Abstract
Polycyclic aromatic hydrocarbons (PAHs) are hazardous environmental contaminants emerging from industrial activities and fossil fuel combustion, posing risks to human health and ecosystems. Biodegradation offers a sustainable approach to mitigating PAH pollution, and here we investigated the efficacy of a peptide hybrid of laccase and O-methyltransferase enzymes from the bacterium Burkholderia cepacia in PAH degradation. Both enzymes demonstrated stability with an instability index below 40, indicating suitability for environmental application. Following active site prediction, the 3D structure of the peptide hybrid, consisting of 71 amino acids, was modelled using trRosetta, achieving a high-quality structure with an ERRAT score above 97%. Further bioinformatic analysis confirmed the hybrid’s non-allergenic and non-virulent properties. Molecular docking studies revealed a robust binding affinity above − 9 kcal/mol, highlighting this peptide hybrid’s potential for effective PAH degradation and suggesting its promise as an eco-friendly bioremediation agent for PAH-contaminated sites.
Similar content being viewed by others
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a class of organic compounds composed of multiple fused aromatic rings, predominantly formed as byproducts of incomplete combustion of organic material, including fossil fuels, wood, and industrial chemicals1. These pollutants are released into the environment through various anthropogenic activities, such as vehicular emissions, industrial discharges, agricultural burning, and the use of petroleum-based products2. Due to their high stability and hydrophobic nature, PAHs accumulate in soil and aquatic systems, leading to widespread contamination. Their persistence and bioaccumulation in the environment pose significant threats to human health, ecosystems, and biodiversity3. The impact of PAHs on human health is particularly concerning, as many PAH compounds are recognized carcinogens, mutagens, and endocrine disruptors. Exposure to PAHs, either through inhalation of polluted air, consumption of contaminated food and water, or dermal contact, has been linked to various adverse health effects, including respiratory issues, developmental disorders, and increased cancer risk4. Additionally, PAHs can exert toxic effects on aquatic organisms, plants, and animals, further disrupting ecosystems and reducing biodiversity5. Given these harmful effects, it is critical to explore effective methods for the degradation and removal of PAHs from the environment.
Biodegradation, a natural process through which microbial organisms break down contaminants into less harmful or non-toxic forms, has emerged as a promising and eco-friendly approach for PAH remediation6. Unlike conventional methods such as incineration or chemical oxidation, biodegradation offers a sustainable and cost-effective means of reducing environmental PAH levels without producing secondary pollutants. Biodegradation is intrinsically a multi-step process that typically requires the synergistic action of multiple enzymes to achieve complete degradation of complex PAH structures. Effective biodegradation involves a diverse array of microbial enzymes that facilitate critical biochemical transformations, including oxidation, methylation, and ring cleavage of PAHs7. The complexity of these transformations can result in a lengthy degradation process, often characterized by inefficiencies due to the intricate interplay of enzymatic activities required for the complete mineralization of PAHs. Consequently, optimizing these enzymatic pathways is essential for enhancing the overall efficiency of PAH biodegradation in contaminated environments.
Burkholderia cepacia, a versatile bacterium known for its enzymatic capabilities and ability to thrive in diverse environments, has been studied extensively for its PAH-degrading potential8. This microorganism’s enzymatic arsenal, particularly its laccase and O-methyltransferase enzymes, plays a crucial role in breaking down complex PAH structures. Laccase, a copper-containing oxidase, initiates the oxidation of PAHs, converting them into intermediate metabolites. The enzyme O-methyltransferase then acts on these intermediates, facilitating their further breakdown into simpler, less toxic compounds.
In this study, the hybrid peptide of laccase and O-methyltransferase from B. cepacia demonstrates enhanced efficiency in PAH degradation by combining the oxidative power of laccase with the methylation activity of O-methyltransferase. This synergistic approach accelerates the biodegradation process, offering a promising, eco-friendly alternative to traditional methods. However, challenges related to scalability and economic feasibility must be addressed. Scaling up production requires optimizing expression systems and purification techniques to reduce costs, and strategies such as enzyme immobilization can enhance reusability and improve cost-effectiveness. Further testing is also needed to ensure stability under varying environmental conditions. With ongoing optimization, this hybrid enzymatic system shows significant potential for application in bioremediation, offering an effective solution to PAH contamination and its harmful impacts on human and ecological health.
