Abstract
Cancer immunotherapy is a beneficial therapy for many cancer types, but predictive pan-tumor biomarkers for clinical benefit are suboptimal. Our study, employing DNA and RNA based analysis, investigated the role of predicted neoantigens in the benefits of immunotherapy within a cohort of 88 patients of European descent with advanced solid tumors. Patients who had a prolonged (> 12 months) duration of immunotherapy exhibited heightened immune responses, characterized by increased levels of predicted neoantigens with strong HLA binding potential, elevated cytotoxic marker levels, and enhanced T cell activity. Furthermore, our analysis revealed associations between prolonged duration of therapy and rare variants, notably within the EPHA8 gene. These variants, exclusive to patients with a prolonged (> 12 months) duration of immunotherapy, suggest potential implications for immunotherapy response. In addition, the evolutionary conservation of these variants across vertebrate species underscores their functional importance in tumor biology and ultimately, treatment outcomes. Despite limitations in sample size and patient homogeneity, our findings emphasize the potential utility of understanding the molecular and immunological mechanisms underlying immunotherapy responses to further refine personalized treatment strategies.
Similar content being viewed by others
Introduction
Cancer remains one of the most formidable challenges in modern medicine, causing significant morbidity and mortality. Despite remarkable advancements in treatment modalities, therapeutic advances are needed. The introduction of immunotherapy has emerged as a major therapeutic advance that has shown benefits across multiple disease types and multiple settings. The use of immune checkpoint therapy by blocking the action of cytotoxic T lymphocyte antigen-4 (CTLA-4)1, programmed death-1 (PD-1)2 and its ligand, PD-L13, has improved cure rates and survival across multiple settings. While several predictive biomarkers have been shown to provide clinical utility, their predictive capacity varies across tumor types and patients4,5,6,7.
Currently, microsatellite instability (MSI-H), tumor mutation burden (TMB), and PD-L1 expression, serve as predictive biomarkers for checkpoint inhibition8. MSI-H and TMB have associated FDA approvals with checkpoint inhibition as pan-tumor biomarkers. PD-L1 expression has demonstrated predictive benefit among many tumor types, but the staining intensity and utility are highly variable9. As an example of variability, patients with metastatic triple negative breast cancer (TNBC) appear to benefit from pembrolizumab only if the PD-L1 CPS score ≥ 10, whereas pembrolizumab appears to confer benefit to patients with non-metastatic TNBC regardless of PD-L1 score10,11,12. While these markers improve personalization, there remains a substantial degree of discordance between biomarker status and benefit from immunotherapy. The complexity of tumor immunity is one explanation for the difficulty in identifying an optimal predictive biomarker.
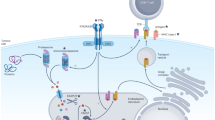
The intricacy of immunotherapy and response to therapy is a function of the tumor, the tumor microenvironment, and its interaction with the host immune system13,14,15,16. Tumors harbor thousands of mutations, and it is well known that tumors with a higher load of genetic alterations have a greater probability to respond well to treatment. Nonsynonymous mutations within tumor cells lead to the formation of altered protein sequences which are proteolytically cleaved to generate short peptides called neoantigens17,18,19. These neoantigens are recognized as non-self by the immune system and are capable of eliciting an immune response, which is the first step in the immune-cancer cycle20. While TMB may not consistently align with immune checkpoint inhibitor (ICI) response21,22, there is a notable correlation between immunogenic neoantigens and the efficacy of ICI treatment19,23,24. The identification and characterization of these neoantigens is of great importance, not only to understand cancer immunity, but also in developing effective therapeutic interventions. The challenge is that not all of them are capable or will be recognized by T cells to initiate the immune response.
In this study, we evaluated for an association between predicted neoantigens with strong potential for binding to HLA molecules and duration of benefit on checkpoint inhibition across tumor types. Furthermore, we evaluated the impact of cytotoxic markers and T-cell activity on duration of therapy.
Results
Patient characteristics
The study cohort comprised 88 patients of European descent (Fig. 1), including 54 males and 34 females. Across this cohort, a total of 21 distinct tumor types were represented, with patients receiving treatment involving PD-1, PD-L1, CTLA4 inhibitors, or combinations thereof. Among these patients, 53 had a treatment duration of less than 3 months (< 3 M), while 35 had therapy for over 12 months (> 12 M), with respective median treatment durations of 42 and 614 days. The median age of patients was 65 years in the < 3 M group and 62 years in the > 12 M group (Table 1).
