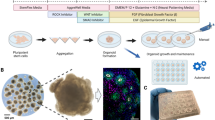
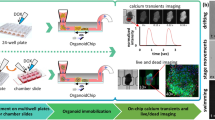
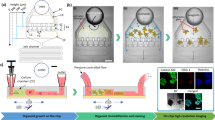
Abstract
Organoids, 3D tissue cultures that mimic real organs, offer valuable models for research. Traditional culture methods rely on manual feeding and orbital shakers, making them labor-intensive and inconsistent. Microfluidic systems have shown their potential to improve reproducibility by controlling media exchange and culture conditions, yet most still require standard incubators, which limit continuous monitoring due to space and humidity constraints. To address this, we developed a modular platform that integrates automated feeding, real-time imaging, and environmental control, eliminating the need for a conventional incubator. A key feature is a vertically oriented PDMS/glass chip that supports precise media delivery and monitoring while preserving incubation conditions, making it ideal for morphological studies. We demonstrated the platform’s ability to maintain metabolic stability and media distribution over time using cerebral organoids. This platform improves organoid research by combining microfluidics, automation, and imaging, enhancing disease modeling, drug testing, and regenerative medicine applications.
Similar content being viewed by others
Data availability
All custom scripts, pH calibration, feeding rates, temperature records, 3D-printed files, microscope images, and CFD videos are available at [[https://github.com/sebtomon89/braingeneersdifussionproject](https:/github.com/sebtomon89/braingeneersdifussionproject)]. Additional modified scripts can be accessed upon request. All other relevant data are available from the corresponding author upon request.
Code availability
Details of publicly available software used in the study are given in the “Data availability” section. Apart from this, no unique custom code or mathematical algorithms were central to reaching the conclusions of this work.
References
Sasai, Y. Next-Generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell. Stem Cell. 12, 520–530 (2013).
Sasai, Y., Eiraku, M. & Suga, H. In vitro organogenesis in three dimensions: self-organising stem cells. Development 139, 4111–4121 (2012).
Mun, S. J. et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 71, 970–985 (2019).
Takasato, M., Er, P. X., Chiu, H. S. & Little, M. H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 11, 1681–1692 (2016).
Völkner, M. et al. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis. Stem Cell. Rep. 6, 525–538 (2016).
Seiler, S. T. et al. Modular automated microfluidic cell culture platform reduces glycolytic stress in cerebral cortex organoids. Sci. Rep. 12, 20173 (2022).
Cho, A. N. et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat. Commun. 12, 4730 (2021).
Schwedhelm, I. et al. Automated real-time monitoring of human pluripotent stem cell aggregation in stirred tank reactors. Sci. Rep. 9, 12297 (2019).
Aguilera-Castrejon, A. & Hanna, J. H. Ex utero culture of mouse embryos from pregastrulation to advanced organogenesis. JoVE 63160 https://doi.org/10.3791/63160 (2021).
Berger, E. et al. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab. Chip. 18, 3172–3183 (2018).
Schuster, B. et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 11, 5271 (2020).
Khan, I. et al. A low-cost 3D printed microfluidic bioreactor and imaging chamber for live-organoid imaging. Biomicrofluidics 15, 024105 (2021).
Maisonneuve, B. G. C. et al. Deposition chamber technology as Building blocks for a standardized brain-on-chip framework. Microsyst. Nanoeng. 8, 86 (2022).
Lee, H. N. et al. Effect of biochemical and Biomechanical factors on vascularization of kidney organoid-on-a-chip. Nano Convergence. 8, 35 (2021).
Castiglione, H. et al. Human brain Organoids-on-Chip: Advances, Challenges, and perspectives for preclinical applications. Pharmaceutics 14, 2301 (2022).
Sanchis-Calleja, F. et al. Decoding morphogen patterning of human neural organoids with a multiplexed single-cell transcriptomic screen. bioRxiv https://doi.org/10.1016/j.jmbbm.2021.105024.
Boussaad, I. et al. Integrated, automated maintenance, expansion and differentiation of 2D and 3D patient-derived cellular models for high throughput drug screening. Sci. Rep. 11, 1439 (2021).
Understrup, K. G. & Programming and control of flow-based microfluidic biochips.
