surface. The mean optical density values were directly measured using Image-Pro Plus 6.0 image analysis system. The data were statistically analyzed using SPSS 25.0 software and visualized using Origin 2021 software. As shown in Fig. 5, the mean optical density values of CAM-related proteins CDH1, CDH11, ITGB6, SELP, and CD44 were significantly higher in yak lung tissues compared to cattle lung tissues at the same altitude (P < 0.05). Among these proteins, CD44 exhibited the highest expression in yak lung tissues, while CDH1 showed the largest expression difference between yaks and cattle, with a difference of 0.039. These findings were largely consistent with the expression trends observed in the qRT-PCR results.
Expression levels of major adhesion molecule proteins in lung tissues of yaks and cattle analyzed by immunohistochemistry. (a) Immunohistochemical staining of cell adhesion-related proteins in lung tissues. (b) Immunohistochemical staining optical density analysis of cell adhesion-related proteins in lung tissues. An asterisk (*) denotes significant difference between the two groups, P < 0.05.
Discussions
Tissue digestion is one of the most commonly used methods for isolating primary cells from animal tissues. It primarily utilizes enzymatic properties to break down the ECM and intercellular junctions, thereby separating individual cells from the tissue structure. Collagenase and trypsin are frequently employed for cell separation experiments. Collagenase acts relatively gently and can specifically hydrolyze the three-dimensional helical structure of native collagen fibers, it can be classified into multiple types based on differences in specificity, with Type I, Type II, and Type III being common43. Trypsin possesses unique proteolytic capabilities, enabling precise degradation of protein components within the ECM, thereby promoting effective cell dissociation44. Our study observed that regardless of whether trypsin or Type I collagenase was used to digest lung tissue, the number of cells isolated per unit area at 1 h, 2 h, and 3 h was significantly lower in yaks compared to cattle. It can be seen that the cell adhesion capacity in lung tissues differs between yaks and cattle, with yak lung tissues demonstrating stronger intercellular or cell-to-matrix adhesion compared to cattle.
Cell adhesion is a process in which cells attach to each other or to the underlying matrix (e.g., the ECM) through CAMs, playing a central role in cell migration45. The adhesive capacity between cells is critical for determining cell polarity and is essential for maintaining the morphology of tissues composed of cells46. Most CAMs are Ca2+-dependent proteins, with calmodulin binding to Ca2+, and the more Ca2+ is bound, the more rigid the CAMs become47. Many CAMs rely on Ca2+ to function, and in the presence of Ca²⁺, they promote cell adhesion and resist hydrolysis by proteases48. Changes in intracellular Ca2+ levels can modulate the expression of immunoglobulin superfamily, cadherin family, integrin family, and other CAMs49. In this study, the Ca2+ content in yak lung tissues was significantly higher than that in cattle. It can be inferred that Ca2+ concentration is associated with the expression of CAMs in yak lung tissues, and yaks exhibit stronger intercellular adhesion within lung tissue compared to cattle.
RNA-seq results revealed that DEGs were significantly enriched in cell adhesion-related pathways, mainly in related GO and KEGG pathways including cell adhesion (GO:0007155), CAMs, and Calcium signaling pathway. qRT-PCR and immunohistochemical analyses further identified changes in the expression of five different types of CAMs, with the expression levels of key CAM-related genes and proteins being significantly higher in yaks compared to cattle. The classical cadherin genes, including CDH1, CDH2, and CDH11, are critical intercellular adhesion molecules that regulate cell movement, clustering, and differentiation, thereby promoting morphogenesis and tissue barrier formation50. These cadherin-related genes play a vital role in regulating lung inflammation and maintaining the alveolar-capillary barrier in yaks. Genes from the integrin family, such as ITGA3, ITGA5, and ITGAV, are involved in endothelial cell migration, cytoskeletal organization, apoptosis initiation, and the regulation of lung inflammation and fibrosis51,52. Members of the immunoglobulin superfamily, including VCAM1, SIGLEC1, and CEACAM1, participate in interactions between cells and pathogens to promote effective phagocytosis and antigen presentation of adaptive immune response, and related genes play a role in regulating the body’s immune functions53,54,55,56. SELP and SELPLG belong to the selectin family. SELP triggers the release of procoagulant microparticles and increases the expression of tissue factor on monocytes, playing a pivotal role in hemostasis and leukocyte recruitment57. SELPLG is essential for leukocyte trafficking during inflammation58. Other CAMs, such as CD44, ESAM, and JAM3, are involved in angiogenesis, leukocyte transendothelial migration, and platelet activation59,60,61. It can be seen that the high expression of CAMs in yak lung tissues likely contributes to their enhanced adaptation to the plateau environment.