CONSORT diagram of patient selection and analysis. This diagram outlines the patient selection and categorization based on duration of therapy and sequencing data availability. The initial cohort consisted of 138 patients. Of these, 50 patients were excluded due to having a duration of therapy between 3 and 12 months. Following these exclusions, 53 patients had a duration of therapy less than 3 months, while 35 patients had a duration exceeding 12 months. DNA sequencing data was available for 88 patients and RNA sequencing data was available for 57 patients.
Distribution of deleterious nonsynonymous exonic mutations and rare variants between the < 3 month and > 12 month groups
A high number of nonsynonymous deleterious mutations was associated with longer duration of therapy (> 12 M), with respective median mutation counts of 87 and 178 in the < 3 M and > 12 M groups, respectively. In the rare variant analysis across the 88 patients, there were a total of 16,807 SNPs within 5,368 genes identified, setting the threshold significance at p < 9.3 × 10− 6. Among the covariates tested, namely age, sex, BMI, ECOG status, TMB, MSI, prior lines of therapy and tumor type, only TMB demonstrated significant association (p ≤ 0.05). Among the genes identified in the rare variant germline analysis, 20 exhibited a significance level of p < 0.01, with the top association in EPHA8 (p = 2.05 × 10− 4; Supplementary Table S1) which was comprised of 3 deleterious variants. The tumor DNA analysis of the EPHA8 gene identified 8 rare exonic variants, 3 of which were also found in the germline analysis. Both germline (n = 3) and somatic(n = 8), were observed only in the patients who had a longer duration of therapy (> 12 M) across several tumor types (Supplementary Fig. S1). In this patient cohort, EPHA8 expression in the tumor was lower in the > 12 M group (p = 0.049; Supplementary Fig. S2).
Conservation of amino acid variants across species
To gauge the evolutionary conservation of amino acid alterations induced by the 8 rare variants in the EPHA8 gene, all vertebrate species listed in the UCSC genome browser25 were scrutinized. This comprehensive analysis encompassed primate (n = 11), euarchontoglires (n = 14), laurasiatherian (n = 25), Afrotheria (n = 6), mammal (n = 5), bird (n = 14), sarcopterygii (n = 8), and fish (n = 16) species. All 8 representative amino acids exhibited high levels of conservation across the 99 species tested, ranging from 84% (83/99) to 98% (97/99), underscoring the evolutionary significance of these amino acids.
T-cell activity and cytotoxic marker levels
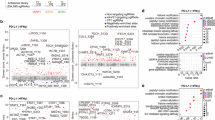
Among the 57 patients with available RNA sequencing data, the expression level of candidate markers for immune activity, GZMA, GZMB, PRF1, NKG7, and CD8A were assessed in the tumor and correlated with duration of therapy. A higher expression of all assessed genes was associated with the longer duration of therapy (> 12 M). (Figures 2 and 3c).
Expression of cytotoxic markers across duration of therapy groups. This figure displays the expression levels of four cytotoxic markers - GZMA, GZMB, PRF1, and NKG7 across two duration of therapy groups. Each panel displays one marker, with the x-axis representing the duration of therapy and the y-axis showing the expression levels (TPM) for each gene.
Deleterious nonsynonymous mutations, predicted neoantigens with strong HLA binding potential and CD8 T Cell expression by Duration of Therapy. Panels (a–c) display the number of deleterious nonsynonymous mutations, the number of strong-binding neoantigens, and CD8 T cell expression levels (TPM) on the y-axis, respectively. The x-axis represents the duration of therapy.
Predicted neoantigen load and HLA binding
Tumors harboring a greater number of mutations tend to yield a higher abundance of predicted neoantigens, thereby increasing tumor immunogenicity. In our study, the median count of 9-mer peptides originating from nonsynonymous deleterious variants was 83 and 177 in the < 3 M and > 12 M groups, respectively, based on analysis of 87 patients. Moreover, the median count of predicted neoantigens demonstrating strong binding affinity for the HLA was 7 and 16 in the < 3 M and > 12 M groups, respectively. These findings are further illustrated in Fig. 3.