Methodology
Sequence retrieval
For the synthesis of peptides aimed at degrading polycyclic aromatic hydrocarbons (PAHs), we selected two enzymes from Burkholderia cepacia: O-methyltransferase and Laccase. The FASTA sequences of these enzymes were retrieved from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). NCBI a division of National Institutes of Health serves as a comprehensive repository, providing detailed information on the primary structures of protein and other biomolecules9.
Physiochemical properties
Expasy ProtParam tool accessible via the Swiss Bioinformatics Resource Portal (https://web.expasy.org/protparam/) was utilized for the sequence characterization of O-methyl transferase and Laccase, provided in FASTA format. ProtParam is a tool that can calculate various chemical and physical properties of proteins10. These proteins include theoretical pI, molecular weight, amino acid composition, instability index, and GRAVY (grand average of hydropathicity). The instability index and GRAVY are particularly important for assessing protein stability.
Prediction of binding sites
The binding sites of the peptide hybrid were predicted using the PrankWeb online tool. PrankWeb (https://prankweb.cz) is a web-based interface that utilizes machine learning algorithms to identify potential ligand-binding sites based on the 3D structure of peptides or proteins11. The peptide hybrid’s 3D structure was uploaded to the tool, which analysed the surface residues and predicted possible binding pockets by assessing geometric and physicochemical features. The results were visualized to highlight the predicted binding sites, providing insights into potential functional regions of the peptide hybrid.
Prediction of 3D structure of peptide hybrid
The 3D structure of the peptide hybrid of two enzymes Lacasse and O-methyltransferase using TrRosetta (https://yanglab.nankai.edu.cn/trRosetta/help/). The trRosetta (transform-restrained Rosetta) algorithm predicts protein structures rapidly and effectively12. It is an online web server that builds the structure of a protein using direct energy minimizations and restrained/constrained Rosetta. The inter-residue geometries, including orientations and distance, are initially predicted using a deep neural network, and then predicted geometries are subsequently converted into restraints /constraints to forecast the structure.
Peptide physiochemical properties
The physicochemical properties of the peptide were predicted by using Expasy ProtParam (https://web.expasy.org/protparam/). This is a tool for predicting the physicochemical properties of protein13. It serves accurately to predict many properties like Molecular weight, Instability Index, GRAVY-Grand Average of Hydropathicity, aliphatic index, extinction coefficient, isoelectric point, molecular weight, and the net charge on protein. It also predicts amino acid composition and a proteins atomic composition.
Assessment of 3D structure quality for peptide hybrid
The validation of 3D structures of peptide hybrid was validated using the Ramachandran plot and ERRAT to assess their quality, with PROCHECK (https://saves.mbi.ucla.edu/). The PDB form of peptide was used as the input for the analysis. The Ramachandran plot visualized the distribution of amino acids into different categories predicting the quality of the 3D structure of the peptide, while ERRAT provided a graphical representation, highlighting amino acids located in either error or non-error regions, helping to evaluate the accuracy of the predicted structures14.
Prediction of allergenicity
By identifying the allergic compounds in the protein molecules, the allergenicity of metalloproteins was determined using AllerTOP v.20 (https://www.ddg-pharmfac.net/AllerTOP/). It is an online server used to predict allergenicity based on the physicochemical properties of proteins15. By comparing the protein sequences in databases, this technique evaluates the ability of the peptide hybrid structure to cause symptoms based on cross-reactivity.
Prediction of virulence
The virulence factor of peptide hybrid was predicted using the Virtual Interactive Chemical Modeling Prediction (VICM-pred) software (https://webs.iiitd.edu.in/raghava/vicmpred/). The tool analyzed the sequence based on specific motifs and structural features associated with pathogenicity16. The output included scores indicating the likelihood of each protein being a virulence factor, which were compared against known virulence proteins in the literature to assess biological relevance and potential pathogenic implications.
Selection of ligands
The ligands selected for degradation are the derivatives of aromatic polycyclic hydrocarbons (PAHs). These are prevalent in the environment as pollutants. Fifteen ligands were selected and their structures were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). PubChem is a comprehensive database maintained by NCBI. It provides an easy access to the information regarding molecular weight, formula and structure of various compounds14.
