Abstract
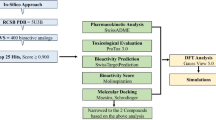
Mirabilis longiflora L. has been used traditionally in some parts of Bangladesh for the treatment of headaches, infectious diseases, painkillers, skin disease, herpes infection, and wound healing, but its effects on multidrug-resistant (MDR) bacteria have remained unidentified. Therefore, we aimed to determine the antibacterial activity of the methanol extract of M. longiflora L. leaves (MEMLL) against MDR Pseudomonas aeruginosa and Bacillus cereus and to recognize possible multitargeting antibacterial phytocompounds through in silico computational approaches targeting the LasR and LpxC proteins in MDR Pseudomonas aeruginosa and the FosB and PlcR proteins in MDR Bacillus cereus. PPS, FT-IR, and GC-MS were used for profiling of the phytocompounds in MEMLL. The antimicrobial activity of MEMLL was evaluated using in vitro agar-well diffusion, MIC, and MBC assays. In silico methods were applied to identify multi-targeting agents from GC-MS-annotated phytocompounds. MEMLL showed dose-dependent antibacterial activity and exposed the presence of 33 phytochemicals in GC-MS analysis. Among these, 6-Hydroxy-4,4,7a-trimethyl-5,6,7,7a-tetrahydrobenzofuran-2(4 H)-one (CID 14334) was identified as a potential antibacterial phytocompound as it exhibited multi-modal and strong binding affinity towards LasR, LpxC, FosB, and PlcR, favorable pharmacokinetics, drug-likeness, physicochemical, and toxicity properties. Finally, Molecular dynamics (MD) simulations demonstrated the structural stability of CID 14,334 within the active sites of LasR, LpxC, FosB, and PlcR. The results of this study offer scientific validation for the traditional use of M. longiflora L. in bacterial infection-related diseases. It also suggests that 6-Hydroxy-4,4,7a-trimethyl-5,6,7,7a-tetrahydrobenzofuran-2(4 H)-one from M. longiflora L. might be responsible for the antibacterial activity and could act as a phytopharmacological lead for the development of LasR and LpxC inhibitors against MDR Pseudomonas aeruginosa and FosB and PlcR inhibitors against MDR Bacillus cereus.
Similar content being viewed by others
Data availability
Available upon reasonable request from the corresponding author.
Abbreviations
- MEMLL:
-
Methanolic Extract of Mirabilis longiflora leaves
- MDR:
-
Multidrug resistance
- FT-IR:
-
Fourier-transform infrared spectroscopy
- PPS:
-
Preliminary phytochemical screening
- GC-MS:
-
Gas chromatography-mass spectrometry
- MIC:
-
Minimum Inhibitory Concentration
- MBC:
-
Minimum Bactericidal Concentration
- OPLS3e:
-
Extending Force Field Coverage for Drug-Like Small Molecules
- TIP3P:
-
Transferable intermolecular potential with 3 points
- ADME:
-
Absorption, Distribution, Metabolism, and Excretion
- SID:
-
Simulation Interaction Diagram
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- Rg:
-
Radius of gyration
- SASA:
-
Solvent accessible surface area
References
Safdar, N. et al. Economic burden of antimicrobial resistance on patients in Pakistan. Front. Public. Health. 13, 1481212 (2025).
Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 12, 3903–3910 (2019).
Global burden of bacterial antimicrobial resistance. In 2019: A systematic analysis. Lancet 399, 629–655 (2022).
Elbaiomy, R. G. et al. Antibiotic resistance: A genetic and physiological perspective. MedComm 6, e70447 (2025).
Elshobary, M. E. et al. Combating antibiotic resistance: Mechanisms, multidrug-resistant pathogens, and novel therapeutic approaches: An updated review. Pharmaceuticals 18, 402 (2025).
Horcajada, J. P. et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32, (2019).
Algammal, A. M. et al. Newly emerging MDR B. cereus in Mugil seheli, as the first report, commonly harbor nhe, hbl, cytK, and pc-plc virulence genes and bla1, bla2, tetA, and erm a resistance genes. Infect. Drug Resist. 15, 2167–2185 (2022).
Swain, J. et al. Pathogenicity and virulence of Pseudomonas aeruginosa: Recent advances and under-investigated topics. Virulence 16, 1 (2025).