In summary, the expression levels of genes and proteins related to cell adhesion were significantly higher in yak lung tissues compared to those cattle. We hypothesize that this enhanced expression in yak lung tissues may strengthen immune responses, improve lung tissue repair capacity, and maintain the structural integrity of the air-blood exchange barrier. Additionally, these molecules may contribute to intrapulmonary angiogenesis, thereby facilitating better adaptation to the hypoxic conditions of the plateau environment. However, this study has several limitations. Due to the challenges associated with obtaining tissue samples from large animals in their natural habitats, we were restricted to a minimal biological sample size for our analysis. This small sample size may limit the generalizability and statistical significance of our findings, making it difficult to fully capture the differences in cell adhesion among different species. Furthermore, the application of our method is not comprehensive and lacks further functional experimental validation. We will increase the sample size in subsequent studies to comprehensively analyze the differences in the expression of CAMs in lung tissues under different conditions and validate the findings with functional experiments to elucidate the relationship between CAMs and adaptation to the plateau environment. Although this study reveals one aspect of plateau adaptation in yaks, further work is needed to refine and extend these findings.
Conclusions
The present study demonstrates that cell adhesion, cell adhesion-related genes and proteins in yak lung tissue were significantly higher than those in cattle. These findings suggest that yaks may enhance the immune effects of lung tissue repair by increasing adhesion ability and upregulating the expression of CAMs, maintaining the integrity of the lung endothelial barrier, and promoting angiogenesis within the lungs, thereby better adapting to the hypoxic conditions of the plateau environment. In the future, the conclusions drawn from this study could be applied to other fields. By elucidating the mechanisms of cell adhesion in yaks’ adaptation to hypoxic environments may provide valuable insights for developing conservation strategies for other plateau-dwelling animals. Genes and proteins associated with cell adhesion in yaks could serve as potential therapeutic targets, offering new avenues for the development of drugs to treat high-altitude-related diseases.
Data availability
Sequence data that support the findings of this study are available in the NCBI SRA with the primary accession code PRJNA1127767.
References
Zeng, J. et al. The fusion of multiple scale data indicates that the carbon sink function of the Qinghai-Tibet plateau is substantial. Carbon Balance Manag. 18, 19 (2023).
Miao, F., Guo, Z., Xue, R., Wang, X. & Shen, Y. Effects of grazing and precipitation on herbage biomass, herbage nutritive value, and Yak performance in an alpine meadow on the Qinghai-Tibetan plateau. PLoS One 10, e0127275 (2015).
Liu, K., Yang, J. & Yuan, H. Recent progress in research on the gut microbiota and Highland adaptation on the Qinghai-Tibet plateau. J. Evol. Biol. 34, 1514–1530 (2021).
Wei, D., Wei, L., Li, X., Wang, Y. & Wei, L. Effect of hypoxia on Ldh-c expression in somatic cells of plateau Pika. Int. J. Environ. Res. Public. Health 13, 773 (2016).
Ma, L. et al. Molecular cloning, characterization and expression of myoglobin in Tibetan antelope (Pantholops hodgsonii), a species with hypoxic tolerance. Gene 533, 532–537 (2014).
Lan, D. et al. Transcriptome profile and unique genetic evolution of positively selected genes in Yak lungs. Genetica 146, 151–160 (2018).
Zhu, Y. et al. Gut microbial adaptation to varied altitudes and temperatures in Tibetan plateau Yaks. Microorganisms 12, 1350 (2024).
Wang, Y. et al. Insight into unique somitogenesis of Yak (Bos grunniens) with one additional thoracic vertebra. BMC Genom. 21, 201 (2020).
Ge, Q. et al. Molecular mechanisms detected in Yak lung tissue via transcriptome-wide analysis provide insights into adaptation to high altitudes. Sci. Rep. 11, 7786 (2021).
Francavilla, C., Maddaluno, L. & Cavallaro, U. The functional role of cell adhesion molecules in tumor angiogenesis. Semin Cancer Biol. 19, 298–309 (2009).
Ren, G., Roberts, A. I. & Shi, Y. Adhesion molecules: Key players in mesenchymal stem cell-mediated immunosuppression. Cell. Adh Migr. 5, 20–22 (2011).
Lefer, A. M., Weyrich, A. S. & Buerke, M. Role of Selectins, a new family of adhesion molecules, in ischaemia-reperfusion injury. Cardiovasc. Res. 28, 289–294 (1994).
Koike, T. et al. Hypoxia induces adhesion molecules on cancer cells: A missing link between Warburg effect and induction of selectin-ligand carbohydrates. Proc. Natl. Acad. Sci. USA 101, 8132–8137 (2004).
Pastwinska, J., Walczak-Drzewiecka, A., Lukasiak, M., Ratajewski, M. & Dastych, J. Hypoxia regulates human mast cell adhesion to fibronectin via the PI3K/AKT signaling pathway. Cell. Adh Migr. 14, 106–117 (2020).
