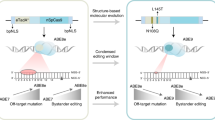
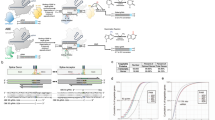
Abstract
Point mutations cause many genetic disorders, but modelling them in organisms is technically challenging. Creating mouse models that mimic these mutations is crucial for establishing a causal relationship between mutations and disease phenotype, thereby supporting the development of therapeutic strategies. Adenine base editors (ABEs) can correct single-nucleotide variants (SNVs) in disease modelling without double-stranded breaks (DSBs) or donor DNA, achieving higher product purity than traditional Cas9 methods. Earlier ABE techniques faced issues like limited targetability, bystander editing, and off-target effects. By combining two editor advancements, we introduced and tested ABE9-SpRY, an improved ABE variant fused with a PAM-flexible SpRY-Cas9 nickase. Our results show that ABE9-SpRY effectively generates three out of four targeted A-to-G mutations in mouse embryos, achieving desired editing efficiencies of up to 96% in individual adult founder mice. Furthermore, we observe fewer off-target events at predicted DNA sites in mouse embryos and in an orthogonal R-loop assay compared with ABE8e-SpRY. ABE9-SpRY also enhances product purity in mouse embryos under pooled sgRNA injections and, as a proof-of-concept, at a single endogenous locus in human induced pluripotent stem cells (hiPSCs), relative to ABE8e-SpRY. Our findings support ABE9-SpRY’s precision at the loci tested and PAM-flexible versatility. Although performance remains sequence-dependent, these data support ABE9-SpRY as a PAM-flexible tool for generating precise point-mutation models where bystander editing is a concern.
Similar content being viewed by others
Data availability
Oligonucleotide and sgRNA sequences used in this study are listed in Supplementary Tables 2, 3, 5, and 6. All raw NGS sequencing data generated in this study were in the NCBI Sequence Read Archive (SRA) database as a BioProject with the accession number PRJNA1291991. Plasmids encoding pCMV_ABE9-SpRY (/#242976) and pEF1α_ABE9-SpRY (#242977) are available at Addgene. Any other data and reagents will be made available upon reasonable request.
References
Williams, R. W. in Principles of Molecular Medicine (eds Runge, M. S. & Patterson, C.) 53–60 (Humana Press, 2006). https://doi.org/10.1007/978-1-59259-963-9_8
Perlman, R. L. Mouse models of human disease: An evolutionary perspective. Evol. Med. Public Health 2016, 170–176. https://doi.org/10.1093/emph/eow014 (2016).
Smith, P., DiLillo, D. J., Bournazos, S., Li, F. & Ravetch, J. V. Mouse model recapitulating human Fcγ receptor structural and functional diversity. Proc. Natl. Acad. Sci. U. S. A. 109, 6181–6186. https://doi.org/10.1073/pnas.1203954109 (2012).
Scherrer, G. et al. Knockin mice expressing fluorescent δ-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc. Natl. Acad. Sci. U. S. A. 103, 9691–9696. https://doi.org/10.1073/pnas.0603359103 (2006).
Lee, H. Genetically engineered mouse models for drug development and preclinical trials. Biomol. Ther. 22, 267–274. https://doi.org/10.4062/biomolther.2014.074 (2014).
Passier, R., Orlova, V. & Mummery, C. Complex tissue and disease modeling using hiPSCs. Cell Stem Cell 18, 309–321. https://doi.org/10.1016/j.stem.2016.02.011 (2016).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. https://doi.org/10.1126/science.1231143 (2013).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506. https://doi.org/10.1038/nrm.2017.48 (2017).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. https://doi.org/10.1038/nbt.2623 (2013).
Hwang, G.-H. et al. Large DNA deletions occur during DNA repair at 20-fold lower frequency for base editors and prime editors than for Cas9 nucleases. Nat. Biomed. Eng. 9, 79–92. https://doi.org/10.1038/s41551-024-01277-5 (2025).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771. https://doi.org/10.1038/nbt.4192 (2018).
Cullot, G. et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 10, 1136. https://doi.org/10.1038/s41467-019-09006-2 (2019).
Weisheit, I. et al. Detection of deleterious on-target effects after HDR-mediated CRISPR editing. Cell Rep. 31, 107689. https://doi.org/10.1016/j.celrep.2020.107689 (2020).
Leibowitz, M. L. et al. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53, 895–905. https://doi.org/10.1038/s41588-021-00838-7 (2021).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. https://doi.org/10.1038/nature24644 (2017).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. https://doi.org/10.1038/nature17946 (2016).
Rees, H. A. & Liu, D. R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. https://doi.org/10.1038/s41576-018-0059-1 (2018).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900. https://doi.org/10.1038/s41587-020-0491-6 (2020).