Discussion
Neoantigens, which arise from somatic mutations within tumor cells, have gained significant attention with regards to cancer immunotherapy due to their potential to elicit robust antitumor immune responses. Our study highlights the dynamic interplay between predicted neoantigens with stronger HLA binding affinity, T-cell activity, cytotoxicity, and treatment duration, shedding light on key determinants of immunotherapy efficacy in a cohort of 88 patients of European descent with various tumor types. These newly formed antigens, recognized as foreign by the immune system, particularly by T-cells, can drive an immune response against the tumor. Analysis of the predicted neoantigens and HLA binding revealed intriguing insights into tumor immunogenicity and potential antigen presentation. Patients who had a longer therapy duration (> 12 M) exhibited a higher number of deleterious nonsynonymous mutations and predicted neoantigens with stronger HLA binding affinity compared to those with shorter therapy duration (< 3 M). This suggests that a higher number of strong HLA binding neoantigens is associated with better outcomes in immunotherapy.
Our study explored the relationship between T-cell activity, cytotoxic marker expression, and treatment duration. The levels of expression of the T-cell marker CD8A and other cytotoxicity markers, such as GZMA, GZMB, PRF1, and NKG7, are established indicators of T-cell activity and pivotal factors in immunotherapy response26,27,28,29,30. Patients with longer therapy durations exhibited elevated expression levels of CD8A and cytotoxic markers, suggesting enhanced T-cell activity and resultant immunotherapy benefit. This aligns with previous research associating higher cytotoxic marker expression with improved treatment outcomes28,29,30 and underscores the importance of monitoring immune cell dynamics during therapy. Furthermore, our analysis of rare variants identified a gene of particular interest, EPHA8, which showed a significant association with longer therapy duration. Notably, EPHA8 expression was found to be lower in patients who were on therapy for a longer duration. Functional analysis revealed that all 8 rare exonic variants, including 3 germline and 5 somatic variants, were highly conserved across species, suggesting their functional relevance. These variants were predicted to be deleterious by bioionformatic tools (SIFT, PolyPhen, and CADD), potentially disrupting normal biological functions, which may further influence therapy outcomes. EPHA8 is an Eph receptor and belongs to the Eph/ephrin receptor tyrosine kinase subfamily. Previous research has demonstrated that mutations in the Ephrin type-A receptors (EPHA) are associated with enhanced efficacy and survival benefits in lung adenocarcinoma patients treated with checkpoint inhibitors31. Additionally, overexpression of the Eph receptor and its ligand ephrin has been implicated in tumorigenesis32,33 and the development of autoimmune diseases, with studies showing increased expression in inflammatory conditions like rheumatoid arthritis, suggesting their potential role in immune cell regulation and inflammation34,35,36. Furthermore, Eph receptor overexpression has also been shown to be a potential therapeutic target in colorectal cancer37. Prior work has shown that high EPHA8 protein expression serves as an independent prognostic marker for poor overall survival in epithelial ovarian cancer patients38.
While our study provides valuable insights into immunotherapy response and tumor biology, it has limitations affecting the generalizability of our findings. Defining precise time points for categorizing responders and non-responders is challenging and somewhat arbitrary, with variable definitions used in other studies to define “significant clinical benefit”. It depends on a number of variables, including disease type and setting39,40,41,42,43. For “clinical futility”, we set a 3-month threshold as this is often the first time point for evaluation of early drug success or futility in routine clinical practice and in clinical trials for advanced cancer. Thus, those with progression at this early time point likely gained no real biological or clinical benefit. For “significant clinical benefit”, we chose 12 months as it would exceed the definition of clinical benefit response rate (defined as disease response or stability for more than six months) and would be less than the median progression free survival (PFS) across many disease types and settings. The retrospective design and small sample size of 88 patients of European descent may limit statistical power and the applicability of our findings to more diverse populations. Larger, prospectively stratified cohorts would help confirm and reinforce our findings, providing more precise patient subgroup characterization. Tumor type did not reach statistical significance in our analysis and was not included as a covariate, though it could influence immunotherapy responsiveness. The impact of intratumor heterogeneity on immune response, which was not accounted for in our study, represents a valuable opportunity for further insight.
