Abstract
Colorectal cancer (CRC) exhibits significant genetic and epigenetic diversity, evolving into sub-clonal populations with varied metastatic potentials and treatment responses. Predicting metastatic disease in CRC patients remains challenging, underscoring the need for reliable biomarkers. While most research on therapeutic targets and biomarkers has focused on proteins, non-coding RNAs such as long non-coding RNAs (lncRNAs) comprise most of the transcriptome and demonstrate superior tissue- and cancer-specific expression. We utilised spatial transcriptomics to investigate lncRNAs in CRC tumours, offering more precise cell-type-specific expression data compared to bulk RNA sequencing. Our analysis identified 301 lncRNAs linked to malignant CRC regions, which we validated with public data. Further validation using RNA–FISH revealed three lncRNAs (LINC01978, PLAC4, and LINC01303) that are detectable in stage II tumours but not in normal epithelium and are upregulated in metastatic tissues. These lncRNAs hold potential as biomarkers for early risk assessment of metastatic disease.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) was the second most common cause of cancer death in the United States in 2022 and made up 10% of all cancer deaths globally1,2. In 2023, there were an estimated 153,000 new CRC cases in the United States alone3, making clinical developments a priority for this disease.
CRC can be divided into three major clinical subtypes based on their genomic or epigenomic status. These are microsatellite instability (MSI) status, chromosomal instability (CIN) status and CpG island methylator phenotype (CIMP)4. A tumour with microsatellite stable (MSS) lesions indicates that microsatellites remain stable and do not vary in length5. Tumours with APC gene mutations (70–90%) are usually MSS6. Tumours with MSI refer to microsatellite sequences that vary in length and have been functionally associated with defects in the DNA mismatch repair machinery7, resulting in a hypermutable phenotype5. The majority of CRC lesions are MSS and develop through CIN pathways, where whole or parts of chromosomes are duplicated or deleted8,9. Finally, CRC tumours can develop following hypermethylation of CpG islands within tumour suppressor genes, resulting in their loss of transcription and, thereby development of CRC10.
With the clinical subtypes described above, as well as more recently classified molecular subtypes11, tumour heterogeneity is a common factor impacting the progression and prognosis of CRC. In addition, inter-tumour variation (such as immune infiltration12) is commonly found between patients. Intra-tumour heterogeneity of cellular populations is also currently a barrier to curative treatment in CRC, as heterogeneous tumours are more likely to develop resistance to therapy or to result in disease recurrence13. The five-year survival rate for stage IV CRC patients is just 10.5%, based on a study of 26,170 patients in the USA Surveillance, Epidemiology, and End Results (SEER) cancer database14.
Therefore, it is vital that optimal preventative treatment is given to those patients most at risk of disease recurrence and metastasis while preventing over-treating patients not at risk. Currently, following screening and removal of polyps and cancers, a patient will receive adjuvant chemotherapy based on the staging of the tumour by a multidisciplinary team, including a pathologist and radiologist. Adjuvant chemotherapy is generally recommended in Stage III and IV patients following full surgical resection of disease (including from metastatic sites)15. It may also be given to stage II patients deemed by clinicians as high-risk (which has many contributing factors), but not to stage II patients with deficiencies in their mismatch repair machinery (associated with MSI), as they receive little benefit from it16,17,18. Overall, disease staging is currently the most important factor when deciding which patients will benefit from adjuvant chemotherapy, but for stage II disease, this can be ambiguous. Progression-free survival could be improved if a highly specific prognostic marker was identified for risk of disease recurrence or metastasis in stage II patients, where there is most benefit for preventative treatment. Combining a risk-specific biomarker with histopathologic disease staging would benefit clinical decision-making substantially, presenting a unique opportunity to reduce patient progression to stage IV disease, with the ultimate goal of reducing disease mortality.
Previous exploration of cancer therapeutics and biomarkers has largely been directed towards protein-coding genes. Since the discovery that the majority of the transcribed genome comprises non-coding RNAs (ncRNAs)19, interest in the identification of ncRNA therapeutic targets and prognostic markers has been growing20,21. The largest class of ncRNAs are long non-coding RNAs (lncRNAs), which are defined as transcripts of >500 nucleotides in length that do not comprise a significant open reading frame22. LncRNAs are primarily transcribed by RNA polymerase II, and so are spliced, capped and polyadenylated22,23, making a large proportion of them detectable using traditional polyadenylation-based mRNA sequencing methods.
