Abstract
The Aedes (Stegomyia) albopictus (Skuse, 1985) (Diptera: Culicidae) is one of the major vectors for Dengue and Chikungunya. However, our study uncovered another mosquito species morphologically similar to Ae. albopictus but is genetically different. The male genitalia of this species possess minute differences in the IX tergum with Ae. albopictus. Nucleotide diversity and mean genetic distance analysis confirmed the genetic difference from Ae. albopictus and other Aedes species. However, this species has a significant degree of genetic similarity with the cryptic species of Ae. albopictus earlier reported from Vietnam and China. The time tree revealed the median divergence time of this species and Ae. albopictus species to be approximately 36.13 million years ago. This study marks the discovery of an Aedes nr. Albopictus species resembling Ae. albopictus in India and third in the world, also reports the distinct morphological feature of the male genitalia for the first time. Our study indicates the sympatric behavior of this species as it shares the breeding habitat of Ae. albopictus. The absence of endosymbiont Wolbachia in this species raises the possibility of reproductive isolation with Ae. albopictus leading to sympatric speciation and increasing virus-carrying capability for this species, having significant implications for vector-borne disease control.
Similar content being viewed by others
Introduction
Currently, within the medically significant Stegomyia subgenus of the genus Aedes there are 132 species recognized globally1. Among these, Aedes (Stegomyia) aegypti (Linnaeus, 1762) and Aedes (Stegomyia) albopictus (Skuse, 1985) are particularly well-known for being primary vectors of Dengue, Chikungunya, and Zika virus. However, other Aedes species within the Stegomyia subgenus, found in Africa and Asia, are also considered potential vectors for these viruses2.
Huang in 1979 included 37 species under the subgenus Stegomyia from the Oriental region and, categorized them into 5 species groups3. Of these, Ae. albopictus belongs to the Albopictus Subgroup under Scutellaris Group. The Scutellaris Group contains 8 species in the Albopictus Subgroup and 7 species in the Scutellaris Subgroup, all in the Oriental region3.
Identification within subgenus Stegomyia mostly relies on morphological characteristics, particularly patterns on the thorax and tarsi in adult mosquitoes. The Scutellaris Group, for instance, is marked by a distinct white stripe on the scutum. However, identifying adult females is challenging due to subtle differences in the morphological characters4. Huang (1975)5 also noted that larvae of many species in this group share similar breeding habitats, which can lead to misidentification, as was the case with Ae. krombeini being mistaken for Ae. albopictus in Sri Lanka. In India, Ae. albopictus, Ae. novalbopictus, Ae. patriciae, Ae. pseudalbopictus, Ae. subalbopictus, Ae. unilineatus, Ae. krombeini, Ae. malayensis, and Ae. scutellaris have been reported under Scutellaris Group6.
Recent studies have identified a novel cryptic species of Ae. albopictus that coexists with Ae. albopictus in the forested regions of Vietnam and China living in sympatry7,8. Although this newly discovered species is morphologically similar to Ae. albopictus, molecular analysis, and cibarial armature morphology revealed this species to be distinct. Cryptic species have also been reported in mosquito genera such as Anopheles and Culex. About 30 Anopheles vector species have been identified as species complexes and the number of cryptic/ isomorphic species identified so far varies in each complex (WHO, 2007)9.
Ae. albopictus commonly hosts Wolbachia endosymbiont, which may cause CI (cytoplasmic incompatibility) in mosquitoes, Wolbachia induced CI may give protection to mosquitoes against various RNA viruses10. This underscores the importance of accurate species identification and also detection of Wolbachia infection in mosquito species.
In our previous investigations in rural forested areas of Tripura, we identified molecularly confirmed Ae. albopictus species, whereas some specimens showed differences in the molecular studies11. In this current study, we have characterized these samples, along with other archived Aedes larva and adult samples from different areas in Tripura, collected between 2019 and 2022.
