Abstract
Apoptotic chromatin condensation inducer 1 (Acin1) is an RNA-binding protein involved in the regulation of alternative splicing, but its physiological function remains unclear. Global deletion of Acin1 causes embryonic lethality around E11.5, with mutants exhibiting developmental delays and increased apoptosis. Using conditional knockout mice, we found that skeletal muscle myofiber-specific Acin1 knockout mice (Acin1 MKO) are viable and fertile and that Acin1 MKO mice show enlarged myofibers and ongoing muscle damage and regeneration, characterized by increased central nuclei and embryonic myosin heavy chain expression. RNA-seq analysis revealed that Acin1 deletion altered the expression and splicing patterns of genes crucial for muscle function. Notable changes included modified splicing of genes associated with muscle disease and mitochondrial function, often resulting in the expression of gene variants typical of immature or diseased muscle. These findings suggest that Acin1 is essential for embryonic development and has limited effects on muscle structure and homeostasis via its regulation of gene expression and alternative splicing.
Similar content being viewed by others
Introduction
Gene expression is regulated not only at the transcriptional level but also via post-transcriptional mechanisms, including splicing, mRNA modifications, transport, stabilization, degradation, and translation. Numerous RNA-binding proteins are involved in these processes. Alternative splicing (AS), a crucial form of post-transcriptional regulation, is the primary mechanism by which multiple protein isoforms with distinct structures, localization, expression levels, and biological functions are generated from a single gene. More than 90% of human genes undergo AS1,2, and dysregulation of AS can lead to severe dysfunction and disease.
Apoptotic chromatin condensation inducer 1 (Acin1), also known as Acinus, was initially identified as a caspase-3 substrate that promotes apoptotic chromatin condensation3. The human or mouse Acin1 gene gives rise to three isoforms (Acin1-L, Acin1-S, and Acin1-S') through alternative splicing mechanisms3. These three isoforms share a common C-terminal region. Within its C-terminus, the Acin1 protein has two structural features typical of SR/SR-related splicing factors: an RNA recognition motif and two arginine/serine (RS) domains, which are rich in RS dipeptides. Acin1 binds to Sin3A-associated protein 18 (SAP18) and RNA-binding protein with serine-rich domain 1 (RNPS1), forming an apoptosis and splicing-associated protein (ASAP) complex that is associated with the exon junction complex (EJC)4,5. Acin1 binds to SAP18 and RNPS1 via the RNSP1–SAP18 binding (RSB) motif, which is located between the two RS domains and the RNA-binding motif of Acin16. Beyond these structural features, the identification of Acin1 as a component of the spliceosome7,8, nuclear speckles (storage sites for pre-mRNA splicing factors)9, and the EJC5 suggests a potential role of Acin1 in pre-mRNA splicing. Studies have reported that the ASAP complex containing Acin1 inhibits in vitro splicing reactions4, and siRNA-mediated depletion of Acin1 in HeLa cells alters gene expression and AS10, suggesting that Acin1 plays a crucial role in mRNA processing. Moreover, Acin1 plays an important role in the physiology and pathology of several tissues and cancer development11,12,13,14,15,16. However, our current understanding of the functions of Acin1 in normal mammalian development and disease progression is still limited.
In this study, we investigated the physiological roles of Acin1 by generating and analyzing conditional knockout mice, focusing on skeletal muscle, which has the highest number of tissue-specific splicing events17,18. Analysis of global and skeletal muscle myofiber-specific Acin1-knockout mice revealed that Acin1 is essential for mouse embryonic development and has limited effects on skeletal muscle structure and homeostasis.
Results
Global Acin1-deficient mice exhibit embryonic lethality
We investigated the role of Acin1 in mouse embryonic development and various tissues by constructing a vector for conditional gene targeting using the loxP system. Our strategy aimed to delete eight exons of Acin1, including four encoding the RNA recognition motif (RRM) (Fig. 1A). Following electroporation of the targeting construct into J1 embryonic stem (ES) cells, of 516 ES clones, we identified two independent clones with successful homologous recombination at the Acin1 locus. These clones were confirmed by Southern blot analysis (Fig. 1B) and subsequently injected into mouse blastocysts to generate chimeric mice. Heterozygous (Acin1N/+) mutant mice were then produced by crossing chimeras with C57BL/6 mice.
Generation and characterization of global Acin1-knockout mice. (A) Schematic representation of the Acin1 gene-targeting strategy. Depicted are the wild-type Acin1 allele ( +), targeting construct, and the following resulting mutant alleles: Acin1Neo (N), Acin1Floxed (F), and Acin1Deleted (D). Exons are shown as black boxes, loxP sites as white arrowheads, and the RNA recognition motif (RRM) as a dotted box. The 5′ and Neo probes used for Southern blotting and the genotyping primers (arrows) are indicated. The diagram includes the diphtheria toxin fragment A (DTA) and neomycin resistance (Neo) genes, with the following relevant restriction sites: AvrII (A), EcoRI (E), HindIII (H), and XbaI (X). Expression of Cre recombinase removes the eight exons, including the four exons that encode RRM, and/or the Neo gene. (B, C) Verification of the Acin1 alleles. (B) Southern blot analysis of AvrII-digested genomic DNA using 5′ and Neo probes, showing wild-type (12.4 kb) and Neo (10.2 kb) allele-specific fragments. (C) PCR genotyping demonstrating allele-specific product sizes. The genotypes are indicated above the lanes in both panels. See also Supplementary Figure S1. (D) Genotype distribution of offspring from Acin1D/+ intercrosses at various developmental stages. The asterisks indicate necrotic embryos.
To analyze global Acin1-deficient mice (Acin1D/D), we crossed Acin1N/+ mice with CAG-Cre transgenic mice, which express Cre recombinase in all tissues19 including germ cells, resulting in the deletion of loxP-flanked DNA (Fig. 1C). While heterozygous Acin1D/+ mice exhibited normal phenotypes, no homozygous Acin1D/D mice were detected among 417 offspring from Acin1D/+ heterozygote crosses (Fig. 1D). To determine the precise timing of embryonic lethality, we sacrificed intercrossed females at various days post coitum and genotyped individual embryos by PCR. The number of homozygous mutant embryos collected between 6.5 and 11.5 days post coitum was in agreement with the expected Mendelian ratio, with all embryos at E11.5 and E12.5 having undergone necrosis (Fig. 1D).
We measured the longitudinal axis length of embryos at E6.5 and E7.5. At E6.5, the lengths of Acin1+/+, Acin1D/+, and Acin1D/D embryos measured 1.04 ± 0.14 mm, 1.08 ± 0.13 mm, and 0.84 ± 0.13 mm, respectively. At E7.5, the lengths were 1.45 ± 0.17 mm, 1.36 ± 0.12 mm, and 0.93 ± 0.26 mm, respectively (Fig. 2A). Acin1D/D homozygous embryos appeared significantly smaller than their Acin1+/+ and Acin1D/+ littermates at E6.5 and E7.5 (Fig. 2A). At E9.5, while Acin1+/+ and Acin1D/+ siblings exhibited dorsal convexity due to complete rotation of the body axis, some Acin1D/D embryos showed either normal development, incomplete dorsal convexity, or abdominal convexity resulting from incomplete body axis rotation (Fig. 2A). Given the smaller size of the mutant embryos, we performed 5-bromo-2'-deoxyuridine (BrdU) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays to examine cell proliferation and apoptosis in embryos lacking the Acin1 gene compared with those with one or two normal alleles (Fig. 2B). Seven days post-mating of heterozygous mice, we injected BrdU intraperitoneally, isolated and fixed the decidua, and prepared paraffin sections for immunostaining using an anti-Acin1 antibody. The staining of small, developmentally delayed embryos was below the detection limit, suggesting that these mice were null mutants of Acin1 (Fig. 2B). In Acin1-negative embryos, we observed no difference in BrdU uptake (Fig. 2B) but an increase in TUNEL-positive cells (Fig. 2B), indicating that loss of Acin1 leads to increased apoptosis rather than decreased proliferation during early embryonic development.
