Abstract
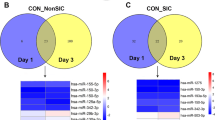
Sudden infant death syndrome (SIDS) remains one of the most common and poorly understood diagnoses of death in infants. In this study, we searched for novel biomarkers to aid in elucidating the pathogenesis of SIDS through a bioinformatics analysis of serum-derived extracellular vesicle (EV) miRNAs using next-generation sequencing. Comparative analyses between infants who died of SIDS and those who died from known causes showed that 15 and 38 miRNAs were significantly up- or down-regulated more than twofold in SIDS, respectively. Myocardial-specific miRNAs, such as miR-1, miR-208, and miR-499, which are known to leak from injured heart, were up-regulated markedly in SIDS EVs. Gene target prediction analyses suggested that the MAP signaling pathway, cardiomyocytes, and cardiac ion channels are involved in the pathogenesis of SIDS. Gene ontology analyses revealed that protein phosphorylation, the actin cytoskeleton and myosin complex, and kinase activity are heavily involved in SIDS. Our results indicate that EV myocardial-specific miRNAs are released into the blood from the heart in SIDS, suggesting the pathogenesis of SIDS is associated with cardiac injury. Studies of EV miRNAs using minimally invasive fluid samples could lead to the discovery of new diagnostic markers for SIDS.
Similar content being viewed by others
Data availability
The data have been deposited with links to BioProject accession number PRJDB37850 in the DDBJ BioProject database.
References
Baruteau, A.-E., Tester, D. J., Kapplinger, J. D., Ackerman, M. J. & Behr, E. R. Sudden infant death syndrome and inherited cardiac conditions. Nat. Rev. Cardiol. 14, 715–726. https://doi.org/10.1038/nrcardio.2017.129 (2017).
Harrington, C. T., Hafid, N. A. & Waters, K. A. Butyrylcholinesterase is a potential biomarker for sudden infant death syndrome. EBioMedicine 80, 104041. https://doi.org/10.1016/j.ebiom.2022.104041 (2022).
Ferrante, L., Opdal, S. H. & Byard, R. W. Further exploration of the influence of immune proteins in sudden infant death syndrome (SIDS). Acta Paediatr. https://doi.org/10.1111/apa.70197 (2025).
Graham, S. F., Chevallier, O. P., Kumar, P., Türkoǧlu, O. & Bahado-Singh, R. O. Metabolomic profiling of brain from infants who died from sudden infant death syndrome reveals novel predictive biomarkers. J. Perinatol. 37, 91–97. https://doi.org/10.1038/jp.2016.139 (2017).
Goldstein, R. D. et al. Inconsistent classification of unexplained sudden deaths in infants and children hinders surveillance, prevention and research: Recommendations from the 3rd international congress on sudden infant and child death. Forensic Sci. Med. Path. 15, 622–628. https://doi.org/10.1007/s12024-019-00156-9 (2019).
Filiano, J. J. & Kinney, H. C. A perspective on neuropathologic findings in victims of the sudden infant death syndrome: the triple-risk model. Biol. Neonate 65, 194–197. https://doi.org/10.1159/000244052 (1994).
Horne, R. S. C., Harrewijn, I. & Hunt, C. E. Physiology during sleep in preterm infants: Implications for increased risk for the sudden infant death syndrome. Sleep Med. Rev. 78, 101990. https://doi.org/10.1016/j.smrv.2024.101990 (2024).
Neubauer, J. et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur. J. Hum. Genet. 25, 404–409. https://doi.org/10.1038/ejhg.2016.199 (2017).
Van Norstrand, D. W. & Ackerman, M. J. Genomic risk factors in sudden infant death syndrome. Genome Med. 2, 86. https://doi.org/10.1186/gm207 (2010).
Hessvik, N. P. & Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 75, 193–208. https://doi.org/10.1007/s00018-017-2595-9 (2018).
Liu, T. et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 47, D89–D93. https://doi.org/10.1093/nar/gky985 (2019).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355. https://doi.org/10.1038/nature02871 (2004).
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandao, B. B. & Kahn, C. R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673. https://doi.org/10.1016/j.cmet.2019.07.011 (2019).
Dash, B. P. et al. Upregulated miR-10b-5p as a potential miRNA signature in amyotrophic lateral sclerosis patients. Front. Cell Neurosci. 18, 1457704. https://doi.org/10.3389/fncel.2024.1457704 (2024).
