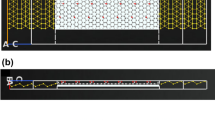
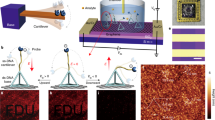
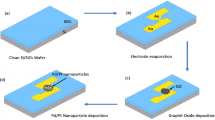
Abstract
The demand for rapid and scalable biosensing technologies has motivated the development of antibody-free platforms capable of operating in complex sample environments. Here, we report an electrochemical biosensor based on engineered M13 bacteriophages displaying a SARS-CoV-2 spike S1–binding peptide immobilized on a reduced graphene oxide (rGO) transducer. The sensor employs a chemiresistive detection mechanism under a fixed low-voltage bias, enabling rapid electrical readout following target binding. Detection of S1 protein was achieved in buffer and in spiked complex matrices, including fetal bovine serum, pasteurized milk, and wastewater, demonstrating matrix tolerance under the tested conditions. The biosensor response is evaluated using a statistically defined binary detection criterion, with an operational limit of detection of 10⁻4 pg/mL in buffer. Compared to a previously reported antibody-functionalized rGO sensor fabricated using the same platform, the phage-based biosensor exhibits comparable sensitivity while offering advantages in genetic tunability and production scalability. While the present study focuses on proof-of-concept validation using spiked samples, these results highlight the potential of engineered phage–graphene interfaces as adaptable biorecognition elements for rapid electrochemical protein sensing in complex environments.
Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Akbari Nakhjavani, S., Mirzajani, H., Carrara, S. & Onbaşlı, M. C. Advances in biosensor technologies for infectious diseases detection. TrAC Trend. Anal. Chem. 180, 117979. https://doi.org/10.1016/j.trac.2024.117979 (2024).
Scott, G. Y. et al. Transforming early microbial detection: Investigating innovative biosensors for emerging infectious diseases. Adv. Biomark. Sci. Technol. 6, 59–71. https://doi.org/10.1016/j.abst.2024.04.002 (2024).
Rasmi, Y., Li, X., Khan, J., Ozer, T. & Choi, J. R. Emerging point-of-care biosensors for rapid diagnosis of COVID-19: Current progress, challenges, and future prospects. Anal. Bioanal. Chem. 413, 4137–4159. https://doi.org/10.1007/s00216-021-03377-6 (2021).
Frigoli, M. et al. Emerging biomimetic sensor technologies for the detection of pathogenic bacteria: A commercial viability study. ACS omega 9, 23155–23171. https://doi.org/10.1021/acsomega.4c01478 (2024).
Sequeira-Antunes, B. & Ferreira, H. A. Nucleic acid aptamer-based biosensors: A review. Biomedicines 11, 3201. https://doi.org/10.3390/biomedicines11123201 (2023).
Guliy, O. I., Evstigneeva, S. S., Khanadeev, V. A. & Dykman, L. A. Antibody phage display technology for sensor-based virus detection: Current status and future prospects. Biosensors (Basel) https://doi.org/10.3390/bios13060640 (2023).
Wang, M., Pang, S., Zhang, H., Yang, Z. & Liu, A. Phage display based biosensing: Recent advances and challenges. TrAC Trend. Anal. Chem. 173, 117629. https://doi.org/10.1016/j.trac.2024.117629 (2024).
Léguillier, V., Heddi, B. & Vidic, J. Recent advances in aptamer-based biosensors for bacterial detection. Biosensors (Basel) https://doi.org/10.3390/bios14050210 (2024).
Kohlberger, M. & Gadermaier, G. SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Biochem. 69, 1771–1792. https://doi.org/10.1002/bab.2244 (2022).
Moon, J. et al. Research progress of M13 bacteriophage-based biosensors. Nanomaterials 9, 1448. https://doi.org/10.3390/nano9101448 (2019).
Smith, G. P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317. https://doi.org/10.1126/science.4001944 (1985).
Rakonjac, J., Gold, V. A. M., León-Quezada, R. I. & Davenport, C. H. Structure, biology, and applications of filamentous bacteriophages. Cold Spring Harbor protoc. https://doi.org/10.1101/pdb.over107754 (2024).