LncRNAs have been shown to play important roles in numerous biological processes, including X-chromosome inactivation24, epigenetic regulation of gene expression25, and post-transcriptional regulation26. Dysregulation of lncRNA expression can result in disease, with lncRNAs characterised as drivers of a variety of cancers27,28,29,30,31, including CRC32,33,34,35,36,37,38,39,40. LncRNAs are highly suited as oncology targets due to their superior tissue- and cancer-specific expression compared to protein-coding genes41,42,43. They have the potential to serve as clinically relevant biomarkers to improve diagnosis, prediction of recurrence, therapeutic targeting, treatment response, and/or disease monitoring. An example of this is PCA3, a lncRNA biomarker which has been clinically implemented as a diagnostic test for prostate cancer44. PCA3 was found to be specifically elevated 34-fold in prostate cancer (but not benign prostate hyperplasia), a significant and measurable increase compared to normal prostate tissue45. The PCA3 test for prostate cancer was highly accurate in comparison to its protein counterpart, prostate-specific antigen46,47, indicating the potential of lncRNAs as clinically significant biomarkers. Other lncRNAs have also been identified that may be translated into diagnostic clinical cancer biomarkers in the future, with some prominent examples being H19 in gastric cancer48, HOTAIR in oral squamous cell carcinoma49, UCA1 in bladder cancer50 and HULC in hepatic metastasis of CRC51.
The potential for highly specific expression patterns, along with straightforward detection using PCR or FISH, make lncRNAs favourable for clinical implementation, as clear signals and accurate predictive qualities directly translate to increased certainty for clinicians. Historically, biomarkers and therapeutic targets have been identified using microarrays and bulk RNA sequencing (RNA-seq). However, these methods can result in a bias towards genes that are expressed at high levels on a population average, masking the expression of genes that have high expression in specific but small subpopulations52. With some notable exceptions (like MALAT153, NEAT154 and XIST55), lncRNAs are generally expressed at lower levels within the cell compared to protein-coding genes41,56 and in a highly specific fashion57, making their detection with bulk RNA-seq suboptimal. In addition, bulk RNA-seq methods often contain a mixture of cell types (including tumour, stroma and adjacent normal tissue), confounding discovery analysis. We used spatial transcriptomics to resolve these issues by allowing for the specific investigation of malignant epithelial populations at a near single-cell resolution, without losing the spatial architecture of the tumour and its surrounding microenvironment58. Spatial transcriptomics is emerging as a powerful tool to study cancers59. Because the spatial context of the tumour is retained, we were able to precisely identify and characterise regions of the tumour which will seed metastasis, guided by both the transcriptome and the histopathology of the tissue.
In this study, we used spatial transcriptomics to identify 301 lncRNAs specifically expressed in malignant regions of CRC patient tissues compared with adjacent normal colon epithelium. We cross-validated these malignancy-associated lncRNAs in publicly available datasets60,61, and prioritised LINC01978, PLAC4 and LINC01303 for further characterisation. We used the publicly available TCGA-COAD cohort61 to assess their correlation with patient survival and expression in disease subtypes and used hybridisation chain reaction (HCR) to amplify RNA fluorescent in situ hybridisation (FISH) signal to evaluate their expression in an additional cohort of patient tissues, including from stage II patients with matched metachronous second primary (“disease recurrence”), from stage IV patients with matched synchronous metastasis, and matched normal tissue from surgical resection specimens distant from the primary tumour. We found that LINC01978, PLAC4 and LINC01303 were associated with patient survival as a signature, and LINC01303 was upregulated in MSI and CIMP disease subtypes. Further, HCR–FISH confirmed all three lncRNAs had negligible expression in normal colon tissue, and confirmed they were detectable in stage II tumours, with elevated expression in metastatic tissues. This expression pattern suggests they may be useful markers for the development of metastatic disease, and provides a premise for future in-depth statistical evaluation of prognostic sensitivity and specificity in a larger cohort of patients.