Overall, these findings provide valuable insights into the complex interplay between tumor genetics, immune response dynamics, and treatment outcomes in patients receiving immunotherapy. Patients who received immunotherapy for longer durations exhibited more favorable molecular and immunological profiles compared to those with shorter treatment durations. This work contributes to the ongoing efforts to optimize personalization of immunotherapeutic strategies and improve patient outcomes in cancer treatment. The identification of EPHA8 variants, along with predicted neoantigens and T-cell activity, highlights key factors influencing immunotherapy effectiveness. Future research on EPHA8 as a therapeutic target, coupled with further studies on HLA-binding neoantigens, could lead to more personalized treatments that better harness the immune system’s ability to target and destroy tumors.
In conclusion, predicting the risk-to-benefit ratio of any drug is critical for informed decision making, particularly with immunotherapy. Unfortunately, biomarkers for immunotherapy are currently suboptimal and improved predictive capacity is essential for better therapy stratification. Immunity is a complex and multifactorial process, influenced by the tumor, the host, and the tumor microenvironment. We believe that our comprehensive approach in a real-world population, across tumor types offers valuable insights into this challenge and might help refine the ability to predict benefit. However, further validation in larger datasets is necessary, and ongoing efforts will be focused on assessing the applicability of these biomarkers across tumor types and disease subtypes.
Methods
Selection of patient cohort
The retrospective study cohort consisted of adult patients of European descent who were diagnosed with advanced solid tumors and who underwent treatment with one or more checkpoint inhibitors from November 19, 2013, to September 25, 2020, and evaluated in the IU Health Precision Genomics Program. We specifically evaluated patients that had not received concurrent cytotoxic therapy to avoid durable responses which may have been due to the chemotherapy. Patient inclusion was contingent upon the availability of either whole-genome or whole-exome sequencing data and comprehensive follow up documentation. Patients were categorized into two groups based on treatment duration: those who received therapy for less than 3 months and those surpassing 12 months. Treatment duration was defined as the period from checkpoint inhibitor initiation to disease progression or a change in treatment course. 12 months was selected as a prolonged duration of therapy that would be uniformly accepted as a time of significant clinical benefit and 3 months as clinical futility, each representing an extreme phenotype to optimize detection of key molecular differences. Demographic and clinical data for these patients were extracted from electronic health records via chart reviews. Approval for this study was granted by the Indiana University Institutional Review Board and all methods were performed in accordance with the relevant guidelines and regulations. Written informed consent was obtained from all patients.
Identification of rare germline variants
The patient germline DNA bam files were assessed for quality and processed following the best practices recommended in the GATK germline short variant discovery pipeline44 to generate a multisample GVCF file. In the selected cohort, 29% of the patients had whole-genome sequence data; to maintain consistency with exome data, variants with over 20% missing data were filtered out. Individual patients were retained if their variant data exhibited less than 30% missingness. Multiallelic variants were segregated into distinct rows, followed by left alignment, normalization, and removal of monomorphic variants. The variants were annotated using ANNOVAR45,46, and then filtered based on exonic function (nonsynonymous SNV, startloss, stopgain, stoploss, frameshift insertion/deletion, unknown) and predicted deleteriousness (SIFT47, Polyphen48, CADD49 > = 20). Variants with a minor allele frequency (MAF) ≤ 0.03 in either of the European populations (NHLBI-ESP650050, ExAC51 and 1000G52) were retained and only genes with two or more retained variants were used. Rare variant association analysis was conducted using the R package SKAT(v2.2.5)53,54.
Analysis of the top gene rare variant
The top hit from the germline SKAT rare variant analysis was further investigated in the tumor DNA. The gene region was extracted, and the methodologies outlined in the identification of germline variants were applied to identify rare variants. Rare germline and somatic variants were scrutinized for amino acid conservation across all species cataloged in the UCSC genome browser25. The distribution of these variants, both germline and somatic, within the two treatment duration groups, as well as their prevalence across different cancer types, was tabulated. Gene expression levels of the identified gene were quantified using TPM (Transcripts Per Million) values and compared between patient groups.
T-cell and cell cytotoxicity markers analysis
Among the 88 patients, 57 also had RNA sequence data available. The featurecount tool55 from the subread package(v2.0.2)56,57 was used to summarize the reads. TPM values were used for comparing marker gene expression between the two treatment duration groups, with significance testing conducted using the Wilcoxon rank sum test.