Molecular Docking between peptide and PAHs derivatives
A total of 15 PAHs derivatives present in the environment were selected and underwent docking analysis with the help of PyRx which generated their complexes with the peptide. PyRx (https://pyrx.sourceforge.io/) is a widely used tool designed for the docking analysis of multiple ligands with the protein in a single run15. The purified PDB files of ligands and the peptide were uploaded as input on PyRx. The peptide was converted to macromolecule and ligands were converted to pdbqt file. After energy minimization and the grid setting, docking was run. The results obtained for each ligand were in the form of binding affinities. The docked complex with the highest energy was downloaded and uploaded on the Discovery Studio where the complex was further analysed for interactions between the amino acid residues of peptide with Benzo[a]pyrene.
Results
Sequence retrieval
The FASTA sequence of O-methyltransferase and Laccase were retrieved from NCBI with accession no. XDR27978.1 and AYA62526.1 respectively. O-methyltransferase consists of 221 amino acids and starts with methionine ending at arginine. Similarly, Laccase consists of 279 amino acids starting with methionine and ending at glycine.
Physiochemical properties
The Expasy ProtParam was utilized to analyse the physiochemical properties of proteins, providing valuable insights into their behaviour in in-vitro conditions as shown in the Table 1. The following parameters were calculated for the proteins: number of amino acids, molecular weights, theoretical pI, molecular formula, total number of atoms, instability index, aliphatic index, and GRAVY. The instability index for O-methyltransferase was found to be 36.43, while that for laccase was 28.44, both of which are less than the threshold value of 40. This indicates that both proteins are stable. The GRAVY score of O-methyltransferase is -0.015, suggesting its hydrophilic nature, whereas the GRAVY score of Laccase is 0.124, indicating its hydrophobic nature.
Prediction of binding sites
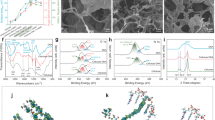
The binding sites of two proteins Lacasse and O-methyltransferase were predicted by using Prankweb Online tool. The result shows that the Lacasse enzyme consist of three binding sites while O-methyltransefrase enzyme consist of two binding sites as shown in Fig. 1. The predicted scores for each binding sites of both the protein is shown in Table 2. These scores highlighting the potential effectiveness of each binding site for ligand binding and providing insights into the functional regions of both enzymes.
The prediction of Binding sites of the Laccase and O-methyltransferase protein through PrankWeb tool. The Figure A shows the Two binding sites of 0-methyltransferase protein (Red shows the binding site 1 and yellow shows the binding site 2). The Figure B shows the three binding sites of Laccase protein (Red shows the binding site1, yellow shows the binding site 2 and orange shows the binding site 3).
Prediction of 3D structure of peptide hybrid
The 3D structure of the peptide hybrid of Lacasse and O-methyltransferase enzyme was predicted by using trRosetta. The 3D structural model generated showed a well-folded peptide with distinct regions corresponding to both laccase and O-methyltransferase domains, indicating proper integration of the hybrid peptide’s features from N terminal to C terminal as shows in Fig. 2.
Peptide physiochemical properties
The physiochemical properties of the peptide hybrid were analyzed through Expasy ProtParam as shown in Table 3. The peptide structure consists of 71 amino acids with a molecular weight of 7798.19. The result shows that the instability index of protein predicted to be 16.99 suggesting that the peptide is stable. Additionally, the Grand Average of Hydropathicity (GRAVY) value is 1.18, indicating that the peptide is hydrophobic in nature. This suggests a strong preference for interacting with membrane proteins, which may enhance its stability and functionality.
Assessment of 3D structure quality for peptide hybrid
The quality assessment of 3D structure of peptide hybrid was predicted through aRAMACHANDRAN plot and ERRAT through PROCHECK server. The results of RAMACHANDRAN plot depicts that the 91.5% of amino acid residues were in most allowed regions, 6.2% in additional regions, 1.7% in the generously allowed region and 0.6% in disallowed regions (Fig. 3). In the case of the ERRAT graph, the overall quality graph was 97.605% (Fig. 4).
Prediction of allergenicity
The allergenicity of peptide hybrid structure was predicted as a probably non-allergen. This suggests that the peptide hybrid is considered to be safe enough to use as a bioremediator due to its low allergenicity and non-allergen activity.
Prediction of virulence
The result suggests that the peptide hybrid is non-virulent with predicted functional class for Metabolism molecule. Further result shows the score of the peptide hybrid structure in different processes as shown in Table 4.
Ligand selection
After selecting 15 ligands PubChem provided their 3D structures along with the information regarding chemical formula and molecular weight which are as follows in Table 5.