Results
Spatial transcriptomics confirms the spatial architecture of cellular tissue components
We set out to profile lncRNA expression in malignant lesions of CRC tumours. We selected stage II (early stage) and stage IV (late stage) CRC patients with fresh frozen, treatment naïve primary tumours, matched metastasis (for stage IV patients) and matched normal tissue (resection ends) from the Dunedin Colorectal Cancer Cohort (DNCRC), a longitudinal biobanking cohort based in Dunedin, New Zealand. We prioritised two patients for characterisation using spatial transcriptomics. These samples were Patient 1 stage II tumour (P1T), Patient 2 stage IV tumour (P2T), and Patient 2 ovarian metastasis (P2M) (Fig. 1A). Additional samples (normal colon from resection ends (P1N1, P1N2) and replicate sections were taken (Supplementary Fig. 1A–E). We annotated general features of the tissue sections prior to collecting gene expression information (Fig. 1B). This was carried out as it would be routinely performed in the clinic, with pathologists (authors CRR and TOH) manually reviewing the H&E stained tissue sections. We observed areas with increased numbers of nuclei (red arrows), which correspond to malignant lesions within the tumours. Adjacent normal tissue (blue arrows) was also present in both tumours. Tissue necrosis was noted in P1T and P2M (yellow arrows), with desmoplastic stroma also observed in P2M (green arrow). No normal ovary tissue was identified in P2M.
A Overview of methodology for identification and characterisation of lncRNA markers with potential clinical relevance. B H&E staining of tumour sections used for Visium spatial gene expression profiling, with pathologist annotations. Left = P1T, Middle = P2T, Right = P2M. Red arrows denote areas with high nuclei density (malignant lesions). Blue arrows denote adjacent normal tissue. Yellow arrows denote necrosis. Green arrows denote desmoplastic stroma (dense, fibrous tissue with low cellularity). Scale bars = 1000 µm. C Unbiased clustering of spots for P1T, P2T and P2M using the FindClusters algorithm (Seurat v4), with assigned cell or tissue types based on transcriptomic profile. Missing spots = removed during the QC process. D Tumour purity analysis based on cell type clusters from C. Adjacent normal = “Colon epithelium” and “Colon epithelium in EMT”. Tumour = all other cell types. E Distribution of cell and tissue types in each tissue, calculated as the sum of spots associated with each cluster, per tissue. F Percentage of total spots associated with each tissue. Image created with Biorender.com.
Following standard quality control and normalisation procedures, our spatial gene expression profiles from CRC patient tumour tissue successfully stratified pathologist-annotated sub-structures within each tumour, with the Seurat v4 FindClusters algorithm classifying spots into clusters that represented the histological morphology of the tissue (Fig. 1C). FindClusters uses the individual gene expression profile and location of each spot to determine how similar they are to other spots. While deep sequencing (up to 300,000 read pairs per tissue-covered spot) resulted in finer resolution of cluster formation, even the minimum recommended sequence depth of 50,000 read pairs per tissue-covered spot was sufficient to identify a variety of different cellular populations (Fig. 1C, Supplementary Fig. 1F). Because CRC tumours contain densely packed cells, we observed expression signal from multiple cell types per spot. To overcome challenges presented by the resolution of the spatial platform, we used a broader annotation of cell and tissue types present in each of the clusters using GPT-4 as described by Hou and Ji62. For each cluster, we provided GPT-4 with a list of upregulated canonical and non-canonical markers and used it to predict which cell types were present in that region of the tissue sample. Because this method used both canonical and non-canonical markers present in the tissue (Supplementary Fig. 2G), we paired GPT-4 cell type predictions with a literature search to validate non-canonical markers. We predicted 9 cell and tissue types across the tissues, including malignant epithelium, necrotic tissue, cancer-associated fibroblasts (CAFs), mesenchymal stromal cells (MSCs), SAMHD1+ stromal cells, desmoplastic stroma, tumour leading edge, colon epithelium undergoing epithelial–mesenchymal transition (EMT) and adjacent normal colon epithelium (Fig. 1C). Tumour purity was evaluated by determining the percentage of spots not covered by adjacent normal colon epithelium (including colon epithelium undergoing EMT) for each sample (Fig. 1D). Each of the samples had at least 80% purity. We also investigated the distribution of cell and tissue types across each sample. Unsurprisingly, this differed based on the patient and tissue, with P1T being the most complex sample and P2M being the least complex (Fig. 1E). We also observed that the two primary tumours, P1T and P2T accounted for 50% of tissue-covered spots, while P2M accounted for the remaining 50% due to its large size (Fig. 1F). Importantly, we were able to precisely identify malignant epithelium within each of the patient samples for further characterisation.