Wu, X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32, 670–676. https://doi.org/10.1038/nbt.2889 (2014).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. https://doi.org/10.1038/nbt.2647 (2013).
Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437. https://doi.org/10.1038/s41586-019-1161-z (2019).
Li, S., Liu, L., Sun, W., Zhou, X. & Zhou, H. A large-scale genome and transcriptome sequencing analysis reveals the mutation landscapes induced by high-activity adenine base editors in plants. Genome Biol. 23, 51. https://doi.org/10.1186/s13059-022-02618-w (2022).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891. https://doi.org/10.1038/s41587-020-0453-z (2020).
Chen, L. et al. Engineering a precise adenine base editor with minimal bystander editing. Nat. Chem. Biol. 19, 101–110. https://doi.org/10.1038/s41589-022-01163-8 (2023).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. https://doi.org/10.1126/science.aba8853 (2020).
Liao, J. et al. Therapeutic adenine base editing of human hematopoietic stem cells. Nat. Commun. 14, 207. https://doi.org/10.1038/s41467-022-35508-7 (2023).
Patel, S. & Kilpatrick, B. S. Two-pore channels and disease. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1865, 1678–1686. https://doi.org/10.1016/j.bbamcr.2018.05.004 (2018).
García-Rúa, V. et al. Increased expression of fatty-acid and calcium metabolism genes in failing human heart. PLoS ONE 7, e37505. https://doi.org/10.1371/journal.pone.0037505 (2012).
Deutsch, R., Kudrina, V., Freichel, M. & Grimm, C. Two-pore channel regulators—Who is in control?. Front. Physiol. https://doi.org/10.3389/fphys.2024.1534071 (2025).
Medert, R. et al. Genetic background influences expression and function of the cation channel TRPM4 in the mouse heart. Basic Res. Cardiol. 115, 70. https://doi.org/10.1007/s00395-020-00831-x (2020).
Stallmeyer, B. et al. Mutational spectrum in the Ca2+-activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum. Mutat. 33, 109–117. https://doi.org/10.1002/humu.21599 (2012).
Tu, T. et al. A precise and efficient adenine base editor. Mol. Ther. 30, 2933–2941. https://doi.org/10.1016/j.ymthe.2022.07.010 (2022).
Jeong, Y. K. et al. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 39, 1426–1433. https://doi.org/10.1038/s41587-021-00943-2 (2021).
Cao, X. et al. Engineering of near-PAMless adenine base editor with enhanced editing activity and reduced off-target. Mol. Ther. Nucleic Acids 28, 732–742. https://doi.org/10.1016/j.omtn.2022.04.032 (2022).
Zhang, Z. et al. Engineering an adenine base editor in human embryonic stem cells with minimal DNA and RNA off-target activities. Mol. Ther. Nucleic Acids 29, 502–510. https://doi.org/10.1016/j.omtn.2022.07.026 (2022).
Li, G. et al. A novel base editor SpRY-ABE8eF148A mediates efficient A-to-G base editing with a reduced off-target effect. Mol. Ther. Nucleic Acids 31, 78–87. https://doi.org/10.1016/j.omtn.2022.12.001 (2023).
Xiong, X. et al. Split complementation of base editors to minimize off-target edits. Nat. Plants 9, 1832–1847. https://doi.org/10.1038/s41477-023-01540-8 (2023).
Zeng, H. et al. A split and inducible adenine base editor for precise in vivo base editing. Nat. Commun. 14, 5573. https://doi.org/10.1038/s41467-023-41331-5 (2023).
Kim, Y.-h et al. Sniper2L is a high-fidelity Cas9 variant with high activity. Nat. Chem. Biol. 19, 972–980. https://doi.org/10.1038/s41589-023-01279-5 (2023).
Wang, Z. et al. Decreasing predictable DNA off-target effects and narrowing editing windows of adenine base editors by fusing human Rad18 protein variant. Int. J. Biol. Macromol. 253, 127418. https://doi.org/10.1016/j.ijbiomac.2023.127418 (2023).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. https://doi.org/10.1038/nature26155 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. https://doi.org/10.1038/nature14592 (2015).
Huang, T. P. et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631. https://doi.org/10.1038/s41587-019-0134-y (2019).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262. https://doi.org/10.1126/science.aas9129 (2018).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. https://doi.org/10.1038/nature14299 (2015).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481. https://doi.org/10.1038/s41587-020-0412-8 (2020).
Zhao, L. et al. PAM-flexible genome editing with an engineered chimeric Cas9. Nat. Commun. 14, 6175. https://doi.org/10.1038/s41467-023-41829-y (2023).