Results
Morphological identification
180 adult mosquitoes were identified as Aedes species. Out of 180, 139 resembled Ae. albopictus, and 15 were damaged Aedes species. So, these 15 damaged Aedes specimens and 139 that resembled Ae. albopictus were subjected for molecular identification.
ITS2 PCR results
In our study, 125 samples showed the typical band size of 550 bp. This particular band size has been previously identified as characteristic of the Ae. albopictus11. Fourteen samples showed band size between 350 and 400 bp and were subjected to sequencing analysis (Fig. 1).
DNA sequence analysis
In this study, we analyzed 14 DNA samples that demonstrated a band size of 350–400 bp in the ITS2 PCR test. These specimens were subsequently sequenced for both the COI and ITS2 genes. Out of the 14 specimens, successful sequencing was achieved for 13 specimens by either COI or ITS2, (5 for COI and 9 for ITS2). One specimen was successfully sequenced for both genes.
The blast similarity search for the 5-COI and 6-ITS2 sequences (Accession numbers: OR139061-OR139064, ON007025, PF741448, OR129842-OR129846) revealed to be similar to Ae. albopictus cryptic species earlier reported from Vietnam and China7,8 (Figs. 2 and 3). The remaining three ITS2 sequences did not match any sequence existing in the GenBank. Two specimens were found morphologically similar to Ae. annandalei, and one was morphologically close to the genus Topomiya (RMRC unpublished data).
Phylogenetic analysis
The phylogenetic analysis of these COI sequences was performed alongside those previously reported Ae. albopictus cryptic species from Vietnam and China7,8.
Additionally, sequences of Ae. albopictus from India and other global regions, Ae. subalbopictus from India, Ae. vittatus and Ae. aegypti were included (Fig. 2).
Similarly, in the ITS2-based phylogenetic tree, our sequences were in the same clade along with the Ae. albopictus cryptic species from Vietnam and China7,8, along with Ae. albopictus, and Ae. aegypti as outgroup. Notably, in both phylogenetic trees, the isolates from this study clustered closely with those from Vietnam and China (Figs. 2 and 3). We have provisionally designated Aedes albopictus cryptic species found in our study as Aedes nr. albopictus.
Nucleotide diversity and mean genetic distance analysis
Very low nucleotide diversity and mean genetic distance of this study population was recorded with the population of Ae. albopictus cryptic species of China and Vietnam respectively. However, the nucleotide diversity and mean genetic distance with Ae. albopictus and other Aedes were comparatively higher (Tables 1 and 2).
Time tree analysis
Time tree analysis gave an approximate date of divergence between the Aedes species included in this study. The median divergence time of Ae. albopictus and Ae. albopictus cryptic species in the analysis was found to be 36.13 million years ago, Ae. subalbopictus diverged from Ae. albopictus around 21.75 million years ago whereas Ae. flavopictus diverged from Ae. subalbopictus around 18.11 million years ago. From the analysis, it was revealed that Ae. aegypti (46.01 million years ago) was the most ancient ancestor among the species included in the time tree estimation. (Fig. 5).
Haplotype analysis
Focusing on the five isolates of Ae. nr. albopictus from our current study, we identified four unique haplotypes. These are designated as Hap_1, Hap_2, Hap_3, and Hap_4, each representing a new haplotype discovered from this study. Meanwhile, Hap_3 aligned with Ae. albopictus cryptic sequences previously reported from China and Vietnam, indicating a shared haplotype with these regions. This analysis contributes to the growing understanding of the genetic diversity within the Ae. albopictus species, particularly highlighting the newly identified haplotypes in the Indian context. It also underscores the importance of continuous monitoring and genetic characterization of mosquito populations and understanding their distribution and evolution.