Small embryos and delayed development of Acin1-deficient mice. (A) Whole-mount images of embryos at E6.5, E7.5, and E9.5, with Acin1-deficient embryos identified by a single PCR product (lower band). Scale bar: 1 mm. The lane numbers correspond to embryo numbers shown in the images. Genotypes are indicated below each lane. At E6.5, the lengths of Acin1+/+, Acin1D/+, and Acin1D/D embryos measured 1.04 ± 0.14 mm, 1.08 ± 0.13 mm, and 0.84 ± 0.13 mm, respectively. At E7.5, the lengths were 1.45 ± 0.17 mm, 1.36 ± 0.12 mm, and 0.93 ± 0.26 mm, respectively. The graphs show quantification of the longitudinal axis length of each embryo at E6.5 and E7.5. N indicates the number of embryos analyzed per group. See also Supplementary Figure S2. (B) Histological and molecular analyses of E7.5 embryos. Sagittal sections show hematoxylin and eosin (HE) staining, ACIN1 immunostaining, BrdU labeling for proliferation assessment, and TUNEL staining for apoptosis assessment. EPC ectoplacental cavity, ECC exocoelomic cavity, AC amniotic cavity. Scale bar: 200 μm. Upper and lower bar graphs show the percentage of BrdU-positive cells (proliferation) and TUNEL-positive cells (apoptosis), respectively, among 300 cells counted per embryo. Data are presented as means ± SD (N = 5). Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant.
Acin1 deficiency in skeletal muscle myofibers alters muscle morphology and increases susceptibility to injury
A high rate of tissue-specific splicing occurs in skeletal muscle17,18. Therefore, we performed a functional analysis of Acin1 in skeletal muscle. First, we conducted immunofluorescence staining of Acin1 in cross-sections from the tibialis anterior (TA) muscle of 12-week-old male mice to determine the expression of ACIN1 protein in adult skeletal muscle. Our findings revealed that the ACIN1 protein was more abundant in nuclei within myofibers than in Pax7-positive satellite cells and cells between the myofibers (Fig. 3A). To evaluate the role of Acin1 in myofibers, we generated skeletal muscle myofiber-specific Acin1-knockout mice by breeding Acin1F/F mice with HSA-Cre transgenic mice, in which Cre recombinase is expressed under the control of the muscle-specific human skeletal actin (HSA) promoter20. Floxed Acin1 (Acin1F/+) mice were created by crossing AcinN/+ mice with EIIa-Cre transgenic mice21,22 and removing the Neor gene flanked by two loxP sequences.
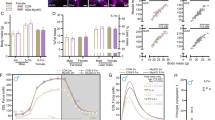
Generation of skeletal muscle-specific Acin1-knockout mice. (A) Immunofluorescence staining of Acin1 in tibialis anterior (TA) muscle cross-sections from 12-week-old wild-type mice. White: laminin α-2, green: Acin1, magenta: Pax7, blue: DAPI. Scale bar: 25 μm. The graph shows the ACIN1 fluorescence intensity (arbitrary unit) of cells between myofibers, Pax7-positive cells and myofibers. Data are presented as means ± SD (N = 4). The fluorescence intensity of ACIN1 staining was calculated by measuring 100 cells each for interstitial cells and myocytes, and 30 Pax7+ cells, for each TA muscle, and then calculating the average. (B) Immunostaining of Acin1 in TA muscle cross-sections from control (Ctrl) and skeletal muscle-specific Acin1-knockout (MKO) mice. The white arrowheads indicate the nuclei within the muscle fibers. Scale bar: 25 μm. The graph shows the percentages of Acin1-positive nuclei inside and outside myofibers in both groups. Data are presented as means ± SD (N = 3). (C) The expression level of Acin1 mRNA in TA muscles from Ctrl and MKO mice. Data are presented as means ± SD (N = 3). (D) Quantitative analysis of body and muscle weights. Bar graphs display the total body weight (TBW), individual muscle weights (TA, gastrocnemius (GA), soleus (SOL)), and muscle-to-TBW ratios. Data are presented as means ± SD (N = 8). (E) Forelimb grip strength test of Ctrl and MKO mice at 12 weeks of age. The two graphs show grip strength (N) and grip strength per total body weight (N/g). Data are means ± SD (N = 3). Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant.
To confirm ACIN1 protein deficiency, we prepared cross-sections of TA muscles from HSA-CreTg/+;Acin1F/F (MKO) mice and HSA-CreTg/+;Acin1+/+ (Ctrl) mice and performed immunofluorescent staining of ACIN1. Compared with Ctrl mice, MKO mice exhibited a significant decrease in the proportion of ACIN1-positive nuclei within myofibers, while no significant difference was observed in nuclei outside of myofibers (Fig. 3B). According to qPCR, the mRNA level of Acin1 in TA muscles was significantly lower in MKO than Ctrl mice (Fig. 3C). MKO mice were viable, fertile, and indistinguishable from Ctrl mice in terms of total body weight (Fig. 3D). However, the weight of skeletal muscles (TA, gastrocnemius, and soleus) and the ratio of skeletal muscle weight to total body weight were significantly increased in MKO mice compared with Ctrl mice (Fig. 3D). Furthermore, despite the increased muscle mass, grip strength measurements in 12-week-old male Ctrl and MKO mice showed no significant difference between the genotypes (Fig. 3E).
In the TA muscles of MKO mice, we observed significant increases in the myofiber cross-sectional area (CSA), minimal Feret’s diameter, number of centrally nucleated myofibers, and ratio of centrally nucleated myofibers to total myofibers (Fig. 4A). Given that an increase in centrally located nuclei suggests muscle regeneration due to disease or injury, we examined the area positive for embryonic myosin heavy chain (eMyHC), a marker of muscle regeneration, and the number of cells positive for Pax7, a muscle satellite cell marker. We found increases in the size of, and percentage of total myofiber CSA in, the eMyHC-positive area of the TA muscle of MKO mice compared with Ctrl mice (Fig. 4B). However, the number of, and percentage of total myofiber count in, Pax7-positive cells were not significantly different between MKO and Ctrl mice (Fig. 4C). To assess whether MKO mice were more susceptible to muscle injury, we intraperitoneally injected the MKO and Ctrl groups with Evans blue dye (EBD), which can enter and be retained only by cells with ruptured membranes23. Our results showed that the number of, and percentage of total myofibers in, EBD-positive myofibers were significantly greater in the TA of MKO mice than Ctrl mice, indicating that the TA of MKO mice experienced substantially more muscle damage compared with Ctrl mice (Fig. 4D). We performed TUNEL staining on TA muscle sections from 12-week-old Ctrl and MKO mice (Fig. 4E). The results showed a significant increase in TUNEL-positive nuclei in MKO mice, indicating enhanced apoptosis. We compared the presence of central nuclei in TA muscle between 12-week-old and 90-week-old mice. In MKO mice, the proportion of muscle fibers containing central nuclei was significantly higher in 90-week-old mice compared to 12-week-old counterparts (Fig. 4F). These results suggest that Acin1 plays an important role in maintaining skeletal muscle homeostasis, with muscle fiber degeneration and regeneration accumulating with age in MKO mice.