Spatial transcriptomics detects spatially distinct expression patterns for mRNAs in patient tissue
We observed spatially distinct expression patterns for various mRNAs across the patient tissues (Fig. 2A, Supplementary Figs. 2–4). In P1T, we observed MMP1 expression specifically in the leading edge of the tumour (Fig. 2A, row 1). Matrix metalloproteinases (MMPs) are a family of endopeptidases, with MMP-1 acting as a collagenase63. MMP1 expression has been correlated with poor prognosis in CRC, and described as a marker for hematogenous metastasis64. We also observed SPP1 expression in stromal regions of P1T, with the majority expressed in predicted MSCs (Fig. 2A, row 1). SPP1 encodes osteopontin, a pro-inflammatory cytokine65. While it has been shown that SPP1 is expressed by tumour cells (as reviewed in ref.66), it can also be expressed in macrophages67. In CRC, SPP1+ macrophages have been associated with FAP+ fibroblasts in an interaction which may play a role in the formation of desmoplastic stroma, thereby increasing resistance to immune therapies60. Since SPP1 expression is localised outside of the malignant epithelium in P1T, it is possible that there is an immune hotspot in the stroma where SPP1+ macrophages are localised.
A Spatial expression patterns for CRC-associated mRNA transcripts. Top panels = P1T, middle panels = P2T, bottom panels = P2M. Scale = normalised, scaled expression values. B Spatial expression patterns for CRC-associated lncRNA transcripts. Top panels = P1T, middle panels = P2T, bottom panels = P2M. Scale = normalised, scaled expression values (P1T = 0–4. P2T, P2M = 0–2). C Gene expression metrics. Top panel = Percentage of tissue-covered spots expressing lncRNAs in P1T, P2T and P2M. Middle panels = Total genes and lncRNA genes captured per spot. Bottom panel = Mean normalised expression levels, stratified by cell type. Image created with Biorender.com.
We observed IGLC2 expression in specific parts of malignant epithelial regions in P1T and P2T (Fig. 2A, rows 1 and 2). IGLC2 encodes the light chain 2 component of immunoglobulins, which are antibodies secreted by B-cells. In normal intestinal mucosa, B-cells respond to dietary antigen challenges68. In CRC, tumour-infiltrating B-cells have been less studied, however, one study profiled the immune environment of CRC patient tumours using single-cell sequencing and found that B-cells expressing IGLC2 correlated with poor prognosis69. Overall, P1T showed higher expression of IGLC2, suggesting high numbers of tumour-infiltrating B-cells present in the tissue.
In P2T and P2M, we observed COL1A1 expression in stromal regions of the tumour (Fig. 2A, rows 2 and 3). COL1A1 encodes a major component of type I collagen and has been shown to promote metastasis in CRC70. While lower in P2T, expression of COL1A1 was very high in the desmoplastic stroma of P2M, as would be expected from tissue of this type. We also observed PIGR expression in the P2T region where normal colon epithelium was undergoing EMT (Fig. 2A, row 2). PIGR encodes the polymeric immunoglobulin receptor and has been shown to have a potential tumour-suppressive effect in CRC71. In our data, this would suggest a possible anti-tumour response in this region of the tissue.
In P2M, we observed expression of CKB and FABP1 in the malignant lesions of the tumour (Fig. 2A, row 3). CKB encodes creatine kinase B, a part of the creatine shuttle72. While the mechanism for CKB’s involvement in cancer is unclear, it is upregulated at metastatic foci, and its expression has been associated with high tumour grade72. This concurred with our own findings (Supplementary Fig. 3B). Meanwhile, FABP1, encoding fatty acid binding protein 1, has been shown to be a marker of enterocytic differentiation expressed in the absorptive cells of CRC patient-derived organoids73. To our knowledge, its role in metastasis has not been characterised.
Spatial transcriptomics detects spatially distinct expression patterns for lncRNAs in patient tissue
We also investigated the spatially distinct expression of several well-characterised lncRNAs in our spatial data (Fig. 2B, Supplementary Figs. 2–4). In all three tissues, the most highly expressed lncRNA was MALAT1. MALAT1 is a regulatory lncRNA which has demonstrated oncogenic involvement in multiple cancers, including CRC, as reviewed in refs. 74 and 75. In P1T, MALAT1 expression was localised outside of the B-cell infiltrated region of the tumour (designated by IGLC2 expression described above) (Fig. 2B, row 1). This expression pattern was marked in P1T only, with more ubiquitous, but lower expression overall observed in P2T and P2M (Fig. 2B, rows 2 and 3), suggesting a degree of patient specificity to this pattern.