Dissection of male terminalia
In our study, the morphology of adult male and female specimens closely resembled those of Ae. albopictus, as evidenced by the characteristic median white stripe on the scutum4,13 [Figure 6 (A), (B)]. A detailed examination of the male terminalia in three male specimens revealed that two were consistent with Ae. albopictus, identified by the horn-like median projection (MP) on their IX tergum [Figure 6 (C), (D)]. However, the IX tergum of the third specimen [Figure 6 (C)] presented slight variations, though it still resembled to Ae. albopictus, rather than any other species in the Scutellaris Group as described by Huang in 1972 for the oriental region4. Specifically, the middle projection of the IX tergum in this third specimen was observed to be narrower and smaller compared to the broader and larger structure typically seen in Ae. albopictus [Figure 6 (D)]. Furthermore, the two side lobes of the IX tergum in this specimen exhibited several prominent hairs, differing from the pattern observed in Ae. albopictus. These morphological observations highlight subtle yet significant variations within the specimens, contributing to our understanding of the diversity and morphological range within the Ae. albopictus species, particularly in the context of the Scutellaris Group in the oriental region. Specific confirmation of these three male specimens were also carried out by molecular methods.
Detection of Wolbachia
Wolbachia was found to be present naturally in the 7 Ae. albopictus specimens out of 9. However, it was absent in all the 10 Ae. nr. albopictus via amplification of the wsp gene (Supplementary Figure).
Discussion
Reproductively isolated isomorphic and sympatric populations under a taxon are called cryptic or sibling species (WHO, 1998)12. Cryptic species are common in insects. It was estimated that for each morphologically identified insect species, there can be on average 3.1 cryptic species (Ni & Weins, 2022)13.
Adults of the Scutellaris Group of mosquitoes are morphologically characterized by having a median longitudinal white stripe of narrow scales from the anterior margin of the scutum to the wing root. However, they are most difficult to differentiate because of subtle differences and variation in their morphological characters (Huang, 1972)4. The presence of isomorphic species like Ae. subalbopictus and Ae. pseudoalbopictus along with Ae. albopictus creates a significant challenge in correctly identifying such species7,8,9,11. In addition, the presence of a cryptic species clearly emphasized the need for a molecular-based species identification in such a situation. In our present study, out of 139 morphologically resembled Ae. albopictus specimens,14 were found to be other species when subjected to molecular methods.
The phylogenetic trees, nucleotide diversity & mean genetic distance analysis confirmed that our study population is genetically different from Ae. albopictus, Ae. subalbopictus and other Aedes species. Although, this species is in the same clade as the Ae. albopictus cryptic species earlier reported from Vietnam and China (Tables 1 and 2), we have discovered three new haplotypes (Hap_1, Hap_2, and Hap_4), (Fig. 4). Also, this is the first time we have shown the sympatrically co-breeding of Ae. nr. albopictus with Ae. albopictus in rubber collection bowls.
The time tree analysis revealed the median divergence time of the Ae. nr. albopictus species and Ae. albopictus species to be approximately 36.13 million years ago and Aedes aegypti was the most ancient ancestor (approximately 46.01 million years ago). The divergent time between Ae. albopictus and Ae. aegypti found in our study is similar to those studies reported earlier by Zhao et al.14 and Nardi et al.15. These findings support the speciation of Ae. nr. albopictus species during the course of evolution.
In our present study the Ae. nr. albopictus specimens were morphologically very similar to those of Ae. albopictus (Fig. 6A). It was reported earlier that adult females of Scutellaris Group are most difficult to identify because of minute differences and variation in their taxonomic characters (Huang, 1972)4. However, the male genitalia of such species contains distinct characters (Huang,1972)4. Considering the same we have tried to examine male specimens of Ae. nr. albopictus species. Even though we could examine only one male specimen, we have observed subtle differences in the IX tergum of male genitalia of the Ae. nr. albopictus with Ae. albopictus [Figure 6 (A), (C)].