Setty, B. N. & Stuart, M. J. Vascular cell adhesion molecule-1 is involved in mediating hypoxia-induced sickle red blood cell adherence to endothelium: Potential role in sickle cell disease. Blood 88, 2311–2320 (1996).
Mathew, S. A., Chandravanshi, B. & Bhonde, R. Hypoxia primed placental mesenchymal stem cells for wound healing. Life Sci. 182, 85–92 (2017).
Ergün, S. et al. CEA-related cell adhesion molecule 1: a potent angiogenic factor and a major effector of vascular endothelial growth factor. Mol. Cell. 5, 311–320 (2000).
Chen, W. J. et al. Gene expression profiles in hypoxic preconditioning using cDNA microarray analysis: altered expression of an angiogenic factor, carcinoembryonic antigen-related cell adhesion molecule 1. Shock 24, 124–131 (2005).
Donahue, T. R., Hiatt, J. R. & Busuttil, R. W. Collagenase and surgical disease. Hernia 10, 478–485 (2006).
Liu, Y. X. et al. Effects of collagenase type I on the structural features of collagen fibres from sea cucumber (Stichopus japonicus) body wall. Food Chem. 301, 125302 (2019).
Taraskin, A. S. et al. Quench me if you can: Alpha-2-macroglobulin trypsin complexes enable serum biomarker analysis by MALDI mass spectrometry. Biochimie 185, 87–95 (2021).
Li, J. et al. Changes of collagen content in lung tissues of plateau Yak and its mechanism of adaptation to hypoxia. PeerJ 12, e18250 (2024).
Cole, H. A., Howard, B. H. & Clark, D. J. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Methods Enzymol. 513, 145–168 (2012).
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. & Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 (2012).
Wolkers, W. F. & Oldenhof, H. Principles underlying cryopreservation and freeze-drying of cells and tissues. Methods Mol. Biol. 2180, 3–25 (2021).
Lange, T. S. et al. Mg2+ and Ca2+ differentially regulate beta 1 integrin-mediated adhesion of dermal fibroblasts and keratinocytes to various extracellular matrix proteins. Exp Cell Res. 214, 381–388 (1994).
Ko, K. S., Arora, P. D., Bhide, V., Chen, A. & McCulloch, C. A. Cell-cell adhesion in human fibroblasts requires calcium signaling. J Cell Sci. 114, 1155–1167 (2001).
Shimaoka, M., Takagi, J. & Springer, T. A. Conformational regulation of integrin structure and function. Annu Rev. Biophys. Biomol. Struct. 31, 485–516 (2002).
Jozefczuk, J. & Adjaye, J. Quantitative real-time PCR-based analysis of gene expression. Methods Enzymol. 500, 99–109 (2011).
Li, X. et al. LGALS1 regulates cell adhesion to promote the progression of ovarian cancer. Oncol. Lett. 26, 326 (2023).
Zhu, X., Luo, X., Jiang, S. & Wang, H. Bone morphogenetic protein 1 targeting COL1A1 and COL1A2 to regulate the epithelial-mesenchymal transition process of colon cancer SW620 cells. J. Nanosci. Nanotechnol. 20, 1366–1374 (2020).
Mao, X. G. et al. CEBPD is a master transcriptional factor for hypoxia regulated proteins in glioblastoma and augments hypoxia induced invasion through extracellular matrix-integrin mediated EGFR/PI3K pathway. Cell. Death Dis. 14, 269 (2023).
Yuan, L. et al. Airway epithelial ITGB4 deficiency induces airway remodeling in a mouse model. J. Allergy Clin. Immunol. 151, 431–446 (2023).
Shen, X. Y. et al. Extracellular calcium ion concentration regulates chondrocyte elastic modulus and adhesion behavior. Int. J. Mol. Sci. 22, 10034 (2021).
Su, L. T. et al. TRPM7 regulates polarized cell movements. Biochem. J. 434, 513–521 (2011).
Obata, A., Ogasawara, T. & Kasuga, T. Combinatorial effects of inorganic ions on adhesion and proliferation of osteoblast-like cells. J. Biomed. Mater. Res. A 107, 1042–1051 (2019).
Fucikova, J., Spisek, R., Kroemer, G. & Galluzzi, L. Calreticulin and cancer. Cell. Res. 31, 5–16 (2021).
Dalal, P. J., Muller, W. A. & Sullivan, D. P. Endothelial cell calcium signaling during barrier function and inflammation. AM. J. Pathol. 190, 535–542 (2020).
Chen, H. et al. Transgenic overexpression of ITGB6 in intestinal epithelial cells exacerbates dextran sulfate sodium-induced colitis in mice. J. Cell. Mol. Med. 25, 2679–2690 (2021).
Blight, B. J. et al. Cell adhesion molecules are upregulated and May drive inflammation in chronic rhinosinusitis with nasal polyposis. J. Asthma Allergy 14, 585–593 (2021).