Enhanced muscle damage and regeneration in skeletal muscle-specific Acin1-knockout mice. (A) Muscle morphometric analysis. Representative images of TA cross-sections from Ctrl and MKO mice immunostained for Acin1, laminin α-2, and DAPI. Arrowheads indicate centrally located nuclei. Scale bar: 50 μm. Graphs shows the total myofiber number, each cross-sectional area (CSA), the minimal Feret diameter, and the centrally nucleated myofibers and their proportions. Data are means ± SD (N = 8). (B) Assessment of myofiber regeneration. Images show embryonic myosin heavy chain (eMyHC)-positive fibers in the TA muscles of Ctrl and MKO mice. Scale bar: 50 μm. Graphs show the total CSA, eMyHC-positive area, and their ratio. Data are means ± SD (N = 6). (C) Satellite cell quantification. Images depict Pax7-positive cells in the TA muscles of Ctrl and MKO mice. Scale bar: 25 μm. Graphs show total myofiber count, Pax7-positive cell number, and Pax7-positive cell-to-myofiber ratio. Data are means ± SD (N = 6). (D) Evaluation of muscle fiber permeability. Images show Evans blue dye (EBD) uptake in the TA myofibers of Ctrl and MKO mice. Scale bar: 25 μm. Graphs show the total myofiber count, EBD-positive myofiber count, and their ratio. Data are means ± SD (N = 6). (E) Detection of apoptosis. Images show TUNEL staining of sections of the TA muscle of Ctrl and MKO mice. Scale bar: 25 μm. Graph shows the ratio of myofibers with TUNEL-positive nuclei to total myofibers in the TA muscle. Data are means ± SD (N = 5). (F) Increased proportion of myofibers with central nuclei in TA muscle of 90-week-old mice compared to 12-week-old mice. Scale bar: 25 μm. Graph shows the percentage of myofibers with central nuclei to total myofibers in the TA muscle of 12-week-old mice and 90-week-old mice. Data are means ± SD (N = 3). In all panels except panel E, white: laminin α-2, magenta: Acin1 (A, F), eMyHC (B), Pax7 (C), or EBD (D), blue: DAPI. In panel E, white: laminin α-2, green: TUNEL staining (FITC), blue: DAPI. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, NS: not significant.
Transcriptome analysis reveals Acin1 deficiency-induced gene expression changes in skeletal muscle
To determine whether transcriptome alterations induced by Acin1 deficiency account for the observed muscle phenotype, we performed RNA-seq analysis in TA muscles obtained from MKO and Ctrl mice. We found 413 differentially expressed genes (q-value < 0.05, |log2FoldChange|> 1.0) between MKO and Ctrl muscles, with 232 upregulated and 181 downregulated genes (Fig. 5A). Gene Ontology (GO) enrichment analysis, conducted using Metascape and DAVID24,25,26, demonstrated significant enrichment of pathways related to muscle contraction, development, and differentiation among the upregulated genes (Fig. 5B). In contrast, the downregulated genes were primarily associated with the spermine metabolic process (Fig. 5B). The RNA-seq data further confirmed differential expression (q-value < 0.05, |log2FoldChange|> 1.0) of several genes involved in muscle morphogenesis and function between MKO and Ctrl muscles (Fig. 5C). These findings suggest that muscle tissue damage due to Acin1 deficiency may induce transcriptional gene changes that promote compensatory muscle hypertrophy, potentially explaining the observed phenotypic alterations in muscle tissue.
Transcriptomic analysis of tibialis anterior muscles from skeletal muscle-specific Acin1-knockout mice. (A) Differential gene expression analysis by RNA-seq. The volcano plot illustrates changes in mRNA abundance between MKO and Ctrl TA muscles. Blue dots represent significantly downregulated genes (q < 0.05, log2FoldChange < − 1), yellow dots significantly upregulated genes (q < 0.05, log2FoldChange > 1), and green dots genes without expression changes. (B) GO enrichment analysis of differentially expressed genes using Metascape (upper) and DAVID (lower). The bar graph displays the top five enriched biological processes for upregulated and downregulated genes, ranked by -log10(q-value). The threshold line indicates p < 0.05. (C) Expression changes in muscle contraction- and muscle organ development-related genes. The bar graph shows the log2FoldChange values of the significantly altered genes (q < 0.05) involved in muscle morphogenesis and function. Positive and negative values indicate upregulation and downregulation, respectively, in MKO compared with Ctrl TA muscles.
Acin1 deficiency alters alternative splicing patterns in skeletal muscle
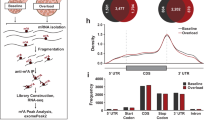
Acin1, an SR-related protein, is involved in AS. To examine the effects of Acin1 deficiency on splicing and AS in skeletal muscle, a comprehensive analysis was performed using rMATS and CASH software (Fig. 6A)27,28. The rMATS analysis revealed that Acin1 regulates various AS events, including skipped exons (SEs), alternative 3′/5′ splice sites (A3SS/A5SS), retained introns (RIs), and mutually exclusive exons. Notably, SEs emerged as the most significantly affected type, accounting for 65.91% of the observed changes (Fig. 6A). Complementing these findings, CASH software detected alterations in single exon cassette (Cassette), A3SS, A5SS, RI, alternative end (AltEnd), and alternative start (AltStart) events due to Acin1 deletion; of these, Cassette events, corresponding to SE in the rMATS analysis, were the most frequently observed (31.25%) (Fig. 6A). The predominance of SE/Cassette events suggests that Acin1 may play a particularly crucial role in modulating exon inclusion or exclusion, potentially influencing the functional diversity of muscle-specific proteins.
AS analysis of TA muscles from skeletal muscle-specific Acin1-knockout mice. (A) Distribution of AS events identified by rMATS (left) and CASH (right). The table shows the total AS events and significant AS events for each AS type. AS types include skipped exons (SE), alternative 5′/3′ splice sites (A5SS/A3SS), retained introns (RI), mutually exclusive exons (MXE), single exon cassettes (Cassette), multiple adjacent cassette exons (Cassette_multi), and alternative start/end exons (AltStart/AltEnd). The numbers in parentheses indicate the percentage of each type of AS event in the Significant AS events. (B) GO enrichment analysis of genes with altered AS events detected by rMATS and CASH analyses. Bar graphs show the enriched GO biological processes identified by Metascape (upper) and DAVID (lower), expressed as -log10(p-value). The threshold line indicates p < 0.05.