Although only one male specimen with differences in genitalia characteristics was found, it does suggest that this is a distinct species different from Ae. albopictus found in the area. Phylogenetic analyses (Figs. 2 and 3) conclusively indicate the presence of this species which is morphologically similar to Ae. albopictus but genetically different from Ae. albopictus, Ae. subalbopictus, Ae. pseudoalbopictus, Ae. flavopictus, Ae. aegypti, Ae. vittatus, while resembling Ae. albopictus cryptic species from China and Vietnam. Hence this is the first study to confirm the presence of a distinct Aedes species. In view of the evidence seen morphologically of male specimen, molecular distinctness between the study population and Ae. albopictus, we provisionally designate this species found in Dhalai, Tripura as ‘Aedes nr. albopictus’ following the nomenclature suggested by Sigovini et al.16, . Here nr. stands for near meaning is that species is close to Ae. albopictus but not identical. Further studies are required to formally describ this species as per the guidelines of International Code of Zoological Nomenclature (ICZN). This discovery marks for the first time such a species has been identified in this subcontinent region, contributing significantly to our understanding of the genetic diversity and distribution of the Aedes species in India.
The specimens of the Ae nr. albopictus in this study were from light trap collection in the huts, situated in the forested region (Fig. 6), thus are similar to collections in Vietnam7.
Several questions are still unanswered or only partially answered because Ae. nr. albopictus species was found for the first time in Tripura state, India. These include whether this species has breeding habitat preference, whether both pre- and post-mating barriers or only one of them responsible for the evolution of Ae. nr. albopictus species found in this study and what are the behavioural differences, like resting and feeding preferences, responses to existing vector control tools, vectorial potential as Wollbachia was not seen in this species, while it is present in Ae. albopictus. Moreover, what evolutionary processes in mosquito cryptic species lead to reproductive isolation in the absence of morphological distinction is yet to be deciphered, though some studies on cryptic species have tried to understand the aspect of reproductive isolation17. While emerging species of the malaria vector Anopheles gambiaen complex, Anopheles culicifacies complex have been shown to exhibit varied ecological preferences and strong pre and / or postzygotic reproductive isolation and cryptic species of Ae. mariae and Ae. zammitii under Mariae species complex have demonstrated post-mating reproductive isolation. However, there is still minimal or no data reporting reproductive isolation in Culex and Aedes albopictus mosquito cryptic species17,18.
Our study clearly indicates that these are sympatric species as both Ae. albopictus and Ae. nr. albopictus species were found in the same breeding habitat in the rubber collection bowls (Fig. 6). Sympatric speciation is a process whereby two closely related species occupying the same ecological niche undergo divergent evolution, leading to genetic differentiation that ultimately prevents them from interbreeding, and/or Wolbachia present in Ae. albopictus played a role in speciation. This sympatric association is an evolutionary mechanism that facilitates the emergence of new cryptic species19. Some studies have tried to answer whether Wolbachia infection has an impact on the reproductive isolation of mosquitoes. Researchers have identified Wolbachia-mediated reproductive isolation, particularly through unidirectional cytoplasmic incompatibility (CI), as a mechanism that could lead to population divergence and potentially contribute to the formation of new species20. Wolbachia infection can cause CI in insects such as mosquitoes, fruit flies, and butterflies. Crosses between infected males and uninfected females result in infertility. This one-way reproductive barrier could eventually lead to genetic divergence and reproductive isolation between populations, potentially driving speciation20. However, it’s important to note that reproductive isolation is often a complex process involving multiple mechanisms. While Wolbachia-mediated CI can be a significant factor, it may not be the sole driver of reproductive isolation between populations. Other factors, such as behavioural isolation, where individuals from different populations have preferences for mating with their own kind, and hybrid inviability, where hybrids between different populations have reduced fitness or viability, can also contribute to reproductive isolation20. In cases where crosses between infected males and uninfected females result in infertility due to Wolbachia-induced CI, the other direction of the cross (infected females with uninfected males) may remain fertile20.
Interestingly while Ae. albopictus is mostly harbouring Wolbachia in the guts, no Wolbachia was found in the Ae. nr. albopictus species in our studies or previous study7,8.
The lack of Wolbachia endosymbiont in our study, its near-absence in China’s Aedes albopictus cryptic species, suggests that Wolbachia may have played a role in its speciation20.