Kostylev, M. et al. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc. Natl. Acad. Sci. USA. 116, 7027–7032 (2019).
Rutherford, S. T. & Bassler, B. L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 (2012).
Zhang, X. et al. A comprehensive review of the pathogenic mechanisms of Pseudomonas aeruginosa: Synergistic effects of virulence factors, quorum sensing, and biofilm formation. Front. Microbiol. 16, 1619626 (2025).
Miranda, S. W., Asfahl, K. L., Dandekar, A. A. & Greenberg, E. P. Pseudomonas aeruginosa quorum sensing. In Advances in Experimental Medicine and Biology 1386 95–115 (Springer, 2022).
Abbas, H. A., Shaldam, M. A. & Eldamasi, D. Curtailing quorum sensing in Pseudomonas aeruginosa by sitagliptin. Curr. Microbiol. 77, 1051–1060 (2020).
Krause, K. M. et al. Potent LpxC inhibitors with in vitro activity against multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, e00977–e00919 (2019).
Alanazi, A. et al. A combined in silico and MD simulation approach to discover novel LpxC inhibitors targeting multidrug-resistant Pseudomonas aeruginosa. Sci. Rep. 15, 16900 (2025).
Wu, Y. et al. Overcoming the drug resistance barrier: Progress in fosfomycin combination therapy against multidrug-resistant pathogens. Front. Microbiol. 16, 1702881 (2025).
Travis, S. et al. Inhibition of fosfomycin resistance protein FosB from Gram-positive pathogens by phosphonoformate. Biochemistry 62, 109–117 (2023).
Qin, J. et al. NupR is involved in the control of plcr: A pleiotropic regulator of extracellular virulence factors. Microorganisms 13, 212 (2025).
Sayem, M. et al. Comprehensive genomic analysis reveals virulence and antibiotic resistance genes in a multidrug-resistant Bacillus cereus isolated from hospital wastewater in Bangladesh. Sci. Rep. 15, 22915 (2025).
Abdallah, E. M., Alhatlani, B. Y., de Paula Menezes, R. & Martins, C. H. G. Back to nature: Medicinal plants as promising sources for antibacterial drugs in the post-antibiotic era. Plants 12, 3077. https://doi.org/10.3390/plants12173077 (2023).
Imon, R. R. et al. Natural defense against multi-drug resistant Pseudomonas aeruginosa: Cassia occidentalis L. in vitro and in silico antibacterial activity. RSC Adv. 13, 28773–28784 (2023).
Malakar Moumita & Biswas, S. Mirabilis: Medicinal uses and conservation. In Floriculture and Ornamental Plants (eds. Datta, S. K. and Gupta, Y. C.) 1–57 (Springer, 2020). https://doi.org/10.1007/978-981-15-1554-5_28-1
Kumar Yadav, S. et al. Extraction, phytochemical investigation, and antiulcer activity of hydroalcoholic extract of Mirabilis longiflora. Indo Am. J. P Sci. 9 (2022).
Rahman, M. M. et al. Mikania cordata leaves extract promotes activity against pathogenic bacteria and anticancer activity in EAC cell-bearing Swiss albino mice. J. Appl. Pharm. Sci. 10, 112–122 (2020).
Zaman, S. B. et al. Evaluation of antibiotic susceptibility in wound infections: A pilot study from Bangladesh. F1000Res 6, (2017).
Rahman, M. M. et al. Unveiling the therapeutic efficacy of extract and multitargeting phytocompounds from Christella dentata (Forssk.) Brownsey & jermy against multidrug-resistant Pseudomonas aeruginosa. RSC Adv. 14, 6096–6111 (2024).
Bhardwaj, S. et al. Molecular simulation-based investigation of thiazole derivatives as potential LasR inhibitors of Pseudomonas aeruginosa. PLoS ONE. 20, e0320841 (2025).
Travis, S. et al. Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus. RSC Med. Chem. 14, 947–956 (2023).
Grenha, R. et al. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. Proc. Natl. Acad. Sci. USA. 110, 1047–1052 (2013).
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013).
Harder, E. et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296 (2016).
Talukder, M. E. K. et al. Molecular docking, QSAR, and simulation analyses of EGFR-targeting phytochemicals in non-small cell lung cancer. J. Mol. Struct. 1321, 139924 (2025).
Banerjee, P., Eckert, A. O., Schrey, A. K. & Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263 (2018).