Further analysis focused on identifying genes subject to AS. By integrating data from both rMATS and CASH, we identified 55 genes exhibiting abnormal splicing patterns in MKO muscles. To elucidate the functional implications of these splicing changes, we performed GO enrichment analysis using Metascape (Fig. 6B). This analysis revealed that the affected genes were enriched in biological processes related to muscle structural development, the transmembrane receptor protein Ser/Thr kinase signaling pathway, regulation of cell morphogenesis, translation, and the MAPK cascade. We conducted a parallel GO enrichment analysis using DAVID to gain additional insight (Fig. 6B). The GO enrichment analysis highlighted the enrichment of categories related to skeletal muscle tissue development, regulation of cell shape, striated muscle cell differentiation, mRNA destabilization, and positive regulation of sodium ion transport. These findings collectively underscore the multifaceted impact of Acin1 on skeletal muscle biology, providing a foundation for further investigation into the molecular mechanisms underlying its role in AS and muscle function.
Subsequent analyses focused on five genes that showed consistent AS changes in the rMATS and CASH analyses. These genes included fragile X mental retardation, autosomal homolog 1 (Fxr1), integrin subunit alpha 7 (Itga7), sarcoglycan gamma (Sgcg), caspase recruitment domain family member 19 (Card19), and mitochondrial elongation factor 2 (Mief2), the first three of which are involved in muscle myopathy and muscular dystrophy29,30,31,32 and the latter two in mitochondrial function33,34 (Fig. 7A). The AS changes observed in these five genes included exon skipping and intron retention. In Fxr1, we detected decreased retention of an intron (87 nt) in exon 12 (Fig. 7B). Itga7 exhibited increased skipping of exon 26 (113 nt) (Fig. 7C), while Card19 showed decreased skipping of exon 6 (60 nt) (Fig. 7D). Mief2 demonstrated increased retention of an intron (194 nt) between exons 2 and 3 (Fig. 7E). Sgcg displayed decreased skipping of exon 2 (167 nt) (Fig. 7F). In Fxr1 and Card19, the observed alterations in AS did not result in a shift of the open reading frame (ORF). Similarly, in Sgcg, the ORF remained unaffected as the start codon is in exon 3. In Itga7, the deletion of exon 26 induces an ORF shift; however, this shift generates a new stop codon in the terminal exon (exon 27), thereby avoiding nonsense-mediated mRNA decay. Consequently, a protein with an altered C-terminus is likely produced from this transcript lacking exon 26. In Mief2, changes in AS lead to an ORF shift, resulting in a premature termination codon. This scenario is highly likely to trigger nonsense-mediated mRNA decay, suggesting that no protein is produced from transcripts containing the insertion between exons 2 and 3. RNA-seq analysis revealed differential gene expression patterns among the investigated genes. Although Fxr1 and Itga7 exhibited no significant changes in their expression levels, Card19 and Mief2 demonstrated significant upregulation, while Sgcg showed significant downregulation (Fig. 7A). To validate these splicing events, we employed RT-PCR and quantified the ratio of the long PCR product to the total PCR product amounts, expressed as a percentage (Fig. 8). These results suggest that Acin1 has diverse and gene-specific roles in splicing regulation in skeletal muscle. The AS alterations identified in genes associated with muscle pathology and mitochondrial function suggest that Acin1 deficiency may affect muscle physiology and energy metabolism.
Five genes commonly detected by rMATS- and CASH-based analyses of AS. (A) Key information for each gene, including gene symbol, full name, splicing type, and genomic location. The metrics of the splicing changes are provided, with IncLevelDiff values from rMATS analysis, deltaPSI values from CASH analysis, and fold changes and FDR p-values from RNA-seq analysis. (B–H) Sashimi plots illustrating read coverage and exon–exon junctions for selected AS genes: (B) Fxr1, (C) Itga7, (D) Card19, (E) Mief2, and (F) Sgcg. Numbers on the arches indicate junction reads in Ctrl and MKO samples.
RT-PCR validation of AS events in the five genes highlighted in Fig. (7B–F). Schematic diagrams show primer locations (arrows), constitutive exons (black and dark gray boxes), and alternative exons (light gray boxes). Bar plots present quantification of RT-PCR results (N = 4), showing the relative abundance of splice variants in Ctrl and MKO samples. Data represent mean ± SD. ***p < 0.001. See also Supplementary Figure S3.
Discussion
The embryonic lethality of global Acin1-KO mice suggests a requirement for Acin1 in early developmental processes that cannot be compensated for by other RNA processing factors. Acin1 forms the ASAP complex, a peripheral component of the EJC, by binding to RNPS1 and SAP18. While global knockout of RNPS1 has been reported to be embryonic lethal35, detailed analyses are lacking, and no reports exist on global knockout of SAP18. Consequently, the functional overlap between these factors and Acin1 in early mouse development remains undetermined. Furthermore, embryos with global knockout of Pinin, which forms the EJC peripheral factor PSAP by binding to RNPS1 and SAP18 (similar to Acin1), fail to survive beyond the implantation period36. The phenotype of such embryos differs significantly from that of Acin1 global knockout embryos. This observation aligns with previous studies demonstrating that Acin1 and Pinin function distinctly as peripheral factors of the EJC37. Acin1 is classified within the SR protein family due to its characteristic RS domain and RRM38. Global knockout studies of classical SR proteins have revealed distinct developmental consequences; knockout of SRSF1 (ASF/SF2), SRSF2 (SC35), or SRSF3 (SRp20) is lethal before implantation or E7.5, while SRSF5 knockout results in perinatal lethality39,40,41,42. These phenotypes differ markedly from the phenotype of Acin1 global knockout embryos. Transformer 2 beta homolog (TRA2B), also known as Splicing factor, arginine/serine-rich 10, contains one RRM flanked by two RS domains and functions as a sequence-specific splicing activator43. Interestingly, the phenotype of TRA2B global knockout embryos closely resembles that of Acin1 global knockout embryos, with both showing growth retardation around E8.5–E9.5 and subsequent lethality44. This phenotypic similarity suggests potential functional overlap between Acin1 and TRA2B in early post-implantation development, although their precise relationship remains to be explored.
In skeletal muscle, Acin1 deficiency suggests that Acin1 has limited effects in maintaining muscle fiber integrity. The increased susceptibility to injury and subsequent compensatory regeneration observed in Acin1-deficient muscles suggests that proper splicing regulation is important for muscle fiber maintenance. The altered splicing patterns in Acin1 MKO mice affected genes crucial for muscle function and mitochondrial dynamics. Most notably, the splicing alterations in Fxr1 and Itga7 resulted in increased expression of variants typically associated with undifferentiated muscle. In Fxr1, decreased retention of the 87-nt insertion in exon 12 was observed in Acin1 MKO mice. This insert is mostly skipped in myoblasts, while it is included in approximately 50% of myotube transcripts45. In Itga7, AS generates two isoforms with distinct intracellular domains: Itga7A, which contains a 113-nt insert (exon 26), and Itga7B, which lacks this insert46. Undifferentiated myoblasts exclusively express Itga7B, whereas Itga7A expression increases during myotube formation47. The increased exon 26 skipping observed in Acin1 MKO mice suggests elevated expression of the undifferentiated Itga7B isoform. Itga7 forms heterodimers with Itgb1 and preferentially binds laminin in skeletal muscle basal lamina48. While the cytoplasmic tail does not affect laminin binding, these isoforms interact with different intracellular proteins: Itga7A binds Def-6, a guanine nucleotide exchange factor for Rac1, while Itga7B interacts with the p130/crk complex and muscle integrin binding protein49,50,51. These findings align with previous observations that misregulation of AS can contribute to muscular dystrophy via expression of developmentally inappropriate isoforms52.