Moreover, the absence of a Wolbachia endosymbiont in this species also raises the possibility of increasing virus-carrying capability21 Viral detection could not be done because samples were used for studies to confirm the new species. Thus, this study calls for future studies that screen for RNA viruses like dengue, chikungunya, and rift valley fever to plan new vector control tools.
Our molecular approach, coupled with morphological studies, reaffirms the importance of such methods in identifying and confirming morphologically similar species, as highlighted in our previous studies11. Molecular identification not only aids in genetic characterization but also helps in accurately identifying breeding habitats shared by isomorphic species which would have otherwise been considered Ae. albopictus, only by morphological examination. The discovery of these species, particularly in the absence of molecular studies in India, suggests the possibility of missing more such isomorphic species in the past, underscoring the need for further research on archived samples.
Conclusion
This study marks the discovery of Ae. nr. albopictus species resembling Ae. albopictus in India. Our findings from Tripura, India highlight the possibility of sympatric speciation of Ae. nr. albopictus species. This is a significant development in understanding the importance of molecular characterization, identification of breeding sites, evolution of Aedes species and transmission dynamics of Dengue and Chikungunya in rural, forested areas of Tripura, India.
Materials and methods
Study area and collection of mosquitoes
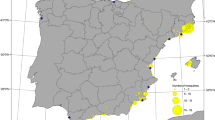
This study was conducted using archived Aedes samples collected from five districts of Tripura: Dhalai, Gomoti, Sepahijala, Unokoti, and West District from 2019 to 2022. Mosquito larvae were collected from various natural and artificial containers and adult mosquitoes were caught in the traps or hand-caught (Fig. 7).
Morphological identification of mosquitoes
For morphological identification, we followed the standard keys4,22. The adults emerging from larvae, along with adults caught in light traps and by hand (Mouth aspirator), were first identified based on their morphological features. Male genitalia of three adult males were examined by dissecting the male terminalia and by mounting them on glass slides following the book (Huang, 1972)4.
Molecular identification of mosquito species
To perform molecular identification, genomic DNA was extracted from individual larvae and the legs of adult mosquitoes. Using a 1.5 mL centrifuge tube, the whole larva or mosquito’s leg was grinded by using a micro pestle and then QIAamp DNA micro kit’s DNA extraction methodology was followed to extract the total nucleic acids. The DNA was stored at −20ºC for further analysis. We targeted the mitochondrial cytochrome oxidase I (COI) and Internal Transcribed Spacer II (ITS2) regions. For COI region, briefly the PCR was performed in 50 µL reaction volume with approximately 50 ng DNA as a template using 2X master mix (Promega, USA) following methodologies described by Kumar et al.23. Additionally, MgCl2 (1.5 mM) was added in the final reaction as part of modification. For ITS2 region amplification a gradient PCR protocol was employed following the primer pairs described by Walton et al.24. 56 ºC was taken as annealing temperature since the annealing temperature described in the reference protocol did not yield positive result. The final PCR was carried out in a 50 µL 2X master mix (GoTaq Promega, USA) and 1.5 mM MgCl2 with approximately 50 ng DNA as a template. Post electrophoretic separation, PCR products were visualized under UV Transilluminator (BioRad XR).
Sequencing of COI and ITS2 genes and phylogenetic analysis
Following post-PCR clean-up, the amplicons were bi-directionally sequenced at Eurofins Genomics, India. Quality value greater than or equal to Qv20 was considered for base calling. Sequence editing was performed in Bio-edit v7.0.5.3 software25and submitted to the GenBank. Similar sequences were retrieved from NCBI, and phylogenetic analysis was done in MEGA X software26. The phylogenetic trees for the COI and ITS2 genes were constructed by using the Maximum Likelihood method based on the best fit model27. For the COI tree construction 12 Ae. aegypti, 12 Ae. vittatus, 10 Ae. albopictus, 3 Ae. flavopictus, 2 Ae. subalbopictus, 8 Ae. albopictus cryptic species of China, 11 Ae. albopictus cryptic species of Vietnam and 5 of this study were used. In the ITS2 phylogenetic tree 2 Ae. albopictus, 2 Ae. aegypti, Ae. albopictus cryptic species of China, 3 Ae. albopictus cryptic species of Vietnam and 6 of this study were used. The length of the alignment used to construct the phylogenetic tree for COI and ITS2 gene were 396 and 331 base pairs respectively.