Kim, S., Heo, H. R., Kim, C. S. & Shin, H. H. Genetically engineered bacteriophages as novel nanomaterials: Applications beyond antimicrobial agents. Front. Bioeng. Biotechnol. 12, 1319830. https://doi.org/10.3389/fbioe.2024.1319830 (2024).
Campuzano, S., Pedrero, M., Barderas, R. & Pingarrón, J. M. Breaking barriers in electrochemical biosensing using bioinspired peptide and phage probes. Anal. Bioanal. Chem. 416, 7225–7247. https://doi.org/10.1007/s00216-024-05294-w (2024).
Hsu, C. et al. Recent progress on phage display-based biosensing systems for detection of pathogenic bacteria in food and water. Microchem. J. 208, 112356. https://doi.org/10.1016/j.microc.2024.112356 (2025).
Zhou, Y. et al. Development of a phage-based electrochemical biosensor for detection of Escherichia coli O157: H7 GXEC-N07. Bioelectrochemistry (Amsterdam, Netherlands) https://doi.org/10.1016/j.bioelechem.2022.108345 (2023).
Sedki, M., Chen, X., Chen, C., Ge, X. & Mulchandani, A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens. Bioelectron. 148, 111794. https://doi.org/10.1016/j.bios.2019.111794 (2020).
Nakama, K., Sedki, M. & Mulchandani, A. Label-free chemiresistor biosensor based on reduced graphene oxide and M13 bacteriophage for detection of coliforms. Anal. Chim. Acta. 1150, 338232. https://doi.org/10.1016/j.aca.2021.338232 (2021).
Shin, J. H. et al. Electrochemical detection of caspase-3 based on a chemically modified M13 phage virus. Bioelectrochemistry (Amsterdam, Netherlands) https://doi.org/10.1016/j.bioelechem.2022.108090 (2022).
Yang, H. J., Raju, C. V., Choi, C. & Park, J. P. Electrochemical peptide-based biosensor for the detection of the inflammatory disease biomarker, interleukin-1beta. Anal. Chim. Acta. 1295, 342287. https://doi.org/10.1016/j.aca.2024.342287 (2024).
Shin, J. H. et al. Quantitative label-free determination of thrombin using a chemically-modified M13 virus-electrode interface. Biotechnol. Bioproc. E. 28, 235–245. https://doi.org/10.1007/s12257-022-0361-9 (2023).
Shin, J. H., Park, T. J., Hyun, M. S. & Park, J. P. A phage virus-based electrochemical biosensor for highly sensitive detection of ovomucoid. Food chem. 378, 132061. https://doi.org/10.1016/j.foodchem.2022.132061 (2022).
Abdelhamied, N., Abdelrahman, F., El-Shibiny, A. & Hassan, R. Y. A. Bacteriophage-based nano-biosensors for the fast impedimetric determination of pathogens in food samples. Sci. Rep. 13, 3498. https://doi.org/10.1038/s41598-023-30520-3 (2023).
Zheng, Z. et al. Sensitive amperometric immunosensor for pathogen antigen based on MoS2@AuNPs assembling dual-peptide as bioprobes with significant dual signal amplification. Anal. Chim. Acta. 1355, 344015. https://doi.org/10.1016/j.aca.2025.344015 (2025).
Yang, F. et al. Phage display-derived peptide for the specific binding of SARS-CoV-2. ACS omega 7, 3203–3211. https://doi.org/10.1021/acsomega.1c04873 (2022).
Seo, G. et al. Ultrasensitive biosensing platform for Mycobacterium tuberculosis detection based on functionalized graphene devices. Front. Bioeng. Biotechnol. 11, 1313494. https://doi.org/10.3389/fbioe.2023.1313494 (2023).
Kadadou, D. et al. Optimization of an rGO-based biosensor for the sensitive detection of bovine serum albumin: Effect of electric field on detection capability. Chemosphere (Oxford) https://doi.org/10.1016/j.chemosphere.2022.134700 (2022).