This research significantly advances our understanding of the tissue-specific functions of Acin1 beyond its role in general splicing mechanisms. The identification of muscle-specific selective splicing events regulated by Acin1 emphasizes its crucial role in tissue-specific functions and its impact on organ homeostasis. The Acin1Flox mouse model established in this study will serve as a valuable tool for future investigations into the role of Acin1 in various tissues, potentially revealing additional tissue-specific functions and therapeutic targets for related diseases.
Limitations of the study
While this study provides significant insights into the role of Acin1 in embryonic development and muscle homeostasis, several limitations should be acknowledged. First, our RNA-seq analysis using short-read sequencing and computational tools such as rMATS and CASH, although widely used for alternative splicing analysis, cannot comprehensively identify all splicing events occurring within a single transcript. Nanopore-based sequencing of full-length transcripts would provide a more complete picture of Acin1-dependent alternative splicing landscape in skeletal muscle.
Second, although we identified several genes with altered splicing patterns in Acin1-deficient muscle, we were unable to validate the consequences of these splicing changes at the protein level. Western blot analysis demonstrating size changes in proteins encoded by alternatively spliced transcripts (Itga7, Card19, and Fxr1) and expression level changes (particularly the downregulation of Sgcg) would strengthen the mechanistic link between Acin1 deficiency and the observed muscle phenotype.
Third, our functional assessment of muscle was limited to grip strength measurements, which showed no significant difference between control and MKO mice despite the evident morphological and molecular changes. More comprehensive functional analyses, including treadmill running tests, ex vivo muscle tension measurements, and electromyography, would provide deeper insights into how Acin1 deficiency affects muscle performance and fatigue resistance.
Fourth, while we observed alterations in genes related to energy metabolism in our transcriptomic analysis, we did not directly assess metabolic parameters such as mitochondrial function, substrate utilization, or energy expenditure. Such analyses would clarify the impact of Acin1 deficiency on muscle metabolism.
Fifth, we did not examine potential changes in muscle fiber type composition, which could contribute to the observed increases in cross-sectional area and muscle weight. Analysis of muscle fiber type distribution would enhance our understanding of how Acin1 deficiency affects muscle physiology.
Sixth, although we demonstrated that MKO mice exhibit increased susceptibility to muscle damage under basal conditions, we did not evaluate their response to acute or chronic exercise challenge. Such studies would further elucidate the role of Acin1 in muscle adaptation and repair following physiological stress.
Finally, we observed alterations in splicing patterns of several sarcomeric proteins, but did not thoroughly investigate how these changes might affect sarcomere structure and function. A more detailed analysis of sarcomeric protein isoforms, including troponin T and I, could provide additional insights into the mechanism underlying muscle fiber vulnerability in Acin1-deficient mice.
Addressing these limitations in future studies will provide a more comprehensive understanding of how Acin1-mediated splicing regulation contributes to muscle development, homeostasis, and function.
Materials and methods
Antibodies and reagents
Rabbit polyclonal anti-Acin1 (Cat# HPA000657, RRID: AB_1078091) was purchased from ATLAS ANTIBODIES. Alexa Fluor 488 goat anti-rabbit IgG (Cat# ab150077, RRID: AB_2630356), Alexa Fluor 488 goat anti-rat IgG (Cat# ab150157, RRID: AB_2722511), Alexa Fluor 555 goat anti-mouse IgG (Cat# ab150114, RRID: AB_2687594), Alexa Fluor 568 goat anti-rabbit IgG (Cat# ab175471, RRID: AB_2576207), Alexa Fluor 568 goat anti-mouse IgG (Cat# ab175473, RRID: AB_2895153), Alexa Fluor 647 goat anti-rat IgG (Cat# ab150159, RRID: AB_256682) and Rat monoclonal anti-BrdU were purchased from Abcam. Mouse monoclonal anti-Pax7 (Cat# pax7, ID: AB_528428) and mouse monoclonal anti-eMyHC (Cat# F1.652, RRID: AB_528358) were purchased from DSHB. Rat monoclonal anti-BrdU was purchased from Santa Cruz Biotechnology. Histofine Simple Stain Mouse Max-PO (R) (Cat# 414341F, RRID: AB_2819094), Histofine Simple Stain Mouse Max-PO (Rat) (Cat# 414,311), and Diaminobenzidine (DAB) substrate kit (Cat# 425,011) were purchased from Nichirei Biosciences. 5-bromo-2'-deoxyuridine (BrdU) (Cat# 027–15,561) and Evans blue dye (Cat# 054–04062) was purchased from Wako. In situ Apoptosis Detection Kit (Cat# MK500) was purchased from Takara Bio.
Animal experiments
CAG-Cre transgenic mice (RBRC09807) were purchased from Riken BioResource Research Center (Ibaraki, Japan). EIIa-Cre transgenic mice (JAX: 003,724), and HSA-Cre transgenic mice (JAX: 006,149) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animals were maintained and used according to a protocol approved by the Animal Experiment Committee of Ehime University (Approval No. 05E32-1,16). All mice were housed in a specific pathogen-free facility under a 12-h light/dark cycle with ad libitum water and food.
Construction of the targeting vector
An ES (R1) cell genomic library was screened by plaque hybridization using a 4.4 kb human Acin1-S’ cDNA probe. Of the 8.2 × 105 clones screened, 5 overlapping clones encompassing the region corresponding to Acin1-S and Acin1-S’, and the 3′ UTR region in the Acin1-L gene, were isolated. The 8.4 kb EcoRI fragment, which contains exons 11–19 of the Acin1-L gene, had a single loxP inserted into the XbaI site between exons 18 and 19 of the Acin1-L gene. This fragment was subcloned as a long arm into the pLox-neo vector. The 1.5 kb HindIII–EcoRI fragment containing exon 10 of the Acin1-L gene was used as a short arm. A DT-A gene was used for negative selection of clones with random integration of the targeting vector.
Generation of Acin1D/+ mutant mice
ES cells (J1) were electroporated with the linearized targeting construct and cultured in the presence of G418. G418-resistant ES cells were screened for homologous recombination by PCR (30 cycles of 94ºC for 30 s, 58ºC for 30 s, and 72ºC for 3 min) using the upstream gene-specific primer and the downstream Neor-specific primer. Correctly targeted ES clones were verified by Southern analysis using the 5′-flanking regions and Neor coding region. Chimeric mice were produced by microinjection of ES clones into C57BL/6J blastocysts. Acin1N/+ mice generated by mating chimeric males with C57BL/6J females were detected by coat color and reconfirmed by PCR analysis of tail DNA. To generate Acin1D/+ mice (B6.129S4-Acin1tm1.2Maot, MGI: 7,711,658), Acin1N/+ mice were mated with CAG-Cre transgenic mice (RBRC09807, Riken BioResource Research Center), which ubiquitously express the Cre transgene19.
