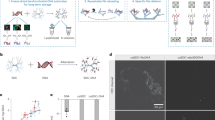
Abstract
We present a DNA extraction protocol for atmospheric bioaerosol samples collected on glass-fiber filters widely used in air quality monitoring. The protocol produces high-quality molecules suitable for third-generation sequencing and other applications. The initial protocol was developed and applied in a Bioaerosol campaign performed in Finland and Lithuania in 2021 using low-volume air samplers, which posed stringent requirements to the method sensitivity. The protocol included a phenol–chloroform step for DNA purification, thus involving aggressive reagents; it was also quite time consuming and laborious. The present study advances this protocol to exclude the use of hazardous chemicals by using the SPRI paramagnetic bead technology for DNA purification and compares it to several commercial extraction methods. Despite trailing in efficiency to the initial method, the new development proved to be more efficient than several column-based commercial kits. The updated protocol was effective for a relatively high mass ratio of biological material to filter material: 70 nanograms of potential DNA on the filter to one milligram of filter fiber, as detected with the initial phenol–chloroform-based method. However, the new approach was not effective for a mass ratio lower than 15 nanograms of potential DNA per milligram of the filter material. The applicability of the new protocol for preparation of samples for the 3rd generation sequencing was confirmed by subsequent processing of the samples with the Oxford Nanopore (ONT) GridION sequencer.
Similar content being viewed by others
Data availability
The DNA sequences obtained within the study have been uploaded to the ENA database as trimmed read data and are publicly available from the project PRJEB76246.
References
Fröhlich-Nowoisky, J. et al. Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos. Res. 182, 346–376 (2016).
Sofiev, M. & Bergmann, K. C. Allergenic Pollen. A Review of the Production, Release, Distribution and Health Impacts (Springer Netherlands, 2012).
Walser, S. M. et al. Evaluation of exposure–response relationships for health effects of microbial bioaerosols – A systematic review. Int. J. Hyg. Environ Health. 218, 577–589 (2015).
Kim, K. H., Kabir, E. & Jahan, S. A. Airborne bioaerosols and their impact on human health. J. Environ. Sci. 67, 23–35 (2018).
Goldman, D. L. & Huffnagle, G. B. Potential contribution of fungal infection and colonization to the development of allergy. Med. Mycol. 47, 445–456 (2009).
Damialis, A., Halley, J. M., Gioulekas, D. & Vokou, D. Long-term trends in atmospheric pollen levels in the City of Thessaloniki, Greece. Atmos. Environ. 41, 7011–7021 (2007).
Buters, J. T. M. et al. Pollen and spore monitoring in the world. Clin. Transl Allergy. 8, 9 (2018).
Stephens, B. What have we learned about the microbiomes of indoor environments? mSystems 1, e00083–e00016 (2016).
Shin, S. K. et al. Metagenomic insights into the bioaerosols in the indoor and outdoor environments of childcare facilities. PLoS ONE. 10, e0126960 (2015).
Serrano-Silva, N. & Calderón-Ezquerro, M. C. Metagenomic survey of bacterial diversity in the atmosphere of Mexico City using different sampling methods. Environ. Pollut. 235, 20–29 (2018).
Spring, A. M. et al. A method for collecting atmospheric microbial samples from set altitudes for use with Next-Generation sequencing techniques to characterize communities. Air Soil. Water Res. 11, 117862211878887 (2018).
Sofiev, M. et al. Bioaerosols in the atmosphere at two sites in Northern Europe in spring 2021: outline of an experimental campaign. Environ. Res. 214, 113798 (2022).
Jiang, W. et al. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nat. Protoc. 10, 768–779 (2015).
Wydro, U. Soil Microbiome study based on DNA extraction: A review. Water 14, 3999 (2022).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Bock, D. G., Liu, J., Novikova, P. & Rieseberg, L. H. Long-read sequencing in ecology and evolution: Understanding how complex genetic and epigenetic variants shape biodiversity. Mol. Ecol. 32, 1229–1235 (2023).
Kim, C., Pongpanich, M. & Porntaveetus, T. Unraveling metagenomics through long-read sequencing: a comprehensive review. J. Transl Med. 22, 111 (2024).
Abdel-Latif, A. & Osman, G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant. Methods. 13, 1 (2017).