Kadadou, D. et al. Detection of SARS-CoV-2 in clinical and environmental samples using highly sensitive reduced graphene oxide (rGO)-based biosensor. Chem. Eng. J. https://doi.org/10.1016/j.cej.2022.139750 (2023).
Figueroa-Miranda, G. et al. Delineating charge and capacitance transduction in system-integrated graphene-based BioFETs used as aptasensors for malaria detection. Biosens. Bioelectron. 208, 114219. https://doi.org/10.1016/j.bios.2022.114219 (2022).
Walters, F. et al. A rapid graphene sensor platform for the detection of viral proteins in low volume samples. Adv. NanoBiomed. Res. (Online) https://doi.org/10.1002/anbr.202100140 (2022).
Zhang, Z., Zhang, L., Huang, Y., Wang, Z. & Ren, Z. A. Planar-gate graphene field-effect transistor integrated portable platform for rapid detection of colon cancer-derived exosomes. Biosensors (Basel) 15, 207. https://doi.org/10.3390/bios15040207 (2025).
Moosa, A. A. & Abed, M. S. Graphene preparation and graphite exfoliation. Turkish J. chem. 45, 493–519. https://doi.org/10.3906/kim-2101-19 (2021).
de Heer, W. A. et al. Large area and structured epitaxial graphene produced by confinement controlled sublimation of silicon carbide. Proc. Natl. Acad. Sci. 108, 16900–16905. https://doi.org/10.1073/pnas.1105113108 (2011).
Zhang, P., Li, Z., Zhang, S. & Shao, G. Recent advances in effective reduction of graphene oxide for highly improved performance toward electrochemical energy storage. Energy Environ. Mater. 1, 5–12. https://doi.org/10.1002/eem2.12001 (2018).
Ozbey, S., Keles, G. & Kurbanoglu, S. Innovations in graphene-based electrochemical biosensors in healthcare applications. Microchim. Acta. 192, 290. https://doi.org/10.1007/s00604-025-07141-w (2025).
Yu, H. et al. Reduced graphene oxide nanocomposite based electrochemical biosensors for monitoring foodborne pathogenic bacteria: A review. Food control. 127, 108117. https://doi.org/10.1016/j.foodcont.2021.108117 (2021).
Sin, M. L., Mach, K. E., Wong, P. K. & Liao, J. C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert. Rev. Mol. Diagn. 14, 225–244. https://doi.org/10.1586/14737159.2014.888313 (2014).
Rodríguez-Franco, P., Abad, L., Muñoz-Pascual, F. X., Moreno, M. & Baldrich, E. Effect of the transducer’s surface pre-treatment on SPR aptasensor development. Sens. Actuator. B. Chem. https://doi.org/10.1016/j.snb.2013.10.046 (2014).
Tai, W. et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 17, 613–620. https://doi.org/10.1038/s41423-020-0400-4 (2020).
Chaibun, T. et al. Highly sensitive and specific electrochemical biosensor for direct detection of hepatitis C virus RNA in clinical samples using DNA strand displacement. Sci. Rep. 14, 23792–10. https://doi.org/10.1038/s41598-024-74454-w (2024).
Poudyal, D. C. et al. Low-volume electrochemical sensor platform for direct detection of paraquat in drinking water. Electrochem 5, 341–353. https://doi.org/10.3390/electrochem5030022 (2024).
Armbruster, D. A. & Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29(1), 49 (2008).
Yang, L., Zhang, L., Jiao, X., Qiu, Y. & Xu, W. The electrochemical performance of reduced graphene oxide prepared from different types of natural graphites. RSC Adv. 11, 442–452. https://doi.org/10.1039/d0ra09684a (2021).
Feizi, S., Mehdizadeh, A., Hosseini, M. A., Jafari, S. A. & Ashtari, P. Reduced graphene oxide/polymethyl methacrylate (rGO/PMMA) nanocomposite for real time gamma radiation detection. Nucl. Instrum. Method. Phys. Res. 940, 72–77. https://doi.org/10.1016/j.nima.2019.06.001 (2019).