Genotyping
For detection of the Acin1N, Acin1F, Acin1+, or Acin1D allele, PCR genotyping (30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min) was performed using a three-primer set: m10F, m18F, and m14R. PCR amplification of the Acin1N, Acin1F, Acin1+, and Acin1D alleles produces 857-bp, 857-bp, 738-bp, and 463-bp bands, respectively. To detect mice carrying the Neor or Cre gene, a Neor-specific or Cre-specific primer set, respectively, was used for PCR (30 cycles of 94ºC for 30 s, 60ºC for 30 s, and 72ºC for 30 s). PCR of Neor or Cre produced a 367-bp or 430-bp band, respectively. Sequences of the primers for genotyping are listed in Table 1.
Histological and immunohistochemical analyses and cell proliferation and apoptosis assays for Acin1-deficient embryos
For histological and immunohistochemical analyses, decidual tissues from pregnant mice were fixed in 4% paraformaldehyde (PFA) overnight at 4°C. After fixation, the decidual tissues were dehydrated in ethanol and then embedded in paraffin after replacing the ethanol with xylene. Tissue sections were cut to a thickness of 4 μm, deparaffinized, and subsequently stained with hematoxylin and eosin. For immunodetection of Acin1, serial sections were subjected to antigen retrieval via heat treatment in citrate buffer solution (pH 6.0) for 10 min at 120°C. Endogenous peroxidase activity was then quenched by incubation in 3% H2O2 for 10 min. Subsequently, the sections were incubated overnight at 4°C with the primary antibody (rabbit anti-Acin1) diluted 1:200 in antibody diluent buffer (Cat# SP-5035, Vector Laboratories). The primary antibody was detected by incubating the sections with Histofine Simple Stain Mouse Max-PO (R) at room temperature for 30 min. To assess cell proliferation, cells in the S phase of the cell cycle were labeled with BrdU. Pregnant mice at 7.5 days post-coitum were administered BrdU intraperitoneally (100 μg/g body weight). The animals were euthanized 45 min post-injection, and decidual tissues were excised from the uterus. Following overnight fixation in 4% PFA, the decidual tissues were dehydrated in ethanol. After replacing the ethanol with xylene, decidual tissues were embedded in paraffin and sectioned at a thickness of 4 μm. After deparaffinization, sections underwent antigen retrieval and endogenous peroxidase quenching procedures, as described for Acin1 immunodetection. Subsequently, the sections were incubated overnight at 4°C with the primary antibody (rat anti-BrdU) diluted 1:500 in antibody diluent buffer. Detection of the primary antibody was achieved using Histofine Simple Stain Mouse Max-PO (Rat) at room temperature for 30 min. Apoptosis was evaluated by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay using the In Situ Apoptosis Detection Kit. Sample preparation was conducted following the protocol described for the BrdU assay. Tissue sections were subjected to proteolytic digestion with Proteinase K (10 μg/mL) for 15 min, followed by a 5-min wash with phosphate-buffered saline (PBS) at room temperature. Endogenous peroxidase activity was then quenched by incubation in 3% H2O2 for 5 min. The TUNEL reaction was performed by incubating sections in a reaction mixture containing 45 μL Labeling Safe Buffer and 5 μL TdT Enzyme at 37°C for 90 min. Following PBS washing, the sections were incubated with 50 μL Anti-FITC HRP Conjugate at 37°C for 30 min. Signal visualization for all immunohistochemical, cell proliferation, and apoptosis assays was accomplished via a horseradish peroxidase reaction using a 3,3′-diaminobenzidine substrate kit. Tissue sections were subsequently counterstained with hematoxylin to provide nuclear contrast. Digital micrographs were acquired using the KEYENCE BZ-X800 fluorescence microscope.
Generation of skeletal muscle myofiber-specific Acin1-knockout mice
The floxed Neor gene cassette was removed from the mouse germline by crossing Acin1N/+ mice with EIIa-Cre transgenic mice (JAX: 003,724, The Jackson Laboratory)21,22. Offspring with various recombination events at loxP sites due to partial excision of Cre recombinase were backcrossed with C57BL/6J to obtain Acin1F/+ mice (B6.129S4(FVB)-Acin1tm1.1Maot, MGI: 7,711,657). Intercrossing of heterozygous Acin1F/+ mice generated homozygous Acin1F/F mice. Acin1F/F mice were crossed with HSA-Cre transgenic mice (JAX: 006,149, The Jackson Laboratory) to generate HSA-CreTg/+;Acin1F/F mice (MKO)20. HSA-CreTg/+;Acin1+/+ mice were used as the control (Ctrl). Unless otherwise noted, all experiments were conducted using 12-week-old male mice.
Immunofluorescence and microscopy
For immunofluorescence (IF) analysis, muscle tissues were rapidly frozen in liquid nitrogen-cooled isopentane, and cryosections of 10 μm thickness were prepared. Sections were fixed using 4% PFA in PBS for 5 min, followed by permeabilization with 0.5% Triton X-100 in PBS for 10 min. Non-specific binding was blocked using 5% goat serum in PBS, after which the sections were incubated with primary antibodies overnight at 4°C. Immunoreactivity was visualized using fluorochrome-conjugated secondary antibodies. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, 1 μg/mL). Fluorescence micrographs were acquired using the KEYENCE BZ-X800 fluorescence microscope and quantitatively analyzed using Fiji software53. The total cross-sectional area of muscle sections and individual muscle fiber areas were quantified based on laminin immunostaining of transverse muscle sections. The number of centrally located nuclei was determined by manual counting using Fiji software. For vital staining, Evans blue dye dissolved in PBS was administered intraperitoneally (1 mg/0.1 mL/10 g body weight). Mice were euthanized using CO2 16 h post-injection, and tibialis anterior (TA) muscle sections were fixed for 10 min in acetone at − 20°C. For embryonic myosin heavy chain (eMyHC) staining, tissue sections were fixed in acetone at − 20°C for 10 min and subjected to heat-induced epitope retrieval by boiling for 30 min before blocking. For Pax7 staining, sections were air-dried for 5 min before PFA fixation. To detect apoptosis, we performed the TUNEL assay (In Situ Apoptosis Detection Kit, Takara bio) on tissue sections. The primary antibodies used for immunofluorescence staining were rabbit anti-Acin1 (1:500), rat anti-laminin α-2 (1:50), mouse anti-Pax7 (1:5), and mouse anti-eMyHC (1:40). The secondary antibodies were used at a 1:1,000 dilution.
Forelimb grip strength test
Forelimb grip strength was assessed by using a grip strength meter (GPM-100B, MELQUEST, Toyama, Japan). Experiments were performed by the same operator, who was blinded to the genotypes of the mice.