Li, Y. et al. Decoding the limits of deep learning in molecular docking for drug discovery. Chem. Sci. 16, 17374–17390 (2025).
De Ferreira, R. & Schapira, M. A systematic analysis of atomic protein-ligand interactions in the PDB. Medchemcomm 8, 1970–1981 (2017).
Shams, W. et al. Pharmacokinetics of drug absorption and metabolism: Mechanisms, influencing factors, and screening strategies in drug development. J. Neonatal Surg. 14, 574–581 (2025).
Ojha, H. et al. Chapter 7 - Chem-bioinformatic approach for drug discovery: In silico screening of potential antimalarial compounds. In Chemoinformatics and Bioinformatics in the Pharmaceutical Sciences (eds Sharma, N., Ojha, H., Raghav, P. K. & Goyal, K.) 207–243 (Academic, 2021). https://doi.org/10.1016/B978-0-12-821748-1.00005-1
Omran, B. A., Tseng, B. S. & Baek, K. H. Nanocomposites against Pseudomonas aeruginosa biofilms: Recent advances, challenges, and future prospects. Microbiol. Res. 282, 127656 (2024).
Fan, Y. et al. Bacillus cereus is a key microbial determinant of intractable otitis media with effusion. Commun. Med. 5, 150 (2025).
Parmanik, A. et al. Current treatment strategies against multidrug-resistant bacteria: A review. Curr. Microbiol. 79, 388 (2022).
Alam, R. et al. GC-MS analysis of phytoconstituents from Ruellia prostrata and Senna Tora and identification of potential anti-viral activity against SARS-CoV-2. RSC Adv. 11, 40120–40135 (2021).
Talukder, M. E. K. et al. Cheminformatics-based identification of phosphorylated RET tyrosine kinase inhibitors for human cancer. Front Chem 12, (2024).
Usha, V., Thangaraj, V. & Thirunarayanan & G. Antimicrobial potent (2E)-3-Chloro-4-Nitro-Phenylchalcones. Indian J. Chem. 56 (2017).
Jayawardena, T. U. et al. Sargassum Horneri and isolated 6-hydroxy-4,4,7a-trimethyl-5,6,7,7a-tetrahydrobenzofuran-2(4H)-one (HTT); LPS-induced inflammation Attenuation via suppressing NF-κB, MAPK, and oxidative stress through Nrf2/HO-1 pathways in RAW 264.7 macrophages. Algal Res. 40 (2019).
Dawra, M. et al. Biological activities and chemical characterization of the Lebanese endemic plant Origanum ehrenbergii Boiss. Flavour. Fragr. J. 36, 339–351 (2021).
Akhter, S. et al. Ruellia prostrata Poir. Activity evaluated by phytoconstituents, antioxidant, anti-inflammatory, antibacterial activity, and in silico molecular functions. J. Saudi Chem. Soc. 26 (2022).
Acknowledgements
The authors are grateful to the Bangladesh Reference Institute for Chemical Measurements (BRICM) for their support in FT-IR and GC-MS analysis. We would like to thank and are grateful to the Laboratory of Computational Biology (BioSol Centre), Bangladesh, for instrumental support in conducting the dynamics simulation.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
Shahina Akhter: investigation, methodology, formal analysis, writing-original draft. Md. Enamul Kabir Talukder: investigation, data curation, methodology, formal analysis, writing-original draft, writing-review & editing. Md. Tarikul Islam: investigation, methodology, formal analysis, writing-original draft, writing-review & editing. Md. Baha Uddin: investigation, methodology, formal analysis. Nafis Fuad Shahir: investigation, methodology, formal analysis. Nazia Islam Rafi: investigation, methodology, formal analysis. Sadia Israt: investigation, methodology, formal analysis. Rahat Alam: resources, software, writing-review & editing. Mohamad Abu Hena Mostofa Jamal: resources, writing-review & editing. Md. Mashiar Rahman: conceptualization, resources, data curation, validation, supervision, funding acquisition, writing-review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Akhter, S., Talukder, M.E.K., Islam, M.T. et al. Uncovering the bactericidal potential of extract and multi-targeting phytochemicals from Mirabilis longiflora L. leaves against multidrug-resistant Pseudomonas aeruginosa and Bacillus cereus. Sci Rep (2026). https://doi.org/10.1038/s41598-026-40444-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-40444-3