Molecular docking between peptide and PAHs derivatives
PyRx was utilized to perform docking between the ligands and the peptide. All the ligands showed excellent docking results. Benzo[a]pyrene, Indeno[1,2,3-cd] pyrene, and Benzo[g, h,i]perylene were the lead ligands that showed the best docking energies of − 9.6, 9.5 and 9.2. Benzo[a]pyrene (B[a]P), Indeno[1,2,3-cd]pyrene (I[1,2,3-cd]P), and Benzo[g, h,i]perylene (B[g, h,i]P) are significant polycyclic aromatic hydrocarbons (PAHs) known for their toxicity, persistence, and prevalence in the environment. B[a]P is classified as a Group 1 carcinogen by the International Agency for Research on Cancer (IARC) and is linked to various cancers, particularly lung and gastrointestinal cancers. I[1,2,3-cd]P is also considered a potential human carcinogen, while B[g, h,i]P, though less frequently highlighted, poses similar toxicity concerns. These compounds are highly persistent in the environment, remaining in soil, water, and air for extended periods and leading to bioaccumulation in organisms. They primarily originate from urban sources such as vehicle emissions and industrial processes and can also be found in contaminated food products17. Energies of the ligands are shown in Table 6.
Among these binding affinities the maximum energy recorded is -9.6 with Benzo[a]pyrene. Discovery Studio was used for the interaction analysis. Benzo[a]pyrene shows conventional hydrogen bonding with Glu87 and Leu84 residues with the bond length of 4.13 Å and 5.35 Å. It also formed one alkyl interaction with Leu165 having bond length of 5.87 Å shown in Figs. 5 and 6.
Discussion
In this study, we explored a novel peptide hybrid composed of laccase and O-methyltransferase enzymes from Burkholderia cepacia for the biodegradation of polycyclic aromatic hydrocarbons (PAHs), evaluating its stability, binding affinity, and structural integrity. PAHs are known for their persistence in the environment and association with human health risks, making their removal essential for mitigating ecological damage18. Conventional methods for PAH degradation, such as chemical oxidation and physical removal, can be costly and generate secondary pollutants19. Consequently, the microbial degradation of PAHs has garnered attention for its efficiency and eco-friendliness. Several studies have reported on various bacteria and enzymes capable of degrading PAHs20, yet the focus on peptide hybrids in enhancing degradation efficiency remains limited. Our approach aimed to bridge this gap by designing a peptide hybrid combining the oxidative capabilities of laccase with the methylation activity of O-methyltransferase to create a more potent biodegradation mechanism. Similarly, O-methyltransferase enzymes play a role in reducing toxicity by methylating these intermediates21. However, most prior research has utilized these enzymes individually, which can limit the degradation efficiency and leave toxic intermediates in the environment. This study developed a hybrid peptide incorporating both enzymatic functions within a single structure, potentially offering more comprehensive PAH breakdown. This hybridization approach aligns with findings from similar studies that report enhanced biodegradation using multifunctional enzyme systems but advances the field by providing a stable, highly effective peptide hybrid for PAH degradation22.
The structural stability of the peptide hybrid is a critical parameter for environmental application. Using instability index analysis, we confirmed that both laccase and O-methyltransferase remained stable within the hybrid structure, each displaying an instability index below 40. This result indicates the hybrid’s potential to remain functional in various environmental conditions, offering advantages over other reported single-enzyme approaches, which may be more susceptible to degradation23. The peptide hybrid’s 3D structure was constructed through trRosetta, yielding a reliable model with an ERRAT score above 97%, indicative of a highly accurate structure. The structure’s stability was further corroborated by allergenicity and virulence predictions, which demonstrated that the peptide hybrid is both non-allergenic and non-virulent, essential for safe application in bioremediation. In contrast, earlier models of peptide structures in PAH biodegradation often lacked stability validation, underlining our study’s contribution to safer environmental applications24. The molecular docking results of our hybrid peptide showed a high binding affinity, exceeding − 9 kcal/mol, with targeted PAH compounds. This affinity suggests a strong interaction between the peptide hybrid and PAHs, providing a foundation for effective degradation at contaminated sites. The high binding affinity observed in this study surpasses many reported values in similar PAH-degrading systems, positioning our hybrid peptide as a promising agent for PAH removal. These findings demonstrate the utility of this engineered peptide hybrid in potentially minimizing the release of harmful intermediates and ensuring thorough biodegradation of PAH contaminants.