Nucleotide difference and diversity analysis
The nucleotide difference and diversity among the different Aedes species were calculated for the COI gene using the software DnaSP v.628.
Mean genetic distance analysis
The mean genetic distance among the different Aedes species was calculated for the COI gene using the MEGA X software26.
Time tree analysis
For the estimation of the time tree, the approximate divergence time between Ae. aegypti and Ae. albopictus was calculated using the time tree of life online database29with a class interval of 15–59 million years ago14,15,30. The time tree was constructed in MEGA X software26 using the mitochondrial COI sequences of Ae. albopictus cryptic (India, China and Vietnam). Ae. albopictus, Ae. subalbopictus, Ae. vittatus and Ae. aegypti. The COI sequence of Culex quinquefasciatus was included as an outgroup. The time tree was inferred by applying the ReLTime method to the constructed phylogenetic tree27. Confidence intervals and the minimum and maximum time boundaries on nodes for which calibration densities were provided, were computed using the Tao et al.31 method. The length of the alignment for inferring the time tree was 367 base pairs.
Haplotype network analysis
For haplotype network analysis, we used PopART v.1.7 software32 and the minimum spanning method for analyzing variation in the mitochondrial COI region. This included sequences from both Ae. albopictus cryptic species sampled in Vietnam and China, as well as other Aedes taxa collected from diverse geographic locations around the globe. A total of 62 COI sequences, including the five detected in our study, were incorporated into the haplotype network analysis.
Detection of Wolbachia in mosquito species
To detect Wolbachia endosymbiont in mosquitoes, we extracted genomic DNA from the abdomen of adult mosquitoes and individual larvae following the DNA extraction protocol of the QIAamp DNA mini kit. The presence of Wolbachia was indicated by a 600 bp amplicon in the PCR analysis. PCR was carried out by targeting the Wolbachia surface protein (wsp) gene using the primer pair wsp81F/wsp691R as previously described by Braig et al.33. The PCR reaction was performed in 50 µL reaction volume using 2X master mix (Promega, USA) with approximately 50 ng of gDNA as template and at an annealing temperature of 55ºC. Known Wolbachia DNA was taken as positive control whereas DNA extracted from the leg of Aedes mosquito was taken as negative control. PCR products were analyzed in ethidium bromide-stained 1.5% agarose gel and viewed under UV Transilluminator (BioRad XR).
Preparation of ecological map with cases and vectors
Utilizing ortho-rectified Indian Remote Sensing satellite data, Cartosat-1 (2.5 m) and LISS-IV (5.8 m), land use and land cover (LULC) mapping were produced utilizing on-screen visual interpretation techniques on the Geographic Information System (GIS) platform. Using the most recent spatial layer data (2019), which was first supplied at 1:10,000 scales by NRSC/ISRO’s Space-based Information Support, major LULC categories and subcategories were delineated and updated. The project team also carried out field verifications to guarantee the accuracy of the data interpretations.
Data availability
Sequences dataset that supports this study (newly discovered cryptic species of Aedes albopictus/ Ae. nr. albopictus), have been submitted to the GenBank database ( NCBI) — Accession Numbers OR139061-OR139064, ON007025, PF741448 and Accession Numbers OR129842-OR129846 respectively.
References
Mosquito Taxonomic Inventory. Available online: (2016). http://mosquito-taxonomic-inventory.info (accessed on 2 March 2024).
Paixão, E. S., Teixeira, M. G. & Rodrigues, L. C. Zika, Chikungunya and dengue: the causes and threats of new and re-emerging arboviral diseases. BMJ Global Health, 3(Suppl 1), e000530. (2018).