RNA-seq and AS analyses
Tissue samples were harvested from the TA muscles of 12-week-old mice (N = 3 per group). Total RNA was extracted using the RNeasy Mini Kit and submitted to the Kazusa DNA Research Institute for RNA-seq analysis. Libraries were prepared using the MGIEasy RNA Directional Library Prep Set in conjunction with the NEBNext Poly(A) mRNA Magnetic Isolation Module. Sequencing was performed on the MGI DNBSEQ-G400RS platform with paired-end 100-bp reads. Raw sequencing data were initially quality-checked and trimmed using FastQC and Trimmomatic54,55. The resulting high-quality reads were aligned to the mouse reference genome (GRCm39/mm39) using STAR56. The mouse reference genome (GRCm39/mm39) was used as downloaded from GENCODE (https://www.gencodegenes.org/mouse/). Subsequently, RSEM57 was employed to quantify gene expression levels. Genes differentially expressed between the two experimental groups were identified using iDEP58 based on the following criteria: q-value (FDR-adjusted p-value) < 0.05 and |log2FoldChange|> 1.0. AS events were identified using two independent software packages: rMATS turbo27 and CASH28. For analysis of AS events, the mouse reference genome (GRCm39/mm39) was used as downloaded from Ensemble (https://www.ensembl.org/Mus_musculus/Info/Index). For rMATS analysis, splicing events were considered significant if they met the following criteria: FDR ≤ 0.05, |IncLevelDifference|≥ 0.2, and average junction read count ≥ 10. For CASH analysis, significant splicing events were defined as those meeting the criteria FDR ≤ 0.05 and |delta PSI| (percentage spliced in) ≥ 0.2. Splice junctions and read densities were visualized by generating Sashimi plots using rmats2sashimiplot27. Gene Ontology (GO) enrichment analysis was performed using Metascape24 and DAVID25,26.
RT-PCR and RT-qPCR
Total RNA was extracted from TA muscles using the RNeasy Mini Kit (Cat# 74,104, QIAGEN), and cDNA was synthesized using FastGene™ Scriptase II (Cat# NE-LS65, NIPPON Genetics) following the manufacturer’s instructions. The resulting cDNA was subjected to reverse-transcription PCR (RT-PCR) to validate AS events identified by RNA-seq analysis. RT-PCR was performed using TaKaRa Ex Premier™ DNA Polymerase (Cat# RR371, Takara Bio). Primers were designed to anneal to flanking constitutive exon regions in Card19, Fxr1, Itga7, Mief2, and Sgcg. PCR products were resolved on 3% agarose gels, and band intensities were quantified using Image Studio Lite software (LI-COR, NE). The relative abundance of the longest isoform was calculated by normalizing its intensity to the total intensity of all isoforms. Sequences of the primers for RT-PCR are listed in Table 2. qPCR for Acin1 using the cDNA mentioned above was performed using ABI 7500 (Applied Biosystems) and KAPA SYBR Fast qPCR Kit (Cat# KK4602, NIPPON Genetics) according to the manufacturer’s protocols. Gene expression levels were normalized to those of the housekeeping gene Rpl13a. Primer sequences used are listed in Table 3.
Statistical analysis
Statistical analyses were conducted using Student’s t-test for pairwise comparisons or one-way analysis of variance followed by Tukey’s post hoc test for multiple comparisons. All analyses were performed using EZR59. Data are presented as the mean ± SD. Statistically significant differences are denoted as follows: *p < 0.05, **p < 0.01, or ***p < 0.001.
Data availability
The RNA-seq datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/, and GSE281879. The additional datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. https://doi.org/10.1038/nature07509 (2008).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415. https://doi.org/10.1038/ng.259 (2008).
Sahara, S. et al. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401, 168–173. https://doi.org/10.1038/43678 (1999).
Schwerk, C. et al. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell Biol. 23, 2981–2990. https://doi.org/10.1128/mcb.23.8.2981-2990.2003 (2003).
Tange, T. O., Shibuya, T., Jurica, M. S. & Moore, M. J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11, 1869–1883. https://doi.org/10.1261/rna.2155905 (2005).
Murachelli, A. G., Ebert, J., Basquin, C., Le Hir, H. & Conti, E. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat. Struct. Mol. Biol. 19, 378–386. https://doi.org/10.1038/nsmb.2242 (2012).
Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245. https://doi.org/10.1101/gr.473902 (2002).
Zhou, Z., Licklider, L. J., Gygi, S. P. & Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185. https://doi.org/10.1038/nature01031 (2002).
Saitoh, N. et al. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890. https://doi.org/10.1091/mbc.e04-03-0253 (2004).
Rodor, J., Pan, Q., Blencowe, B. J., Eyras, E. & Cáceres, J. F. The RNA-binding profile of Acinus, a peripheral component of the exon junction complex, reveals its role in splicing regulation. RNA 22, 1411–1426. https://doi.org/10.1261/rna.057158.116 (2016).
Lin, Y. C., Lu, Y. H., Lee, Y. C., Hung, C. S. & Lin, J. C. Altered expressions and splicing profiles of Acin1 transcripts differentially modulate brown adipogenesis through an alternative splicing mechanism. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194601. https://doi.org/10.1016/j.bbagrm.2020.194601 (2020).
Vucetic, Z. et al. Acinus-S’ represses retinoic acid receptor (RAR)-regulated gene expression through interaction with the B domains of RARs. Mol. Cell Biol. 28, 2549–2558. https://doi.org/10.1128/mcb.01199-07 (2008).
Wang, F., Soprano, K. J. & Soprano, D. R. Role of Acinus in regulating retinoic acid-responsive gene pre-mRNA splicing. J. Cell Physiol. 230, 791–801. https://doi.org/10.1002/jcp.24804 (2015).
Jang, S. W. et al. Serine/arginine protein-specific kinase 2 promotes leukemia cell proliferation by phosphorylating acinus and regulating cyclin A1. Cancer Res. 68, 4559–4570. https://doi.org/10.1158/0008-5472.can-08-0021 (2008).
Pincez, T. et al. Cryptic recurrent ACIN1-NUTM1 fusions in non-KMT2A-rearranged infant acute lymphoblastic leukemia. Genes Chromosomes Cancer 59, 125–130. https://doi.org/10.1002/gcc.22808 (2020).
Tang, Y., Ni, A., Sun, L., Li, S. & Li, G. Analysis of the upregulated expression mechanism of apoptotic chromatin condensation inducer 1 in hepatocellular carcinoma based on bioinformatics. Turk. J. Gastroenterol. 35, 307–315. https://doi.org/10.5152/tjg.2024.23454 (2024).
Merkin, J., Russell, C., Chen, P. & Burge, C. B. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338, 1593–1599. https://doi.org/10.1126/science.1228186 (2012).
Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768. https://doi.org/10.1101/gr.220962.117 (2017).
Sakai, K. & Miyazaki, J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 237, 318–324. https://doi.org/10.1006/bbrc.1997.7111 (1997).
Miniou, P. et al. Gene targeting restricted to mouse striated muscle lineage. Nucleic Acids Res. 27, e27. https://doi.org/10.1093/nar/27.19.e27 (1999).
Lakso, M. et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 93, 5860–5865. https://doi.org/10.1073/pnas.93.12.5860 (1996).
Holzenberger, M. et al. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 28, E92. https://doi.org/10.1093/nar/28.21.e92 (2000).
Matsuda, R., Nishikawa, A. & Tanaka, H. Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J. Biochem. 118, 959–964. https://doi.org/10.1093/jb/118.5.959 (1995).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 (2019).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. https://doi.org/10.1038/nprot.2008.211 (2009).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216-w221. https://doi.org/10.1093/nar/gkac194 (2022).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U. S. A. 111, E5593-5601. https://doi.org/10.1073/pnas.1419161111 (2014).