Another lncRNA highly expressed in all three tissues was NEAT1, which was expressed in a similar pattern to MALAT1, but at slightly lower levels (Fig. 2B, rows 1–3). In CRC, upregulated expression of NEAT1 has been shown to activate the Wnt/β catenin signalling pathway, promoting the proliferation, migration and invasion of CRC cell lines in vitro and in vivo76. Conversely, the lncRNA CYTOR was upregulated in the B-cell infiltrated region of P1T, with low expression in other regions of the tumour (Fig. 2B, row 1). CYTOR has been shown to promote disease progression and metastasis in CRC via regulation of the NFkB signalling pathway77. Its localisation in P1T agrees with this finding, as NFkB signalling plays a major role in the regulation of inflammation (as reviewed in ref.78).
We also saw ZFAS1 expression in P2T and P2M, generally localised to malignant regions, with some expression in stromal areas, but not adjacent normal tissue (Fig. 2B, rows 2 and 3). ZFAS1 was also upregulated in P1T, but not clearly localised to any specific region (Supplementary Fig. 2C). ZFAS1 has previously been characterised as a major regulator of EMT and carcinogenesis in CRC37.
To broadly understand the distribution of lncRNA expression within the patient tissues, we investigated the percentage of spots where annotated lncRNAs were expressed (normalised counts value > 0). We extracted this expression information for all annotated lncRNAs using the GENCODE v41 lncRNA subset annotation file19. We found that in both P1T and P2T, approximately 50% of the tissue-covered spots captured annotated lncRNAs, while in P2M, only approximately 8% of tissue-covered spots captured lncRNAs (Fig. 2C, top panel). This is likely due to the large size of the tissue and the correspondingly high number of spots covered by desmoplastic stroma, which contains little to no gene expression information.
We found that the gene expression patterns for both total RNA and lncRNAs followed non-normal distributions, where the majority of spots captured expression between 100 and 1000 genes, with a long tail of spots which captured expression of higher numbers of genes (Fig. 2C, middle panels). We also observed different cell types within the tissues expressing genes at different mean levels (Fig. 2C, bottom panel) depending on their role in the tumour or tumour microenvironment.
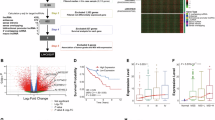
Next, we further focused on spots covering malignant epithelium to assess which lncRNAs were specifically upregulated in these regions of the tissue. To identify a list of potential biomarkers, we compiled a list of annotated lncRNAs expressed in the malignant epithelium and found 301 lncRNAs to be significantly upregulated compared to all other regions of the tissues (>1.5-fold upregulation, padj < 0.05) (Fig. 3A, Step 1). We will refer to this list as “Dunedin cohort lncRNAs” (DCLs).
A Prioritisation matrix of lncRNAs for further investigation. B Venn diagram of lncRNAs identified from Dunedin cohort malignant tissue (DCLs) (yellow), Shanghai cohort malignant cells (SCLs) (purple) and TCGA-COAD tumour tissue (green). C Principal component analysis (PCA) plot of variance within the TCGA-COAD bulk RNA-seq dataset (n = 473 tissues) and matched normal tissue (n = 41). D LncRNA candidate expression in bulk TCGA-COAD RNA-seq data (two-tailed, unpaired t-test, **** = p-value < 0.0001). E High expression (upper quartile) of the three lncRNAs of interest as a signature was significantly correlated with poorer overall survival compared to low expression (lower quartile, log-rank test). F LINC01303 is expressed at significantly higher levels in microsatellite instability (MSI) or CpG island methylator phenotype (CIMP) subtypes, compared to chromosomal instability (CIN), an invasive phenotype (INV), or normal colon epithelium (TCGA-COAD RNA-seq data stratified by clinical subtype, tested using an ordinary one-way ANOVA, with Tukey’s post-hoc test for multiple comparisons, *** = padj < 0.001, **** = padj < 0.0001). Image created with Biorender.com.