Huang, Y. M. Medical entomology studies-XI. The subgenus stegomyia of Aedes in the Oriental region with keys to the species (Diptera: Culicidae). Contrib. Am. Entomol. Inst. 15 (6), 1–79 (1979).
Huang, Y. M. Contributions to the mosquito fauna of Southeast Asia XIV: The subgenus Stegomyia of Aedes in Southeast Asia I (The scutellaris group of species, 1972).
Huang, Y. M. A new species of Aedes (Stegomyia) from Sri Lanka (Ceylon) (Diptera: Culicidae). Mosq. Syst. 7 (4), 345–356 (1975).
Bhattacharyya, D. R. et al. Faunal richness and the checklist of Indian mosquitoes (Diptera: Culicidae). Check List. 10 (6), 1342–1358 (2014).
Minard, G., Tran Van, V., Tran, F. H., Melaun, C., Klimpel, S., Koch, L. K., … Valiente Moro, C. (2017). Identification of sympatric cryptic species of Aedes albopictus subgroup in Vietnam: new perspectives in phylosymbiosis of insect vector. Parasites& vectors,10, 1–14.
Guo, Y. et al. Molecular evidence for new sympatric cryptic species of Aedes albopictus (Diptera: Culicidae) in China: A new threat from Aedes albopictus subgroup? Parasites Vectors. 11, 1–14 (2018).
World Health Organization. Regional Office for South-East Asia. Anopheline species complexes in South and South-east Asia Vol. 57 (World Health Organization, 2007).
Johnson, K. N. The impact of wolbachia on virus infection in mosquitoes. Viruses 7 (11), 5705–5717 (2015).
Biswas, S. et al. First report of rubber collection bowls & plastic and bamboo water containers as the major breeding source of Aedes albopictus with the Indigenous transmission of dengue and Chikungunya in rural forested malaria-endemic villages of Dhalai district, Tripura, India: the importance of molecular identification. Biomedicines 11 (8), 2186 (2023).
Subbarao, S. K. Anopheline species complexes in south-east Asia Vol. 18 (WHO Regional Offices for South-East Asia, 1998).
Li, X. & Wiens, J. J. Estimating global biodiversity: the role of cryptic insect species. Syst. Biol. 72 (2), 391–403 (2023).
Zhao, Z., Su, T. J., Chesters, D., Wang, S. D., Ho, S. Y., Zhu, C. D., … Zhang, C.T. (2013). The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae)and the evolutionary timescale of tachinid flies. PLoS One, 8(4), e61814.
Nardi, F. et al. Domestication of Olive fly through a multi-regional host shift to cultivated Olives: comparative dating using complete mitochondrial genomes. Mol. Phylogenet. Evol. 57 (2), 678–686 (2010).
Sigovini, M., Keppel, E. & Tagliapietra, D. Open nomenclature in the biodiversity era. Methods Ecol. Evol. 7 (10), 1217–1225 (2016).
Zheng, X. L. Unveiling mosquito cryptic species and their reproductive isolation. Insect Mol. Biol. 29 (6), 499–510 (2020).
Subbarao, S. K., Vasantha, K. & Sharma, V. Studies on the crosses between the sibling species of the Anopheles culicifacies complex. J. Hered. 79, 300–303 (1988).
Les, D. H. et al. Through Thick and Thin: cryptic sympatric speciation in the submersed genus Najas (Hydrocharitaceae). Mol. Phylogenet. Evol. 82, 15–30 (2015).
Bruzzese, D. J. et al. Testing the potential contribution of wolbachia to speciation when cytoplasmic incompatibility becomes associated with host-related reproductive isolation. Mol. Ecol. 31 (10), 2935–2950 (2022).
Huang, E. Y., Wong, A. Y., Lee, I. H., Qu, Z., Yip, H. Y., Leung, C. W., … Hui, J.H. (2020). Infection patterns of dengue, Zika and endosymbiont Wolbachia in the mosquitoAedes albopictusin Hong Kong. Parasites & vectors, 13, 1–10.