Overall, our study indicates that the peptide hybrid combining laccase and O-methyltransferase is a robust, stable, and effective approach for PAH biodegradation. Through stability assessments, 3D modeling, and molecular docking studies, we have established a solid foundation for the application of this peptide hybrid in environmental cleanup, offering a promising bioremediation solution for PAH contamination.
The engineered peptide hybrid is a new way of degrading PAHs with synergistic enzyme synergistic properties to overcome the shortcomings of conventional single-enzyme systems. Also, structural stability and high specific binding affinity of the hybrid is also shown, as well as its non-allergenic and non- virulent properties aiming the safe application to the environment. For further studies, this method should be upscaled in order to evaluate its efficacy in real-life scenarios operating in multi-faceted soil or water conditions contaminated with varying PAH substances. Further, investigation into the kinetic parameters of hybrid peptide under varied pH, salinity and temperature also could prove useful in guiding its utility for various ecologies.
Peptide hybrid could be redefined in the molecular scaffold or nano carriers systems to improve the stability of enzymes and increase the number of reuses of the enzyme within the site of contamination. Also the integration of the hybrid peptide with microbial consortia or bioaugmentation techniques would broaden its use, particularly in sites poorly supplied with bioavailable PAHs.
This bioremediation approach improves upon conventional chemical and physical methods for PAH removal by offering an eco-friendly, efficient, and specific solution. Unlike chemical methods, which may generate secondary pollutants, or physical methods, which often require extensive energy and resources, the hybrid peptide provides targeted degradation of PAHs with minimal environmental impact. Furthermore, the hybrid peptide’s high specificity and stability under diverse conditions make it suitable for targeted applications in both controlled systems, such as bioreactors, and natural environments, such as soil or water. These features not only reduce the environmental footprint but also make the process more energy-efficient and scalable for widespread use.
Conclusion
This research introduces an innovative approach for the biodegradation of polycyclic aromatic hydrocarbons (PAHs) through a hybrid peptide that integrates the enzymatic functions of laccase and O-methyltransferase derived from Burkholderia cepacia. By combining the oxidative capabilities of laccase with the methylation potential of O-methyltransferase, the hybrid peptide demonstrated improved degradation efficiency, structural integrity, and environmental suitability. Structural modeling confirmed the hybrid peptide’s stability, achieving an ERRAT score above 97%, while assessments for allergenicity and virulence indicated its safety for potential environmental applications. Additionally, molecular docking studies showed significant binding affinities, with binding energies greater than − 9 kcal/mol, highlighting the peptide’s strong interactions with PAH compounds and its promising role in bioremediation initiatives. This study emphasizes the potential of multifunctional peptide hybrids in tackling persistent organic pollutants, presenting a sustainable and effective alternative to traditional remediation methods. Future studies should aim to validate the performance of the hybrid peptide under various environmental conditions, such as changes in pH, temperature, and pollutant concentrations. Moreover, evaluating the scalability and cost-effectiveness of this bioremediation technique is crucial for its practical application. Such research could lead to groundbreaking, sustainable strategies for addressing PAH contamination on a global scale.
Data availability
All the data has been included in the manuscript.
References
Sahoo, B. M. et al. Polyaromatic hydrocarbons (PAHs): structures, synthesis and their biological profile. Curr. Org. Synth. 17(8), 625–640 (2020).
Mallah, M. A. et al. Polycyclic aromatic hydrocarbon and its effects on human health: an overeview. Chemosphere 296, 133948 (2022).
Girardin, V., Grung, M. & Meland, S. Polycyclic aromatic hydrocarbons: bioaccumulation in dragonfly nymphs (Anisoptera), and determination of alkylated forms in sediment for an improved environmental assessment. Sci. Rep. 10(1), 10958 (2020).
da Silva Junior, F. C. et al. A look beyond the priority: A systematic review of the genotoxic, mutagenic, and carcinogenic endpoints of non-priority PAHs. Environ. Pollut. 278, 116838 (2021).
Honda, M. & Suzuki, N. Toxicities of polycyclic aromatic hydrocarbons for aquatic animals. Int. J. Environ. Res. Public Health. 17(4), 1363 (2020).
Xu, M. et al. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by bacterial mixture. Int. J. Environ. Sci. Technol. 1–12 (2022).
Zhao, H. et al. Assessment on the rings cleavage mechanism of polycyclic aromatic hydrocarbons in supercritical water: A ReaxFF molecular dynamics study. J. Mol. Liq. 415, 126311 (2024).