Dauphin, L. A., Stephens, K. W., Eufinger, S. C. & Bowen, M. D. Comparison of five commercial DNA extraction kits for the recovery of yersinia pestis DNA from bacterial suspensions and spiked environmental samples: DNA extraction from Y. pestis. J. Appl. Microbiol. 108, 163–172 (2010).
Rostami, H., Firoozeh, F., Zibaei, M. & Salahshoorifar, I. Evaluation of genomic DNA extraction using monolayer and bilayer magnetic nanoparticles. Int. J. Enteric Pathog. 8, 51–54 (2020).
Ma, C. et al. Magnetic Nanoparticles-Based extraction and verification of nucleic acids from different sources. J. Biomed. Nanotechnol. 9, 703–709 (2013).
Kumaishi, K. et al. High throughput method of 16S rRNA gene sequencing library Preparation for plant root microbial community profiling. Sci. Rep. 12, 19289 (2022).
Kumaishi, K. et al. Simple Amplicon Sequencing Library Preparation for Plant Root Microbial Community Profiling. http://biorxiv.org/lookup/doi/ (2021). https://doi.org/10.1101/2021.04.14.439905 doi:10.1101/2021.04.14.439905.
Tilak, M. K., Botero-Castro, F., Galtier, N. & Nabholz, B. Illumina library Preparation for sequencing the GC-Rich fraction of heterogeneous genomic DNA. Genome Biol. Evol. 10, 616–622 (2018).
Van Dijk, E. L., Jaszczyszyn, Y. & Thermes, C. Library Preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 322, 12–20 (2014).
Butler, C., Matsumoto, A., Rutherford, C. & Lima, H. K Comparison of the effectiveness of four commercial DNA extraction kits on fresh and frozen human milk samples. MPs 5, 63 (2022).
Esfandani-Bozchaloyi, S., Sheidai, M. & Kalalegh, M. H. Comparison of DNA extraction methods from geranium (Geraniaceae). Acta Bot. Hungarica. 61, 251–266 (2019).
Rohland, N., Siedel, H. & Hofreiter, M. A rapid column-based ancient DNA extraction method for increased sample throughput. Mol. Ecol. Resour. 10, 677–683 (2010).
Wu, L. H., Liu, J. L., Zeng, J. & Zhao, J. Comparison of DNA Extraction and Purification Methods from Different Soils for Metagenomic Sequencing. AMR 955–959, 306–309 (2014).
Kalisa, E., Kuuire, V. & Adams, M. A preliminary investigation comparing high-volume and low-volume air samplers for measurement of PAHs, NPAHs and airborne bacterial communities in atmospheric particulate matter. Environ. Sci. : Atmos. 2, 1120–1131 (2022).
Paatero, J., Hatakka, J. & Viisanen, Y. Concurrent Measurements of Airborne Radon-222, Lead-210 and Beryllium-7 at the Pallas-Sodankylä Gaw Station, Northern Finland (Ilmatieteen laitos, 1998).
Dunn, O. J. Multiple comparisons using rank sums. Technometrics 6, 241–252 (1964).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Completing bacterial genome assemblies with multiplex minion sequencing. Microbial Genomics 3, (2017).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, 10 (2011).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to Single-Cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
FastQC. (2015).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Kallio, M. A. et al. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genom. 12, 507 (2011).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with kraken 2. Genome Biol. 20, 257 (2019).
Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104 (2017).
Acknowledgements
The study was performed within Horizon project SYLVA (grant 101086109) and supported by Academy of Finland projects PS4A (grant 318194), SPORELIFE (grant 355851), DASI (grant 479507) and the ACCC Flagship funding (grant 337552).
Funding
The study was performed within the Horizon project SYLVA (grant 101086109) and supported by the Academy of Finland project PS4A (grant 318194), SPORELIFE (grant 355851), DASI (grant 479507) and ACCC Flagship funding (grant 337552).
Author information
Authors and Affiliations
Contributions
All authors participated in writing and editing of the manuscript and approved its final version. J.S. and S.S. jointly designed the experiment, developed the initial and updated protocol versions and applied them. M.S. participated in the study design. J.P., E.A., A.K., and M.S. provided material, financial, and organizational support for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Salokas, J., Sofieva-Rios, S., Paatero, J. et al. Evaluation of commercial kits and purification approaches for DNA extraction from atmospheric samples for 3rd generation sequencing without amplification. Sci Rep (2026). https://doi.org/10.1038/s41598-026-38534-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-38534-3