Tumor-only nonsynonymous deleterious exonic variants, HLA typing, neoantigen identification, and HLA binding affinity
The patients’ tumor DNA bam files were assessed for quality in conjunction with previously processed normal DNA bam files, and variants exclusive to the tumor were extracted utilizing Mutect2 software(v4.1.9.0)44,58 with the analysis performed on 87 of the 88 tumor bam files, as one from the less than 3 month group was corrupted and could not be used. The variants were annotated using ANNOVAR, and then filtered based on exonic function (nonsynonymous SNV, startloss, stopgain, stoploss, frameshift insertion/deletion, unknown) and predicted deleteriousness (SIFT, Polyphen, CADD > = 20). A custom python script was used to retrieve the 9-mer peptide sequences (neoantigens) corresponding to the filtered exonic variants for each patient. For HLA allele typing, normal bam files from each patient were utilized and the Optitype software(v1.3.3)59,60 was used to call the HLA alleles, following the protocol and guidelines provided by the authors. Finally, the binding affinity of these 9-mer peptides to each patient’s specific HLA alleles was predicted using NetMHCpan software(v4.1b)61,62, ensuring accurate pairing of neoantigens with the relevant HLA for each patient, following the authors protocol and guidelines.
Statistical analysis
Statistical analyses were conducted to evaluate the association between rare variants, cytotoxic activity markers, T-cell activity markers, and predicted neoantigens with duration of therapy. The duration of therapy was categorized as either significant clinical benefit (> 12 M) or clinical futility (< 3 M). A gene-based case-control analysis (SKAT) was employed to identify genes associated with significant clinical benefit, including only covariates, with p ≤ 0.05. The significance threshold was established by applying Bonferroni correction to account for multiple genes tested. The Wilcoxon rank sum test was employed to compare marker gene expression, as well as nonsynonymous deleterious variants and potential neoantigen, between treatment duration groups.
Data availability
The data used during the current study is available from the corresponding author on reasonable request. [https://doi.org/10.7912/6JTE-HS52]
References
Leach, D. R., Krummel, M. F. & Allison, J. P. Enhancement of antitumor immunity by CTLA-4 Blockade. Science 271, 1734–1736. https://doi.org/10.1126/science.271.5256.1734 (1996).
Ishida, Y., Agata, Y., Shibahara, K. & Honjo, T. Induced expression of PD-1, a novel member of the Immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11, 3887–3895. https://doi.org/10.1002/j.1460-2075.1992.tb05481.x (1992).
Cha, J. H., Chan, L. C., Li, C. W., Hsu, J. L. & Hung, M. C. Mechanisms controlling PD-L1 expression in cancer. Mol. Cell. 76, 359–370. https://doi.org/10.1016/j.molcel.2019.09.030 (2019).
Bai, R., Lv, Z., Xu, D. & Cui, J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 8, 34. https://doi.org/10.1186/s40364-020-00209-0 (2020).
Kovacs, S. A., Fekete, J. T. & Gyorffy, B. Predictive biomarkers of immunotherapy response with Pharmacological applications in solid tumors. Acta Pharmacol. Sin. 44, 1879–1889. https://doi.org/10.1038/s41401-023-01079-6 (2023).
Sankar, K. et al. The role of biomarkers in personalized immunotherapy. Biomark. Res. 10, 32. https://doi.org/10.1186/s40364-022-00378-0 (2022).
Wang, D. R., Wu, X. L. & Sun, Y. L. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal. Transduct. Target. Ther. 7, 331. https://doi.org/10.1038/s41392-022-01136-2 (2022).
Sharma, P. et al. Immune checkpoint therapy-current perspectives and future directions. Cell 186, 1652–1669. https://doi.org/10.1016/j.cell.2023.03.006 (2023).
Vranic, S. & Gatalica, Z. PD-L1 testing by immunohistochemistry in immuno-oncology. Biomol. Biomed. 23, 15–25. https://doi.org/10.17305/bjbms.2022.7953 (2023).
Badve, S. S. et al. Determining PD-L1 status in patients with Triple-Negative breast cancer: lessons learned from IMpassion130. J. Natl. Cancer Inst. 114, 664–675. https://doi.org/10.1093/jnci/djab121 (2022).
Schmid, P. et al. Pembrolizumab for early Triple-Negative breast cancer. N Engl. J. Med. 382, 810–821. https://doi.org/10.1056/NEJMoa1910549 (2020).