Wu, W. et al. CASH: a constructing comprehensive splice site method for detecting alternative splicing events. Brief Bioinform. 19, 905–917. https://doi.org/10.1093/bib/bbx034 (2018).
Davidovic, L. et al. Alteration of expression of muscle specific isoforms of the fragile X related protein 1 (FXR1P) in facioscapulohumeral muscular dystrophy patients. J. Med. Genet. 45, 679–685 (2008).
Estañ, M. C. et al. Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy. Nat. Commun. 10, 797. https://doi.org/10.1038/s41467-019-08548-9 (2019).
Hayashi, Y. K. et al. Mutations in the integrin alpha7 gene cause congenital myopathy. Nat Genet 19, 94–97. https://doi.org/10.1038/ng0598-94 (1998).
McNally, E. M. et al. Mutations that disrupt the carboxyl-terminus of gamma-sarcoglycan cause muscular dystrophy. Hum. Mol. Genet. 5, 1841–1847. https://doi.org/10.1093/hmg/5.11.1841 (1996).
Rios, K. E. et al. CARD19 interacts with mitochondrial contact site and cristae organizing system constituent proteins and regulates cristae morphology. Cells https://doi.org/10.3390/cells11071175 (2022).
Palmer, C. S. et al. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573. https://doi.org/10.1038/embor.2011.54 (2011).
Zhong, X. et al. RNPS1 inhibits excessive tumor necrosis factor/tumor necrosis factor receptor signaling to support hematopoiesis in mice. Proc. Natl. Acad. Sci. U. S. A. 119, e2200128119. https://doi.org/10.1073/pnas.2200128119 (2022).
Joo, J. H. et al. Role of Pinin in neural crest, dorsal dermis, and axial skeleton development and its involvement in the regulation of Tcf/Lef activity in mice. Dev. Dyn. 236, 2147–2158. https://doi.org/10.1002/dvdy.21243 (2007).
Wang, Z., Ballut, L., Barbosa, I. & Le Hir, H. Exon Junction Complexes can have distinct functional flavours to regulate specific splicing events. Sci. Rep. 8, 9509. https://doi.org/10.1038/s41598-018-27826-y (2018).
Bartsakoulia, M. et al. A novel mechanism causing imbalance of mitochondrial fusion and fission in human myopathies. Hum. Mol. Genet. 27, 1186–1195. https://doi.org/10.1093/hmg/ddy033 (2018).
Jumaa, H., Wei, G. & Nielsen, P. J. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9, 899–902. https://doi.org/10.1016/s0960-9822(99)80394-7 (1999).
Ding, J. H. et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. Embo J. 23, 885–896. https://doi.org/10.1038/sj.emboj.7600054 (2004).
Xu, X. et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120, 59–72. https://doi.org/10.1016/j.cell.2004.11.036 (2005).
Zhang, X. et al. Splicing factor Srsf5 deletion disrupts alternative splicing and causes noncompaction of ventricular myocardium. iScience 24, 103097. https://doi.org/10.1016/j.isci.2021.103097 (2021).
Tacke, R., Tohyama, M., Ogawa, S. & Manley, J. L. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93, 139–148. https://doi.org/10.1016/s0092-8674(00)81153-8 (1998).
Mende, Y. et al. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum. Mol. Genet. 19, 2154–2167. https://doi.org/10.1093/hmg/ddq094 (2010).
Smith, J. A. et al. FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. J. Cell Biol. https://doi.org/10.1083/jcb.201911129 (2020).
Collo, G., Starr, L. & Quaranta, V. A new isoform of the laminin receptor integrin alpha 7 beta 1 is developmentally regulated in skeletal muscle. J. Biol. Chem. 268, 19019–19024 (1993).
Song, W. K., Wang, W., Sato, H., Bielser, D. A. & Kaufman, S. J. Expression of alpha 7 integrin cytoplasmic domains during skeletal muscle development: alternate forms, conformational change, and homologies with serine/threonine kinases and tyrosine phosphatases. J. Cell Sci. 106(Pt 4), 1139–1152. https://doi.org/10.1242/jcs.106.4.1139 (1993).
Boppart, M. D. & Mahmassani, Z. S. Integrin signaling: linking mechanical stimulation to skeletal muscle hypertrophy. Am. J. Physiol. Cell Physiol. 317, C629-c641. https://doi.org/10.1152/ajpcell.00009.2019 (2019).
Samson, T. et al. Def-6, a guanine nucleotide exchange factor for Rac1, interacts with the skeletal muscle integrin chain alpha7A and influences myoblast differentiation. J. Biol. Chem. 282, 15730–15742. https://doi.org/10.1074/jbc.M611197200 (2007).
Mielenz, D., Hapke, S., Pöschl, E., von Der Mark, H. & von Der Mark, K. The integrin alpha 7 cytoplasmic domain regulates cell migration, lamellipodia formation, and p130CAS/Crk coupling. J. Biol. Chem. 276, 13417–13426. https://doi.org/10.1074/jbc.M011481200 (2001).
Li, J., Rao, H., Burkin, D., Kaufman, S. J. & Wu, C. The muscle integrin binding protein (MIBP) interacts with alpha7beta1 integrin and regulates cell adhesion and laminin matrix deposition. Dev. Biol. 261, 209–219. https://doi.org/10.1016/s0012-1606(03)00304-x (2003).
Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451. https://doi.org/10.1038/nrm.2017.27 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. https://doi.org/10.1038/nmeth.2019 (2012).
Andrews, S. FastQC. A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323. https://doi.org/10.1186/1471-2105-12-323 (2011).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 19, 534. https://doi.org/10.1186/s12859-018-2486-6 (2018).
Kanda, Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant 48, 452–458. https://doi.org/10.1038/bmt.2012.244 (2013).
Acknowledgements
We extend our gratitude to Dr. Yoshihide Tsujimoto for his valuable contributions during the initial stages of this project. We are also indebted to Dr. Kazunori Kon for his continuous support throughout the project. We thank Drs. Masaru Okabe and Masahito Ikawa for their efforts in generating the chimeric mice essential for this study. We also thank Ms. Miki Shinoda for her helpful support. This research was supported by the Division of Medical Research Support, the Advanced Research Support Center (ADRES) at Ehime University. We would like to thank JAM Post (www.jamp.com) for English language editing.
Funding
This work was financially supported by JSPS KAKENHI Grant Number JP22K09425.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.A., H.S. Y.I., and N.M.; Data Curation, M.A.; Investigation, M.A., H.S., and N.O.; Methodology, M.A., H.S., T.K., and N.O.; Resources, M.A., H.S., and Y.I.; Software, M.A., N.T., and M.M.; Writing – Original Draft Preparation, M.A.; Writing – Review & Editing, H.S., N.O., Y.I., and N.M.; Supervision, Y.I., and N.M.; Project administration, M.A.; Funding acquisition, M.A. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
The animal experiments were approved by the Animal Experiment Committee of Ehime University (Approval No. 05E32-1,16). All methods are reported in accordance with Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. We confirmed that all experiments in this study were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aoto, M., Sakai, H., Tokunaga, N. et al. The splicing factor Acin1 is essential for embryonic development but has limited effects on muscle structure and homeostasis. Sci Rep 15, 14017 (2025). https://doi.org/10.1038/s41598-025-98851-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-98851-x