Li, J. et al. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants. Mol. Plant 14, 352–360. https://doi.org/10.1016/j.molp.2020.12.017 (2021).
Villiger, L. et al. In vivo cytidine base editing of hepatocytes without detectable off-target mutations in RNA and DNA. Nat. Biomed. Eng. 5, 179–189. https://doi.org/10.1038/s41551-020-00671-z (2021).
Liu, Z. et al. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat. Commun. 9, 2338. https://doi.org/10.1038/s41467-018-04768-7 (2018).
Ryu, S.-M. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539. https://doi.org/10.1038/nbt.4148 (2018).
Yang, L. et al. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell 9, 814–819. https://doi.org/10.1007/s13238-018-0568-x (2018).
Fu, J. et al. Human cell based directed evolution of adenine base editors with improved efficiency. Nat. Commun. 12, 5897. https://doi.org/10.1038/s41467-021-26211-0 (2021).
Qian, Y. et al. A new compact adenine base editor generated through deletion of HNH and REC2 domain of SpCas9. BMC Biol. 21, 155. https://doi.org/10.1186/s12915-023-01644-9 (2023).
Zhao, D. et al. Engineered domain-inlaid Nme2Cas9 adenine base editors with increased on-target DNA editing and targeting scope. BMC Biol. 21, 250. https://doi.org/10.1186/s12915-023-01754-4 (2023).
Cornean, A. et al. Precise in vivo functional analysis of DNA variants with base editing using ACEofBASEs target prediction. Elife 11, e72124. https://doi.org/10.7554/eLife.72124 (2022).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628. https://doi.org/10.1038/s41587-020-0414-6 (2020).
Joshi, J., Albers, C., Smole, N., Guo, S. & Smith, S. A. Human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) for modeling cardiac arrhythmias: Strengths, challenges and potential solutions. Front. Physiol. https://doi.org/10.3389/fphys.2024.1475152 (2024).
Cerneckis, J., Cai, H. & Shi, Y. Induced pluripotent stem cells (iPSCs): Molecular mechanisms of induction and applications. Signal Transduct. Target. Ther. 9, 112. https://doi.org/10.1038/s41392-024-01809-0 (2024).
Brookhouser, N. et al. A Cas9-mediated adenosine transient reporter enables enrichment of ABE-targeted cells. BMC Biol. 18, 193. https://doi.org/10.1186/s12915-020-00929-7 (2020).
Rosello, M. et al. Disease modeling by efficient genome editing using a near PAM-less base editor in vivo. Nat. Commun. 13, 3435. https://doi.org/10.1038/s41467-022-31172-z (2022).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292. https://doi.org/10.1126/science.aav9973 (2019).
Lee, S.-H. et al. Bystander editing by adenine base editors impairs vision restoration in a mouse model of Leber congenital amaurosis. Mol. Ther. Methods Clin. Dev. https://doi.org/10.1016/j.omtm.2025.101461 (2025).
Tuladhar, R. et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 10, 4056. https://doi.org/10.1038/s41467-019-12028-5 (2019).
Höijer, I. et al. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 13, 627. https://doi.org/10.1038/s41467-022-28244-5 (2022).
Perrotta, R. M. et al. Engineered base editors with reduced bystander editing through directed evolution. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02937-w (2025).
Xiao, Y.-L., Wu, Y. & Tang, W. An adenine base editor variant expands context compatibility. Nat. Biotechnol. 42, 1442–1453. https://doi.org/10.1038/s41587-023-01994-3 (2024).
Qin, W. et al. ABE-ultramax for high-efficiency biallelic adenine base editing in zebrafish. Nat. Commun. 15, 5613. https://doi.org/10.1038/s41467-024-49943-1 (2024).
Zhao, N. et al. Evolved cytidine and adenine base editors with high precision and minimized off-target activity by a continuous directed evolution system in mammalian cells. Nat. Commun. 15, 8140. https://doi.org/10.1038/s41467-024-52483-3 (2024).
Chen, Q. et al. Engineering of peptide-inserted base editors with enhanced accuracy and security. Small 21, 2411583. https://doi.org/10.1002/smll.202411583 (2025).
Liao, J. et al. Sequential amino acid mutagenesis-driven de novo evolution of adenine deaminases enables efficient in vivo base editing in primate. bioRxiv. 2025.2005.2014.653640 (2025). https://doi.org/10.1101/2025.05.14.653640
Silverstein, R. A. et al. Custom CRISPR–Cas9 PAM variants via scalable engineering and machine learning. Nature https://doi.org/10.1038/s41586-025-09021-y (2025).
Fiumara, M. et al. Genotoxic effects of base and prime editing in human hematopoietic stem cells. Nat. Biotechnol. 42, 877–891. https://doi.org/10.1038/s41587-023-01915-4 (2024).