Novel lncRNAs are associated with CRC patient disease and survival
We cross-validated the identified DCLs against a publicly available dataset of single-cell information for five CRC patients and spatial information for four patients, from here on referred to as “Shanghai cohort lncRNAs” (SCLs)60. We extracted lncRNA expression information as described previously, using the GENCODE v41 lncRNA annotation file, selecting upregulated lncRNAs from cells and spots annotated as malignant (>1.5-fold upregulation, padj < 0.05). We detected 204 upregulated SCLs, of which 86 lncRNAs were common between the two cohorts (Fig. 3B).
To increase statistical power, we also analysed RNA-seq data from the TCGA-COAD cohort61. We performed differential expression analysis of genes upregulated in malignant (n = 473) vs. normal colon epithelium (n = 41) using DESeq2 (Fig. 3C), resulting in 1138 differentially upregulated lncRNAs (log2FC > 2, padj < 0.05). Of these, 34 lncRNAs were also identified in either DCLs or SCLs and seven lncRNAs common between all three datasets. In total, we identified 120 lncRNAs present in two or more analysed datasets (“Combined cohort lncRNAs”, CCLs), which we characterised further (Fig. 3A, Step 2).
We next investigated the expression of the CCLs in healthy tissues using the Genotype-Tissue Expression (GTEx) Project portal79, to prioritise lncRNAs with a cancer-specific signal. We eliminated lncRNAs exhibiting a median expression of ≥10 transcripts per million (TPM) in any healthy tissue type, narrowing down our candidate list to 26 lncRNAs (Fig. 3A, step 3).
We further filtered the CCLs by eliminating lncRNAs that overlapped with other ncRNA or mRNA genes to ensure specific detection by in situ hybridisation methods, narrowing down our dataset to 11 lncRNAs (Fig. 3A, step 4). Finally, we prioritised CCLs, which were previously uncharacterised in CRC to investigate further. This step resulted in three lncRNA candidates with potential novel clinical relevance in CRC (“Prioritised candidate lncRNAs”): ENSG00000262188.2 (LINC01978), ENSG00000280109.3 (PLAC4) and ENSG00000250548.7 (LINC01303) (Fig. 3A, step 5). All three lncRNAs were significantly upregulated in the TCGA-COAD dataset. LINC01978 and LINC01303 were also significantly upregulated in the DCLs, while PLAC4 was also significantly upregulated in the SCLs.
We performed further analysis on the TCGA-COAD cohort to more deeply characterise the expression patterns and correlation with patient survival for our top three lncRNAs. While their expression was low on a population level (as could be expected for lncRNA signal detected using bulk RNA-seq), there was significantly higher expression in the tumour tissue compared to normal colon tissue in all cases (Fig. 3D). In addition, high expression (= upper quartile) of all three lncRNAs as a signature was significantly correlated with poorer overall survival (Fig. 3E). We further stratified the cohort by MSI status and observed that high expression of the lncRNAs individually was correlated with survival. LINC01978 was significantly correlated with poorer 5-year survival in patients with MSS tumours (Supplementary Fig. 5). Conversely, high expression of PLAC4 was significantly correlated with improved 5-year survival in patients with MSS tumours, however, there was a trend towards poorer survival in MSI tumours (Supplementary Fig. 5). LINC01303 expression was not correlated with patient survival, however, when stratified by clinical subtype, it was expressed at significantly higher levels in MSI or CIMP tumours, compared to CIN tumours, those with an invasive phenotype (INV), or normal colon epithelium (Fig. 3F).
Finally, we used the TCGA-COAD cohort to investigate the prognostic efficacy of the three lncRNAs as a signature, compared to MSS status, which is known to correlate with a higher risk of recurrence80). We focused on early-stage disease (= TMN tumour grades T1 and T2), as these patients would most likely benefit from a prognostic marker for metastatic disease. In the absence of recurrence-free survival data, we evaluated the expression of the lncRNA genes to predict overall survival status. While small cohort size limited data interpretation, PLAC4 and LINC01303 individually, as well as our 3-lncRNA signature, all performed better than MSS status at predicting patient survival (Supplementary Fig. 6).
Prioritised candidate lncRNAs are detectable in early-stage disease
To investigate the potential of the prioritised candidate lncRNAs as predictors of metastatic disease, we further assessed their expression using HCR-FISH. First, to assess expression in a model system, we performed HCR-FISH in two commonly used epithelial CRC cell lines, HCT116 and HT29. We observed similar expression levels of LINC01978 and PLAC4 in both cell lines but saw that LINC01303 expression was higher in HCT116 cells compared to HT29 cells (Supplementary Figs. 7 and 8). This corresponds with our findings in TCGA-COAD data (Fig. 3F), as HCT116 cells are widely described as MSI positive (as reviewed in ref.81). PLAC4 expression appeared to localise to the outside edge of the nucleus in HT29 cells but not HCT116 cells, and we detected diffuse signal with some areas of more punctate expression in both cell lines for LINC01978.