Maurer, B. et al. Inkube: an all-in-one solution for neuron culturing, electrophysiology, and fluidic exchange. Preprint Biorxiv Doi. https://doi.org/10.1101/2024.12.06.627248 (2024).
Voitiuk, K. et al. A feedback-driven IoT microfluidic, electrophysiology, and imaging platform for brain organoid studies. Internet Things. 33, 101671 (2025).
Talebipour, A., Saviz, M., Vafaiee, M. & Faraji-Dana, R. Facilitating long-term cell examinations and time-lapse recordings in cell biology research with CO2 mini-incubators. Sci. Rep. 14, 3418 (2024).
Wiggins, L. et al. The cellphe toolkit for cell phenotyping using time-lapse imaging and pattern recognition. Nat. Commun. 14, 1854 (2023).
Svensson, C., Medyukhina, A., Belyaev, I., Al-Zaben, N. & Figge, M. T. Untangling cell tracks: quantifying cell migration by time lapse image data analysis. Cytometry Pt A. 93, 357–370 (2018).
Hashimoto, H. et al. Time-lapse imaging of cell cycle dynamics during development in living cardiomyocyte. J. Mol. Cell. Cardiol. 72, 241–249 (2014).
Ly, V. T. et al. Picroscope: low-cost system for simultaneous longitudinal biological imaging. Commun. Biol. 4, 1261 (2021).
Paulsen, B. et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022).
Mostajo-Radji, M. A., Schmitz, M. T., Montoya, S. T. & Pollen, A. A. Reverse engineering human brain evolution using organoid models. Brain Res. 1729, 146582 (2020).
Xinaris, C., Brizi, V. & Remuzzi, G. Organoid models and applications in biomedical research. Nephron 130, 191–199 (2015).
Rajan, D. K. et al. IEEE,. Monitoring pH, temperature and humidity in long-term stem cell culture in CO 2 incubator. In 2017 IEEE International Symposium on Medical Measurements and Applications (MeMeA) 470–474. https://doi.org/10.1109/MeMeA.2017.7985922 (2017).
Smith, R. L., Demers, C. J. & Collins, S. D. Microfluidic device for the combinatorial application and maintenance of dynamically imposed diffusional gradients. Microfluid Nanofluid. 9, 613–622 (2010).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 16, 255–262 (2019).
Rosen, Y. et al. Incubator-free organoid culture in a sealed recirculatory system. bioRxiv Preprint Server for Biology https://doi.org/10.1101/2025.09.03.673593 (2025).
Brunner, M., Doppler, P., Klein, T., Herwig, C. & Fricke, J. Elevated pCO 2 affects the lactate metabolic shift in CHO cell culture processes. Eng. Life Sci. 18, 204–214 (2018).
Miura, Y. & Pașca, S. P. Mapping human brain organoids on a Spatial atlas. Cell. Stem Cell. 28, 983–984 (2021).
Chu, J. & Anderson, S. A. Dev. Cortical Interneurons Neuropsychopharmacol. 40, 16–23 (2015).
Pavon, N. et al. Patterning ganglionic eminences in developing human brain organoids using morphogen gradient inducing device. Cell. Rep. Methods. 4 (1), 100689 (2024).
Kazim, S. F., Enam, S. A. & Shamim, M. S. Possible detrimental effects of neurosurgical irrigation fluids on neural tissue: an evidence based analysis of various irrigants used in contemporary neurosurgical practice. Int. J. Surg. 8, 586–590 (2010).
AF4115T-GRFBY — Dino-Lite Digital Microscope | Americas. https://www.dinolite.us/products/usb-microscopes/af4115t-grfby/
Hernandez, S. et al. Self-organizing neural networks in organoids reveal principles of forebrain circuit assembly. bioRxiv https://doi.org/10.1101/2025.05.01.651773 (2025).
Lei, Y., Shkolnikov, V. & Xin, D. 3D biological cell reconstruction with multi-view geometry. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 495–498. https://doi.org/10.1109/ISBI45749.2020.9098564 (IEEE, 2020).
Tiburcius, S. et al. Egg-yolk core-shell mesoporous silica nanoparticles for high doxorubicin loading and delivery to prostate cancer cells. Nanoscale 14, 6830–6845 (2022).