Chalmers, S. A. et al. The CD6/ALCAM pathway promotes lupus nephritis via T cell-mediated responses. J. Clin. Invest. 132, e147334 (2022).
Gupta, P., Choudhari, V. & Kumar, R. Exploring the genetic mechanisms: SELP gene’s contribution to alleviating vaso-occlusive crisis in sickle cell disease. Gene 928, 148805 (2024).
Wu, S. J., Zhou, X. W., Xu, K. & Cheng, H. M. The role of extraction method to collagen substrates in enzymolysis of type I collagenase. Int. J. Biol. Macromol. 283, 138086 (2024).
Sorensen, A. B. et al. A systematic approach for evaluating the role of surface-exposed loops in trypsin-like Serine proteases applied to the 170 loop in coagulation factor VIIa. Sci. Rep. 12, 3747 (2022).
Huttenlocher, A., Sandborg, R. R. & Horwitz, A. F. Adhesion in cell migration. Curr. Opin. Cell. Biol. 7, 697–706 (1995).
Hirohashi, S. & Kanai, Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 94, 575–581 (2003).
Kong, F. Y., You, H. J., Zheng, K. Y., Tang, R. X. & Zheng, C. F. The crosstalk between pattern-recognition receptor signaling and calcium signaling. Int. J. Biol. Macromol. 192, 745–756 (2021).
Kemler, R., Ozawa, M. & Ringwald, M. Calcium-dependent cell adhesion molecules. Curr. Opin. Cell. Biol. 1, 892–897 (1989).
Sheng, L. F., Leshchyns’ka, I. & Sytnyk, V. Cell adhesion and intracellular calcium signaling in neurons. Cell. Commun. Signal. 11, 94 (2013).
Nguyen, H. T. & Martin, L. J. Classical cadherins in the testis: How are they regulated? Reprod. Fertil. Dev. 35, 641–660 (2023).
Zhang, D. J. et al. Min pig skeletal muscle response to cold stress. PloS One 17, e0274184 (2022).
Madala, S. K. et al. Inhibition of the Αvβ6 integrin leads to limited alteration of TGF-α-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 306, L726–L735 (2014).
Troncoso, M. F. et al. VCAM-1 as a predictor biomarker in cardiovascular disease. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166170 (2021).
Singh, V., Kaur, R., Kumari, P., Pasricha, C. & Singh, R. ICAM-1 and VCAM-1: Gatekeepers in various inflammatory and cardiovascular disorders. Clin. Chim. Acta. 548, 117487 (2023).
Klaas, M., Dubock, S., Ferguson, D. J. P. & Crocker, P. R. Sialoadhesin (CD169 / Siglec-1) is an extended molecule that escapes inhibitory cis-interactions and synergizes with other macrophage receptors to promote phagocytosis. Glycoconj. J. 40, 213–223 (2023).
Jones, C., Virji, M. & Crocker, P. R. Recognition of sialylated meningococcal lipopolysaccharide by Siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol. Microbiol. 49, 1213–1225 (2003).
André, P. P-selectin in haemostasis. Br. J. Haematol. 126, 298–306 (2004).
Fenoglio, C. et al. SELPLG and SELP single-nucleotide polymorphisms in multiple sclerosis. Neurosci. Lett. 394, 92–96 (2006).
Senbanjo, L. T. & Chellaiah, M. A. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell. Dev. Biol. 5, 18 (2017).
Stalker, T. J. et al. Endothelial cell specific adhesion molecule (ESAM) localizes to platelet-platelet contacts and regulates thrombus formation in vivo. J. Thromb. Haemost. 7, 1886–1896 (2009).
Bazzoni, G. The JAM family of junctional adhesion molecules. Curr. Opin. Cell. Biol. 15, 525–530 (2003).
Acknowledgements
We are grateful to our laboratory colleagues for their assistance during the sample collection process.
Funding
This study was supported by the Basic Research Program of Qinghai Provincial Department of Science and Technology (grant number: 2023-ZJ-708). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization and methodology, J.C. and Q.W.; software, N.H., M.T. and X.Z.; formal analysis, H.W. and N.H.; investigation, H.W.; resources, J.C. and Q.W.; data curation, H.W., N.H., M.T. and X.Z.; writing-original draft preparation, H.W. and N.H.; writing—review and editing, J.C. and Q.W.; project administration and funding acquisition, Q.W. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All animal experiments in this study were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, and were approved by the Animal Welfare and Ethics Committee of Qinghai University, Xining, China. All experimental methods adhered to the ARRIVE Guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, H., Huang, N., Tan, M. et al. Characteristics of cell adhesion molecules expression and environmental adaptation in yak lung tissue. Sci Rep 15, 10914 (2025). https://doi.org/10.1038/s41598-025-95882-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95882-2