Peña-Gutiérrez, I., Olalla-Sastre, B., Río, P. & Rodríguez-Madoz, J. R. Beyond precision: Evaluation of off-target clustered regularly interspaced short palindromic repeats/Cas9-mediated genome editing. Cytotherapy 27, 279–286. https://doi.org/10.1016/j.jcyt.2024.10.010 (2025).
Liang, P. et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 10, 67. https://doi.org/10.1038/s41467-018-07988-z (2019).
Kim, D., Kim, D.-e, Lee, G., Cho, S.-I. & Kim, J.-S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 37, 430–435. https://doi.org/10.1038/s41587-019-0050-1 (2019).
Yuan, K. et al. Selict-seq profiles genome-wide off-target effects in adenosine base editing. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaf281 (2025).
Turchiano, G. et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell 28, 1136-1147.e1135. https://doi.org/10.1016/j.stem.2021.02.002 (2021).
Shi, Y., Inoue, H., Wu, J. C. & Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 16, 115–130. https://doi.org/10.1038/nrd.2016.245 (2017).
Heinzelmann, E. et al. iPSC-derived and patient-derived organoids: Applications and challenges in scalability and reproducibility as pre-clinical models. Curr. Res. Toxicol. 7, 100197. https://doi.org/10.1016/j.crtox.2024.100197 (2024).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. https://doi.org/10.1038/s41586-019-1711-4 (2019).
Alves, C. R. R. et al. Optimization of base editors for the functional correction of SMN2 as a treatment for spinal muscular atrophy. Nat. Biomed. Eng. 8, 118–131. https://doi.org/10.1038/s41551-023-01132-z (2024).
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635-5652.e5629. https://doi.org/10.1016/j.cell.2021.09.018 (2021).
Medert, R. et al. Efficient single copy integration via homology-directed repair (scHDR) by 5′modification of large DNA donor fragments in mice. Nucleic Acids Res. 51, e14–e14. https://doi.org/10.1093/nar/gkac1150 (2022).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226. https://doi.org/10.1038/s41587-019-0032-3 (2019).
Doman, J. L., Sousa, A. A., Randolph, P. B., Chen, P. J. & Liu, D. R. Designing and executing prime editing experiments in mammalian cells. Nat. Protoc. 17, 2431–2468. https://doi.org/10.1038/s41596-022-00724-4 (2022).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686. https://doi.org/10.21105/joss.01686 (2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016). https://doi.org/10.18637/jss.v035.b01.
Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots (2022). https://doi.org/10.32614/CRAN.package.ggpubr
Wilke, C. O. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. (2024). https://doi.org/10.32614/CRAN.package.cowplot
Acknowledgements
Sayari Bhunia is a member of HBIGS, the Heidelberg Biosciences International Graduate School. We thank the entire Freichel lab for their constructive feedback on the manuscript. We thank Maren Schneider for help with cloning pEF1α_ABE9-SpRY and XMAS-TREE sgRNAs. We thank the whole team from the Interfakultäre Biomedizinische Forschungseinrichtung (IBF) at Heidelberg University for their expert technical assistance. We thank Angela Wirth for writing the animal proposal. While preparing this work, the authors used AI-assisted technologies (Grammarly and ChatGPT) to improve readability and language. We acknowledge financial support from the Heidelberg University Publikationsfonds for the open-access publication fee.
Funding
Open Access funding enabled and organized by Projekt DEAL. This research was funded by the German Research foundation (DFG) through the Collaborative Research Centres CRC1550 (FKZ 464424253, projects B09 and S01), CRC1328 (335447717, project A21), DFG project FR 1638/5-1 (531047917), the DZHK (German Centre for Cardiovascular Research) and the BMBF (German Ministry of Education and Research); and The Health + Life Science Alliance Heidelberg Mannheim.
Author information
Authors and Affiliations
Contributions
J.K.O. and A.C. designed the study and developed the methodology. J.K.O. and A.C. curated the data and performed the formal analysis. J.K.O., S.B., B.H., V.K., S.D. and F.Z. performed the investigation. J.K.O., S.B. and A.C. conducted validation. J.K.O. and A.C. visualised the data. J.K.O. and A.C. wrote the original draft of the manuscript. J.K.O., S.B., V.K., M.F. and A.C. reviewed and edited the manuscript. M.F. acquired funding and provided resources. A.C. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
Alex Cornean is a co-founder and shareholder of DataHarmony Ltd, a company that provides gene editing-related services and software solutions.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ong, J.K., Bhunia, S., Hilbert, B. et al. ABE9 fused to SpRY Cas9 nickase enables precise generation of bystander free mouse models. Sci Rep (2026). https://doi.org/10.1038/s41598-026-40642-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-40642-z