Cauduro, G. P. et al. Burkholderia Vietnamiensis G4 as a biological agent in bioremediation processes of polycyclic aromatic hydrocarbons in sludge farms. Environ. Monit. Assess. 195(1), 116 (2023).
Schoch, C. L. et al. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database 2020, pbaaa062 (2020).
Naveed, M. et al. An in Silico approach Uncovering the competency of oncolytic human adenovirus 52 for targeted breast cancer virotherapy. Sci. Rep. 14(1), 26405 (2024).
Jakubec, D. et al. PrankWeb 3: accelerated ligand-binding site predictions for experimental and modelled protein structures. Nucleic Acids Res. 50(W1), W593–W597 (2022).
DiMaio, F. Rosetta structure prediction as a tool for solving difficult molecular replacement problems. Protein Crystallogr. 455–466 (2017).
Naveed, M. et al. Design of a novel multiple epitope-based vaccine: an immunoinformatics approach to combat SARS-CoV-2 strains. J. Infect. Public Health. 14(7), 938–946 (2021).
Naveed, M. et al. The natural breakthrough: phytochemicals as potent therapeutic agents against spinocerebellar ataxia type 3. Sci. Rep. 14(1), 1529 (2024).
Naveed, M. et al. Assessment of melia Azedarach plant extracts activity against hypothetical protein of Mycobacterium tuberculosis via GC-MS analysis and in-silico approaches. J. Comput. Biophys. Chem. 23(3), 299–320 (2024).
Kashyap, D., Khan, A. & Lahare, B. Pathogenic protein identification and localization prediction in Pseudomonas fuscovaginae: A study on sheath brown rot in rice. Rev. Argentina Clín. Psicol. 29, 150–160 (2020).
Femi-Oloye, O. P. et al. An assessment of polycyclic aromatic hydrocarbons using Estimation programs. Toxics 12(8), 592 (2024).
Feng, Y., Li, Z. & Li, W. Polycyclic aromatic hydrocarbons (PAHs): environmental persistence and human health risks. Nat. Prod. Commun. 20(1), 1934578X241311451 (2025).
Rubio-Clemente, A., Torres-Palma, R. A. & Peñuela, G. A. Removal of polycyclic aromatic hydrocarbons in aqueous environment by chemical treatments: a review. Sci. Total Environ. 478, 201–225 (2014).
Zhang, L. et al. Microbial degradation of multiple PAHs by a microbial consortium and its application on contaminated wastewater. J. Hazard. Mater. 419, 126524 (2021).
Jaffar, S., Ahmad, S. & Lu, Y. Contribution of insect gut microbiota and their associated enzymes in insect physiology and biodegradation of pesticides. Front. Microbiol. 13, 979383 (2022).
Tesfaye, E. L., Bogale, F. M. & Aragaw, T. A. Biodegradation of polycyclic aromatic hydrocarbons: the role of ligninolytic enzymes and advances of biosensors for in-situ monitoring. Emerg. Contam. 100424 (2024).
Jeon, Y. et al. Unraveling the mechanisms of benzo [a] pyrene degradation by pigmentiphaga Kullae strain KIT-003 using a multi-omics approach. Ecotoxicol. Environ. Saf. 281, 116665 (2024).
Achife, C. E. et al. Assessment and biodegradation of polycyclic aromatic hydrocarbons in soil and water around petroleum products depot Suleja, Nigeria. Appl. Biochem. Biotechnol. 196(5), 2819–2838 (2024).
Acknowledgements
The authors greatly acknowledge and express their gratitude to the Researchers Supporting Project number (RSP2025R462), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization, Muhammad Naveed; methodology, Ayesha Saleem; software, Khadija Khatoon; validation, Maida Salah Ud Din; formal analysis, Ahiba Adil.; investigation, Ayesha Saleem; resources, Tariq Aziz.; data curation, Mitu Al-harbi; writing—original draft preparation, Khadija Khatoon.; writing—review and editing, Tariq Aziz; visualization, Abdullah F Alasmari; supervision, Muhammad Naveed.; project administration, Tariq Aziz.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Naveed, M., Saleem, A., Aziz, T. et al. Elucidating the synergistic role of hybrid peptide from Burkholderia cepacia enzymes in biodegradation of polycyclic aromatic hydrocarbons. Sci Rep 15, 12603 (2025). https://doi.org/10.1038/s41598-025-97007-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97007-1