Barraud, P. J. The Fauna of British India, Including Ceylon and Burma. Diptera. Vol V. Family Culicidae. Tribes Megarhinini and Culicini 463 (Taylor and Francis, 1934).
Kumar, N. P., Rajavel, A. R., Natarajan, R. & Jambulingam, P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 44 (1), 01–07 (2007).
Walton, C., Somboon, P., O’Loughlin, S. M., Zhang, S., Harbach, R. E., Linton, Y.M., … Butlin, R. K. (2007). Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infection,Genetics and Evolution, 7(1), 93–102.
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids symposium series (Vol. 41, No. 41, pp. 95–98). (1999), January.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547 (2018).
Nei, M. & Kumar, S. Molecular evolution and phylogenetics (Oxford University Press, 2000).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302 (2017).
Time Tree. The Timescale of Life. Available online: March (2024). http://www.timetree.org/ (accessed on 27.
Moreno, M. et al. Complete MtDNA genomes of Anopheles Darlingi and an approach to anopheline divergence time. Malar. J. 9, 1–13 (2010).
Tao, Q., Tamura, K., Mello, B. & Kumar, S. Reliable confidence intervals for RelTime estimates of evolutionary divergence times. Mol. Biol. Evol. 37 (1), 280–290 (2020).
Leigh, J. W. & Bryant, D. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 (9), 1110–1116 (2015).
Braig, H. R., Zhou, W., Dobson, S. L. & O’Neill, S. L. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont wolbachia Pipientis. J. Bacteriol. 180 (9), 2373–2378 (1998).
Acknowledgements
We acknowledge the help of Ujjal Sarkar, Chelapro Mog, Prosenjit Malakar, Sangit Debnath, Bidyakanta Das and Kamaljoy Tripura for larva and adult mosquito collection, Vijay Vettri for support in experimentation, Abhijit Das, Ashit Paul, Satyajit Sen, Siddhartha Jaiswal, Mayur Govekar Ratilal, Tapas Bhattacharya and Tanu Jain for support from the vector-borne disease program and State and District Health Department and administration. We also acknowledge Model Rural Health Research Unit (MRHRU) Tripura and Multidisciplinary Research Unit (MRU) Agartala for the logistic support. Prof Subrata Baidya, Agartala Govt Medical College, Tripura for the logistic support.
Funding
This work was funded by the Indian Council of Medical Research (ICMR) extramural grant NER/65/2018-ECD-I to IPB.
Author information
Authors and Affiliations
Contributions
Conceptualization, I.P.B., D.R.B., K.N., and S.K.S.; methodology, S.B., J.R., I.P.B., D.R.B., B.J.B., P.G., T.N., S.R.J., S.K.V., R.P., G.H., A.K.S. and B.B; Software, J.R., S.B., T.N., R.P. and S.R.J.; validation, I.P.B., S.K.S., D.R.B., K.N. and B.J.B.; investigation, S.B., J.R., P.G., I.P.B.,D.R.B., S.R.J. and B.B.; writing original draft preparation, I.P.B., S.B., J.R., D.R.B., T.N., S.K.V., S.R.J and S.K.S.; writing review and editing, D.R.B., S.K.S., K.N., K.B., B.J.B. and I.P.B.; visualization, I.P.B., J.R., S.B., B.J.B., D.R.B., R.P., and S.R.J.; supervision, I.P.B., D.R.B., B.J.B., K.N. and S.K.S.; data curation, S.B., S.R.J., P.G., T.N., J.R; A.K.S., G.H. and B.B.; project administration, K.N. and I.P.B.; funding acquisition, I.P.B., K.N. and H.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Biswas, S., Rajkonwar, J., Jena, S.R. et al. Detection of a sympatric cryptic species mimicking Aedes albopictus (Diptera: Culicidae) in dengue and Chikungunya endemic forest villages of Tripura, India, posing a daunting challenge for vector research. Sci Rep 15, 14237 (2025). https://doi.org/10.1038/s41598-025-96146-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-96146-9