Hidayah, N. M. S. et al. Comparison on graphite, graphene oxide and reduced graphene oxide: Synthesis and characterization. AIP conf. proc. https://doi.org/10.1063/1.5005764 (2017).
Ni, J., Liu, R. & Yan, C. Facile construction of poly(styrene-acrolein)/reduced graphene oxide nanocomposites via in-situ reduction and its corrosion resistance properties in waterborne acrylic resin coating. Chem. Phys. Lett. 772, 138570. https://doi.org/10.1016/j.cplett.2021.138570 (2021).
Khan, M. U. & Shaida, M. A. Reduction mechanism of graphene oxide including various parameters affecting the C/O ratio. Mater. Today Commun. 36, 106577. https://doi.org/10.1016/j.mtcomm.2023.106577 (2023).
Lavín, Á. et al. On the determination of uncertainty and limit of detection in label-free biosensors. Sensors 18, 2038. https://doi.org/10.3390/s18072038 (2018).
Machera, S. J., Niedziółka-Jönsson, J. & Szot-Karpińska, K. Phage-based sensors in medicine: A review. Chemosensors 8, 61. https://doi.org/10.3390/chemosensors8030061 (2020).
Yuan, J. et al. Truncated M13 phage for smart detection of E. coli under dark field. J. nanobiotechnol. https://doi.org/10.1186/s12951-024-02881-y (2024).
Aslan, B. C. et al. Bacteriophage-gated optical sensor for bacteria detection. Anal. Chem. (Washington) https://doi.org/10.1021/acs.analchem.5c00780 (2025).
Miranda, O. R. et al. Colorimetric bacteria sensing using a supramolecular enzyme-nanoparticle biosensor. J. Am. Chem. Soc. 133, 9650–9653. https://doi.org/10.1021/ja2021729 (2011).
Jiang, C., Mu, X., Du, B. & Tong, Z. A review of electrochemical biosensor application in the detection of the SARS-COV-2. Micro. Nano. Lett. 17, 49–58. https://doi.org/10.1049/mna2.12101 (2022).
Patel, S. K. et al. Recent advances in biosensors for detection of COVID-19 and other viruses. RBME 16, 1–16. https://doi.org/10.1109/RBME.2022.3212038 (2023).
Lee, D. Y. et al. Analysis of commercial fetal bovine serum (FBS) and its substitutes in the development of cultured meat. Food Res. Int. 174, 113617. https://doi.org/10.1016/j.foodres.2023.113617 (2023).
Arain, M. A. et al. A review on camel milk composition, techno- functional properties and processing constraints. Food Sci. Anim. Res. 44, 739–757. https://doi.org/10.5851/kosfa.2023.e18 (2024).
Ke, Z. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502. https://doi.org/10.1038/s41586-020-2665-2 (2020).
Laue, M. et al. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci. Rep. 11, 3515. https://doi.org/10.1038/s41598-021-82852-7 (2021).
Kyosei, Y. et al. Ultrasensitive detection of SARS-CoV-2 spike proteins using the Thio-NAD cycling reaction: A preliminary study before clinical trials. Microorganisms 9, 2214. https://doi.org/10.3390/microorganisms9112214 (2021).
Acer, P. T., Kelly, L. M., Lover, A. A. & Butler, C. S. Quantifying the relationship between SARS-CoV-2 wastewater concentrations and building-level COVID-19 prevalence at an isolation residence using a passive sampling approach. medRxiv https://doi.org/10.1101/2022.04.07.22273534 (2022).
Sharma, P. K. et al. Ultrasensitive and reusable graphene oxide-modified double-interdigitated capacitive (DIDC) sensing chip for detecting SARS-CoV-2. ACS Sensor. 6, 3468–3476. https://doi.org/10.1021/acssensors.1c01437 (2021).
Sharma, P. K. et al. Ultrasensitive probeless capacitive biosensor for amyloid beta (Aβ1-42) detection in human plasma using interdigitated electrodes. Biosensor. Bioelectron. 212, 114365. https://doi.org/10.1016/j.bios.2022.114365 (2022).