Sher, J., Miller, C. & Sharma, D. Effect of bisphosphonates on the osteogenic activity of osteoprogenitor cells cultured on titanium surfaces. Int. J. Oral Maxillofac. Implants. 35, 939–947 (2020).
Munro, T., Miller, C. M., Antunes, E. & Sharma, D. Interactions of osteoprogenitor cells with a novel zirconia implant surface. J. Funct. Biomater. 11, 50 (2020).
Bhaduri, A. et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020).
Mazziotta, M. & Pareto, A. Normalization methods for spatio-temporal analysis of environmental performance: revisiting the Min-Max method. Environmetrics 33, e2730 (2022).
Xiang, Y. et al. Generation and fusion of human cortical and medial ganglionic eminence brain organoids. Curr. Protoc. Stem Cell. Biol. 47, e61 (2018).
Giandomenico, S. L., Sutcliffe, M. & Lancaster, M. A. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat. Protoc. 16, 579–602 (2021).
Sohn, L. L. et al. How can microfluidic and microfabrication approaches make experiments more physiologically. Relevant? Cell. Syst. 11, 209–211 (2020).
Amirifar, L. et al. Brain-on-a-chip: recent advances in design and techniques for microfluidic models of the brain in health and disease. Biomaterials 285, 121531 (2022).
Rifes, P. et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat. Biotechnol. 38, 1265–1273 (2020).
Suong, D. N. A. et al. Induction of inverted morphology in brain organoids by vertical-mixing bioreactors. Commun. Biol. 4, 1213 (2021).
Salek, M. M., Sattari, P. & Martinuzzi, R. J. Analysis of fluid flow and wall shear stress patterns inside partially filled agitated culture well plates. Ann. Biomed. Eng. 40, 707–728 (2012).
Goto-Silva, L. et al. Computational fluid dynamic analysis of physical forces playing a role in brain organoid cultures in two different multiplex platforms. BMC Dev. Biol. 19, 3 (2019).
Murasiewicz, H. et al. Engineering considerations on the use of liquid/liquid two-phase systems as a cell culture platform: engineering considerations on the use of liquid/liquid two-phase systems. J. Chem. Technol. Biotechnol. 92, 1690–1698 (2017).
Demers, C. J. et al. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development 143, 1884–1892 (2016).
Amin, N. D. et al. Generating human neural diversity with a multiplexed morphogen screen in organoids. Cell. Stem Cell. 31 (12), 1831–1846E9 (2024).
Wolak, D. J. & Thorne, R. G. Diffusion of macromolecules in the brain: implications for drug delivery. Mol. Pharm. 10, 1492–1504 (2013).
Puschhof, J. et al. Derivation of snake venom gland organoids for in vitro venom production. Nat. Protoc. 16, 1494–1510 (2021).
Liu, Y. et al. Intracellular pH dynamics regulates intestinal stem cell lineage specification. Nat. Commun. 14, 3745 (2023).
Borello, U. & Pierani, A. Patterning the cerebral cortex: traveling with morphogens. Curr. Opin. Genet. Dev. 20, 408–415 (2010).
Kutys, M. L. et al. Uncovering mutation-specific morphogenic phenotypes and paracrine-mediated vessel dysfunction in a biomimetic vascularized mammary duct platform. Nat. Commun. 11, 3377 (2020).
Xue, X. et al. A patterned human neural tube model using microfluidic gradients. Nature 628, 391–399 (2024).
Mostajo-Radji, M. A. et al. Fate plasticity of interneuron specification. iScience 28 (4), 112295. https://doi.org/10.1016/j.isci.2025.112295 (2025).
Rodríguez, C. F. et al. Breaking the clean room barrier: exploring low-cost alternatives for microfluidic devices. Front. Bioeng. Biotechnol. 11, 1176557 (2023).
Rickert, C. A., Bauer, M. G., Hoffmeister, J. C. & Lieleg, O. Effects of sterilization methods on the integrity and functionality of covalent mucin coatings on medical devices. Adv. Mater. Interfaces. 9 (3), 2101716 (2022).
Millet, L. J., Jain, A. & Gillette, M. U. Less is More: Oligomer extraction and hydrothermal annealing increase PDMS bonding forces for new microfluidics assembly and for biological studies. bioRxiv 150953. https://doi.org/10.1101/150953v1.full (2017).