Kanno, S., Sakamoto, T., Fukuta, M., Kato, H. & Aoki, Y. Stability of exosomes in the postmortem serum and preliminary study on exosomal miRNA expression profiling in serum from myocardial infarction cadavers. Int. J. Legal Med. 137, 825–834. https://doi.org/10.1007/s00414-022-02913-y (2023).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672-d677. https://doi.org/10.1093/nar/gkae909 (2025).
Deddens, J. C. et al. Circulating extracellular vesicles contain miRNAs and are released as early biomarkers for cardiac injury. J. Cardiovasc. Transl. Res. 9, 291–301. https://doi.org/10.1007/s12265-016-9705-1 (2016).
Cheng, M. et al. Circulating myocardial microRNAs from infarcted hearts are carried in exosomes and mobilise bone marrow progenitor cells. Nat. Commun. 10, 959. https://doi.org/10.1038/s41467-019-08895-7 (2019).
Chistiakov, D. A., Orekhov, A. N. & Bobryshev, Y. V. Cardiac extracellular vesicles in normal and infarcted heart. Int. J. Mol. Sci. 17, 63 (2016).
Crouser, E. D. et al. Circulating exosomal microRNA expression patterns distinguish cardiac sarcoidosis from myocardial ischemia. PLoS ONE 16, e0246083. https://doi.org/10.1371/journal.pone.0246083 (2021).
Zhang, L. et al. Circulating miR-499 are novel and sensitive biomarker of acute myocardial infarction. J. Thorac. Dis. 7, 303–308. https://doi.org/10.3978/j.issn.2072-1439.2015.02.05 (2015).
Courts, C., Grabmüller, M. & Madea, B. Dysregulation of heart and brain specific micro-RNA in sudden infant death syndrome. Forensic Sci. Int. 228, 70–74. https://doi.org/10.1016/j.forsciint.2013.02.032 (2013).
Yang, B. et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13, 486–491. https://doi.org/10.1038/nm1569 (2007).
Yang, D. et al. MicroRNA-1 deficiency is a primary etiological factor disrupting cardiac contractility and electrophysiological homeostasis. Circ. Arrhythm. Electrophysiol. 17, e012150. https://doi.org/10.1161/CIRCEP.123.012150 (2024).
Xu, H. F. et al. MicroRNA- 1 represses Cx43 expression in viral myocarditis. Mol. Cell Biochem. 362, 141–148. https://doi.org/10.1007/s11010-011-1136-3 (2012).
Liu, X. et al. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: An independent study of Han population. Exp. Gerontol. 72, 230–238. https://doi.org/10.1016/j.exger.2015.10.011 (2015).
Huang, S. et al. Plasma extracellular vesicles microRNA-208b-3p and microRNA-143-3p as novel biomarkers for sudden cardiac death prediction in acute coronary syndrome. Mol. Omics 19, 262–273. https://doi.org/10.1039/d2mo00257d (2023).
Zhao, X., Wang, Y. & Sun, X. The functions of microRNA-208 in the heart. DRCP 160, 108004. https://doi.org/10.1016/j.diabres.2020.108004 (2020).
LaRocca, T. J. et al. CXCR4 cardiac specific knockout mice develop a progressive cardiomyopathy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20092267 (2019).
Garofolo, M. C., Seidler, F. J., Auman, J. T. & Slotkin, T. A. beta-Adrenergic modulation of muscarinic cholinergic receptor expression and function in developing heart. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, R1356-1363. https://doi.org/10.1152/ajpregu.00598.2001 (2002).
D’Alessandra, Y. et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773. https://doi.org/10.1093/eurheartj/ehq167 (2010).
Sequeira, V., Nijenkamp, L. L. A. M., Regan, J. A. & van der Velden, J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. BBA Biomembranes 1838, 700–722. https://doi.org/10.1016/j.bbamem.2013.07.011 (2014).
Bers, D. M. Cardiac excitation-contraction coupling. Nature 415, 198–205. https://doi.org/10.1038/415198a (2002).
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042. https://doi.org/10.1038/nature06418 (2007).
Scranton, K. et al. Cardiac function is regulated by the sodium-dependent inhibition of the sodium-calcium exchanger NCX1. Nat. Commun. 15, 3831. https://doi.org/10.1038/s41467-024-47850-z (2024).
Zhang, L., Kelley, J., Schmeisser, G., Kobayashi, Y. M. & Jones, L. R. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor: Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 272, 23389–23397. https://doi.org/10.1074/jbc.272.37.23389 (1997).