Cortes, J. et al. Pembrolizumab plus chemotherapy in advanced Triple-Negative breast cancer. N Engl. J. Med. 387, 217–226. https://doi.org/10.1056/NEJMoa2202809 (2022).
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330. https://doi.org/10.1038/nature21349 (2017).
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80. https://doi.org/10.1126/science.aaa6204 (2015).
Pitt, J. M. et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 27, 1482–1492. https://doi.org/10.1093/annonc/mdw168 (2016).
Baghban, R. et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell. Commun. Signal. 18 https://doi.org/10.1186/s12964-020-0530-4 (2020).
Lang, F., Schrors, B., Lower, M., Tureci, O. & Sahin, U. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat. Rev. Drug Discov. 21, 261–282. https://doi.org/10.1038/s41573-021-00387-y (2022).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74. https://doi.org/10.1126/science.aaa4971 (2015).
Xie, N. et al. Neoantigens: promising targets for cancer therapy. Signal. Transduct. Target. Ther. 8 https://doi.org/10.1038/s41392-022-01270-x (2023).
Chen, D. S. & Mellman, I. Oncology Meets immunology: the cancer-immunity cycle. Immunity 39, 1–10. https://doi.org/10.1016/j.immuni.2013.07.012 (2013).
Chan, T. A. et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 30, 44–56. https://doi.org/10.1093/annonc/mdy495 (2019).
McGrail, D. J. et al. High tumor mutation burden fails to predict immune checkpoint Blockade response across all cancer types. Ann. Oncol. 32, 661–672. https://doi.org/10.1016/j.annonc.2021.02.006 (2021).
McGranahan, N. et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint Blockade. Science 351, 1463–1469. https://doi.org/10.1126/science.aaf1490 (2016).
Ott, P. A. et al. An Immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. https://doi.org/10.1038/nature22991 (2017).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006. https://doi.org/10.1101/gr.229102 (2002).
Chowdhury, D. & Lieberman, J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol. 26, 389–420. https://doi.org/10.1146/annurev.immunol.26.021607.090404 (2008).
Cullen, S. P., Brunet, M. & Martin, S. J. Granzymes in cancer and immunity. Cell. Death Differ. 17, 616–623. https://doi.org/10.1038/cdd.2009.206 (2010).
Hurkmans, D. P. et al. Granzyme B is correlated with clinical outcome after PD-1 Blockade in patients with stage IV non-small-cell lung cancer. J. Immunother Cancer. 8 https://doi.org/10.1136/jitc-2020-000586 (2020).
Fan, C. et al. PRF1 is a prognostic marker and correlated with immune infiltration in head and neck squamous cell carcinoma. Transl Oncol. 14, 101042. https://doi.org/10.1016/j.tranon.2021.101042 (2021).
Wen, T. et al. NKG7 is a T-cell-Intrinsic therapeutic target for improving antitumor cytotoxicity and cancer immunotherapy. Cancer Immunol. Res. 10, 162–181. https://doi.org/10.1158/2326-6066.CIR-21-0539 (2022).
Bai, H. et al. EPHA mutation as a predictor of immunotherapeutic efficacy in lung adenocarcinoma. J. Immunother Cancer. 8 https://doi.org/10.1136/jitc-2020-001315 (2020).
Arora, S., Scott, A. M. & Janes, P. W. Eph receptors in cancer. Biomedicines 11 https://doi.org/10.3390/biomedicines11020315 (2023).
Surawska, H., Ma, P. C. & Salgia, R. The role of Ephrins and Eph receptors in cancer. Cytokine Growth Factor. Rev. 15, 419–433. https://doi.org/10.1016/j.cytogfr.2004.09.002 (2004).
Coulthard, M. G. et al. Eph/Ephrin signaling in injury and inflammation. Am. J. Pathol. 181, 1493–1503. https://doi.org/10.1016/j.ajpath.2012.06.043 (2012).
Darling, T. K. & Lamb, T. J. Emerging roles for Eph receptors and Ephrin ligands in immunity. Front. Immunol. 10, 1473. https://doi.org/10.3389/fimmu.2019.01473 (2019).