Heo, Y. S. et al. Characterization and resolution of Evaporation-Mediated osmolality shifts that constrain microfluidic cell culture in Poly(dimethylsiloxane) devices. Anal. Chem. 79, 1126–1134 (2007).
Chuah, Y. J. et al. Simple surface engineering of polydimethylsiloxane with polydopamine for stabilized mesenchymal stem cell adhesion and multipotency. Sci. Rep. 5, 18162 (2015).
Park, Y. et al. Modulation of neuronal activity in cortical organoids with bioelectronic delivery of ions and neurotransmitters. Cell. Rep. Methods. 4, 100686 (2024).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA. 110, 20284–20289 (2013).
Rosch, J. C. et al. CRISPR-Mediated isogenic Cell-SELEX approach for generating highly specific aptamers against native membrane proteins. Cel Mol. Bioeng. 13, 559–574 (2020).
Theory of Nonparametric Tests. (Springer Berlin Heidelberg, 2018).
McMurtrey, R. J. Analytic models of oxygen and nutrient Diffusion, metabolism Dynamics, and architecture optimization in Three-Dimensional tissue constructs with applications and insights in cerebral organoids. Tissue Eng. Part. C Methods. 22, 221–249 (2016).
Patrachari, A. R., Podichetty, J. T. & Madihally, S. V. Application of computational fluid dynamics in tissue engineering. J. Biosci. Bioeng. 114, 123–132 (2012).
Ramanujan, S. et al. Diffusion and convection in collagen gels: implications for transport in the tumor interstitium. Biophys. J. 83, 1650–1660 (2002).
Shin, W. et al. Spatiotemporal gradient and instability of Wnt induce heterogeneous growth and differentiation of human intestinal organoids. iScience 23, 101372 (2020).
Saglam-Metiner, P. et al. Spatio-temporal dynamics enhance cellular diversity, neuronal function and further maturation of human cerebral organoids. Commun. Biol. 6, 173 (2023).
Lomax, H. et al. Fundamentals of computational fluid dynamics. Appl. Mech. Rev. 55 (4), B61–B61 (2002).
Andersson, B. et al. Computational Fluid Dynamics for Engineers (Cambridge University Press, 2011).
Acknowledgements
We thank the UCSC Life Sciences Microscopy Center, RRID: SCR_021135, for providing the confocal microscope to acquire the images. Some illustrations were generated using Biorender. We gratefully acknowledge contributions from Dr. Sofie R. Salama and Dr. Kateryna Voitiuk for the feedback on the preparation of this manuscript. During the preparation of this work, the authors used ChatGPT and Grammarly to improve clarity and sentence structure. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Funding
This work was supported by the Schmidt Futures Grant SF857 (D.H., and M.T.); the National Human Genome Research Institute Grant 1RM1HG011543 (D.H. and M.T.); National Science Foundation Grants 2134955 (to D.H. and M.T.), 2034037 (to M.T.), and 2515389 (to D.H., M.A.M.-R. and M.T.); the National Institute of Mental Health Grant 1U24MH132628 and U24NS146314 (both to D.H. and M.A.M.-R.); the California Institute for Regenerative Medicine DISC4-16285 (to M.A.M.-R. and M.T.), and DISC4-16337 (to M.A.M.-R).; by the University of California Office of the President M25PR9045 (to M.A.M.-R. and M.T.). H.E.S. is a National Science Foundation Graduate Student Research Fellowship grantee. S.H. received support from the UC Doctoral Diversity Initiative (DDI-UCSC-IBSC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the National Science Foundation, the University of California, CIRM or any other agency of the State of California.
Author information
Authors and Affiliations
Contributions
S.T.-M. and S.T.S. worked on hardware, software, and platform assembly. S.T.-M., S.H., H.E.S and S.V-C. worked in cell culture and cell staining. S.T.-M., S.H., H.E.S., and G.K. performed biological experiments. S.T.-M., M.A.M.-R., and M.T. conceived the experiments. D.H., M.A.M.-R., and M.T. supervised the team and secured funding. S.T.-M., H.E.S., M.A.M.-R., S.T.S., and M.T. wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Torres-Montoya, S., Hernandez, S., Seiler, S.T. et al. A modular platform for automated organoid culture and longitudinal imaging. Sci Rep (2026). https://doi.org/10.1038/s41598-026-40231-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-40231-0