Chopra, N. et al. Ablation of triadin causes loss of cardiac Ca2+ release units, impaired excitation-contraction coupling, and cardiac arrhythmias. Proc. Natl. Acad. Sci. U. S. A. 106, 7636–7641. https://doi.org/10.1073/pnas.0902919106 (2009).
Fan, G.-C., Yuan, Q., Zhao, W., Chu, G. & Kranias, E. G. Junctin is a prominent regulator of contractility in cardiomyocytes. Biochem. Biophys. Res. Commun. 352, 617–622. https://doi.org/10.1016/j.bbrc.2006.11.093 (2007).
Son, M. J., Kim, M. K. & Yoo, S. H. Identification of mutations of the RYR2 in sudden infant death syndrome. J. Korean Med. Sci. 40, e17. https://doi.org/10.3346/jkms.2025.40.e17 (2025).
Shooshtarian, A. K., O’Gallagher, K., Shah, A. M. & Zhang, M. SERCA2a dysfunction in the pathophysiology of heart failure with preserved ejection fraction: a direct role is yet to be established. Heart Fail. Rev. 30, 545–564. https://doi.org/10.1007/s10741-025-10487-1 (2025).
Park, J. H. & Kho, C. MicroRNAs and calcium signaling in heart disease. Int. J. Mol. Sci. https://doi.org/10.3390/ijms221910582 (2021).
Jaalouk, D. E. & Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 10, 63–73. https://doi.org/10.1038/nrm2597 (2009).
Ottaviani, G., Leonardi, P., Del Fabbro, M. & Ramos, S. G. Sudden unexpected infant and perinatal death: pathological findings of the cardiac conduction system. Diagnostics 15, 1637 (2025).
Dettmeyer, R. B. & Kandolf, R. Cardiomyopathies–misdiagnosed as sudden infant death syndrome (SIDS). Forensic Sci. Int. 194, e21-24. https://doi.org/10.1016/j.forsciint.2009.10.010 (2010).
Bach, V. & Libert, J. P. Hyperthermia and heat stress as risk factors for sudden infant death syndrome: A narrative review. Front Pediatr. 10, 816136. https://doi.org/10.3389/fped.2022.816136 (2022).
Köffer, J., Scheiper-Welling, S., Verhoff, M. A., Bajanowski, T. & Kauferstein, S. Post-mortem genetic investigation of cardiac disease-associated genes in sudden infant death syndrome (SIDS) cases. Int. J. Legal Med. 135, 207–212. https://doi.org/10.1007/s00414-020-02394-x (2021).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. https://doi.org/10.1186/gb-2009-10-3-r25 (2009).
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W. & Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. https://doi.org/10.1093/nar/gkr688 (2012).
Wen, M., Shen, Y., Shi, S. & Tang, T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 13, 140. https://doi.org/10.1186/1471-2105-13-140 (2012).
Zhou, L. et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 5, e15224. https://doi.org/10.1371/journal.pone.0015224 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Kruger, J. & Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451-454. https://doi.org/10.1093/nar/gkl243 (2006).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
Acknowledgements
This work was supported by a Grant-in-Aid for Research from Nagoya City University (grant number 2412004). We acknowledge the assistance of the Research Equipment Sharing Center at Nagoya City University. This work was the result of using research equipment shared with the MEXT Project for Promoting Public Utilization of Advanced Research Infrastructure (Program for Supporting Construction of Core Facilities), grant number JPMXS0441500025. We would also like to thank Dr. Hiroshi Takase of the Research Equipment Sharing Center of Nagoya City University for assistance with scanning electron microscopy observations.
Author information
Authors and Affiliations
Contributions
SK: conceived and designed the experiments. SK, MF, JM, YN: performed the experiments and analyzed the data. MF, HK, TO: performed the autopsies and examinations. SK, MF: wrote the manuscript with contributions from co-authors. TO: project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Institutional Review Board for the Protection of Human Subjects in Research at Nagoya City University (approval number: 60-24-0135, approval date: Jan. 25, 2022). All the procedures were approved and performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki. This study was conducted based on the “Ethical Guidelines for Medical Research Involving Human Subjects (enacted by the Ministry of Health, Labor, and Welfare in Japan).” Information on the implementation of the study was posted on clinical Research Management Center, Nagoya City University Hospital website (https://ncu-cr.jp/patient/clinical_research/clinical_research_cont-2). If bereaved family requested to refuse not be used for research purposes, they were excluded from the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kanno, S., Fukuta, M., Kato, H. et al. Comparative analysis of microRNA expression in serum-derived extracellular vesicles from sudden infant death syndrome cases. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38034-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38034-4