Kitamura, T. et al. Enhancement of lymphocyte migration and cytokine production by ephrinB1 system in rheumatoid arthritis. Am. J. Physiol. Cell. Physiol. 294, C189–196. https://doi.org/10.1152/ajpcell.00314.2007 (2008).
DiPrima, M. et al. Identification of Eph receptor signaling as a regulator of autophagy and a therapeutic target in colorectal carcinoma. Mol. Oncol. 13, 2441–2459. https://doi.org/10.1002/1878-0261.12576 (2019).
Liu, X. et al. EphA8 is a prognostic marker for epithelial ovarian cancer. Oncotarget 7, 20801–20809. https://doi.org/10.18632/oncotarget.8018 (2016).
Pokorny, R. et al. Real-world experience with elective discontinuation of PD-1 inhibitors at 1 year in patients with metastatic melanoma. J. Immunother Cancer. 9 https://doi.org/10.1136/jitc-2020-001781 (2021).
Amiot, M. et al. When to stop immunotherapy for advanced melanoma: the emulated target trials. EClinicalMedicine 78, 102960. https://doi.org/10.1016/j.eclinm.2024.102960 (2024).
Jansen, Y. J. L. et al. Discontinuation of anti-PD-1 antibody therapy in the absence of disease progression or treatment limiting toxicity: clinical outcomes in advanced melanoma. Ann. Oncol. 30, 1154–1161. https://doi.org/10.1093/annonc/mdz110 (2019).
Kaufman, H. L. et al. Durable response rate as an endpoint in cancer immunotherapy: insights from oncolytic virus clinical trials. J. Immunother Cancer. 5 https://doi.org/10.1186/s40425-017-0276-8 (2017).
Kuah, C. Y., Monfries, R., Quartagno, M., Seckl, M. J. & Ghorani, E. What is the optimal duration, dose and frequency for anti-PD1 therapy of non-small cell lung cancer? Ther. Adv. Med. Oncol. 15, 17588359231210271. https://doi.org/10.1177/17588359231210271 (2023).
McKenna, A. et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. https://doi.org/10.1101/gr.107524.110 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. https://doi.org/10.1093/nar/gkq603 (2010).
ANNOVAR. https://annovar.openbioinformatics.org/en/latest/user-guide/download/
Ng, P. C. & Henikoff, S. S. I. F. T. Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814. https://doi.org/10.1093/nar/gkg509 (2003).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods. 7, 248–249. https://doi.org/10.1038/nmeth0410-248 (2010).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894. https://doi.org/10.1093/nar/gky1016 (2019).
Fu, W. et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493, 216–220. https://doi.org/10.1038/nature11690 (2013).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. https://doi.org/10.1038/nature19057 (2016).
Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74. https://doi.org/10.1038/nature15393 (2015).
Wu, M. C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89, 82–93. https://doi.org/10.1016/j.ajhg.2011.05.029 (2011).
SKAT. https://cran.r-project.org/web/packages/SKAT/index.html
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. https://doi.org/10.1093/bioinformatics/btt656 (2014).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108. https://doi.org/10.1093/nar/gkt214 (2013).
SUBREAD. https://subread.sourceforge.net/.
Mutect2. https://gatk.broadinstitute.org/hc/en-us/articles/360051306691-Mutect2
Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316. https://doi.org/10.1093/bioinformatics/btu548 (2014).
OptiType. https://github.com/FRED-2/OptiType
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif Deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48, W449–W454. https://doi.org/10.1093/nar/gkaa379 (2020).
netMHCpan. https://services.healthtech.dtu.dk/cgi-bin/sw_request?software=netMHCpan&version=4.1&packageversion=4.1b&platform=Linux.
Acknowledgements
We sincerely thank the IU Health Precision Genomics Program for generously providing the data essential for our analysis.
Author information
Authors and Affiliations
Contributions
S.P. and B.P.S. conceived and designed the study. S.P. collected data, performed analysis, and wrote the manuscript. P.L. performed chart reviews. C.F. and T.W. helped with tumor classification. S.P., P.L., C.F., T.W., G.J., F.S., E.C., M.T., L.M.R., B.P.S reviewed, provided feedback and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Philips, S., Lu, P., Fausel, C. et al. Association of heightened host and tumor immunity with prolonged duration of response to checkpoint inhibition across solid tumors. Sci Rep 15, 13195 (2025). https://doi.org/10.1038/s41598-025-96925-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-96925-4