Verma, M. K. et al. Rapid diagnostic methods for SARS-CoV-2 (COVID-19) detection: An evidence-based report. J. med. Life. https://doi.org/10.25122/jml-2021-0168 (2021).
Huang, L. et al. Capacitive biosensors for label-free and ultrasensitive detection of biomarkers. Talanta 266, 124951. https://doi.org/10.1016/j.talanta.2023.124951 (2024).
Torres, M. D. T., de Araujo, W. R., de Lima, L. F., Ferreira, A. L. & de la Fuente-Nunez, C. Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter 4, 2403–2416. https://doi.org/10.1016/j.matt.2021.05.003 (2021).
Fabiani, L. et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosensor. Bioelectron. 171, 112686. https://doi.org/10.1016/j.bios.2020.112686 (2021).
Rahmati, Z., Roushani, M., Hosseini, H. & Choobin, H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Microchim. Acta. 188, 105. https://doi.org/10.1007/s00604-021-04762-9 (2021).
Ali, M. A. et al. An advanced healthcare sensing platform for direct detection of viral proteins in seconds at femtomolar concentrations via aerosol jet 3D-printed nano and biomaterials. Adv. Mater. Interfac. https://doi.org/10.1002/admi.202400005 (2024).
Yakoh, A. et al. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosensor. Bioelectron. 176, 112912. https://doi.org/10.1016/j.bios.2020.112912 (2021).
Zhang, Z. et al. High‐affinity dimeric aptamers enable the rapid electrochemical detection of wild‐type and B.1.1.7 SARS‐CoV‐2 in unprocessed saliva. Angew. Chem. Int. Edit. https://doi.org/10.1002/anie.202110819 (2021).
Rahmati, Z., Roushani, M., Hosseini, H. & Choobin, H. Label-free electrochemical aptasensor for rapid detection of SARS-CoV-2 spike glycoprotein based on the composite of Cu(OH)2 nanorods arrays as a high-performance surface substrate. Bioelectrochemistry 146, 108106. https://doi.org/10.1016/j.bioelechem.2022.108106 (2022).
Idili, A., Parolo, C., Alvarez-Diduk, R. & Merkoçi, A. Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sensor. 6, 3093–3101. https://doi.org/10.1021/acssensors.1c01222 (2021).
Acknowledgement
The authors would like to acknowledge Dr. Chia-Yun Lai (Postdoctoral Fellow, Mechanical & Nuclear Engineering, Khalifa University of Science and Technology) for training and assisting HYA to carry out the atomic force microscopy (AFM) measurements. We also acknowledge Professor Matteo Chiesa (Mechanical & Nuclear Engineering, Head of the Laboratory for Energy and Nano Science (LENS) at Khalifa University of Science and Technology) for facilitating access to and providing training at Khalifa University’s AFM facility. Figures in this work were created with BioRender.com.
Funding
This work was supported by a Research Innovation Student Grant (RIG-S) by Khalifa University (RIG-2023-032), the Center for Membranes and Advanced Water Technology (CMAT) at Khalifa University (Award No. RC2-2018-009), and the Center for Biotechnology (BTC) at Khalifa University.
Author information
Authors and Affiliations
Contributions
A.F.Y., H.A.S., and S.W.H. conceived and supervised the project. H.Y.A. performed the phage construction, rGO synthesis, biosensor fabrication, and materials characterization. S.P. and L.T. assisted in biosensor fabrication and data analysis. M.I.H. and L.T. contributed to SEM, EDS, and XRD imaging and analysis. H.Y.A. and L.T. carried out biosensor testing and electrochemical measurements. Figures were prepared by H.Y.A. and A.F.Y. The initial manuscript draft was written by H.Y.A., and all authors except M.I.H. contributed to revisions. A.F.Y., S.W.H., and H.A.S. secured project funding. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alshehhi, H.Y., Tizani, L., Palanisamy, S. et al. An engineered M13 phage–rGO electrochemical biosensor for rapid detection of viral protein in complex matrices. Sci Rep (2026). https://doi.org/10.1038/s41598-026-37008-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-37008-w