To understand expression patterns in patient tissue, we selected a second cohort of stage II and stage IV CRC patients (n = 7) from the DNCRC, using formalin-fixed, paraffin-embedded (FFPE) samples from the normal colon, primary tumour (including metachronous second primaries), and matched metastasis tissues (n = 17) (Fig. 4A). Due to the highly specific nature of signal amplification achieved using HCR-FISH (Fig. 4B) we were able to observe a range of expression levels in patient tissues (Fig. 4C–E, Supplementary Fig. 9). Overall, as is expected for lncRNA expression, abundance was ~50 normalised spots per assessed tissue area, with a greater number of spots observed in some tissue areas. However, PLAC4 appeared to have very patient-specific expression with very high levels in some stage II tumours and metastatic tissues. This corresponds well to the TCGA-COAD data, where PLAC4 had a small group of patients with log2 fold change values of 5–10.
A Overview of patient cohort for HCR-FISH. Patient 1 = Sigmoid colon primary tumour (stage IIA at first diagnosis), metachronous second primary in ascending colon with associated synchronous metastasis to right lung, upper lobe. Patient 2 = Sigmoid colon primary tumour (stage IVa), synchronous metastasis to para-aortic lymph node. Patient 3 = Recto-sigmoid colon primary tumour (stage IIA at first diagnosis), metachronous second primary in descending colon with associated synchronous metastasis to right lung, middle lobe. Patient 4 = Rectal tumour (stage IIA), metachronous metastasis to the left lung, lower lobe. Patient 5 = Ascending colon primary tumour (stage IV), synchronous metastasis to retroperitoneum. Patient 6 = Mid-ascending colon primary tumour (stage IVb), synchronous metastasis to omentum. Patient 7 = Caecal primary tumour (stage IVa), synchronous metastasis to Liver segments 5 and 6. B Overview of amplification of FISH signal using HCR. C Representative HCR–FISH signal for LINC01978 in Patient 7 primary tumour (left panel = DAPI nuclear staining, left-middle panel = RNA–FISH signal, right-middle panel = MALAT1 positive control expression, right panel = merge). D Representative HCR–FISH signal for PLAC4 in Patient 2 omentum metastasis (left panel = DAPI nuclear staining, left-middle panel = RNA-FISH signal, right-middle panel = MALAT1 positive control expression, right panel = merge). E Representative HCR-FISH signal for LINC01303 in Patient 6 liver metastasis (left panel = DAPI nuclear staining, left-middle panel = RNA–FISH signal, right-middle panel = MALAT1 positive control expression, right panel = merge). F Quantification of HCR-FISH signal for LINC01978. G Quantification of HCR-FISH signal for PLAC4. H Quantification of HCR-FISH signal for LINC01303. Scale bar = 100 µm for all images. DAPI stain denotes nuclei. Met = Metastasis. Rec = Disease recurrence (second metachronous primary). Error bars = mean ± SEM. Tested using a Kruskal–Wallis test for non-parametric data, with Dunn’s post-hoc test for multiple comparisons. * = padj < 0.05. Image created with Biorender.com.
We observed LINC01978 to be expressed in the muscularis mucosae surrounding the crypts in patient 4’s stage IV caecal tumour (Fig. 4C), while PLAC4 exhibited more diffuse expression in the metastatic omental tissue of patient 2. LINC01303 was highly upregulated in some but not all cells of patient 6’s metastatic liver tissue. Across all patients and tissues that were analysed, we observed some variation in expression (particularly for PLAC4, as described above), but found that all three lncRNAs were detectable in both stage II and metastatic tissues, and showed negligible expression in normal colon tissues, confirming our findings from the transcriptomic studies described above (Fig. 4F-H). Stage II tumours had higher expression of LINC01978, PLAC4 and LINC01303 compared to stage IV and metachronous second primaries (“disease recurrence”), however, lncRNA expression was always highest in metastatic tissue.
In summary, we have outlined methods for specific detection of lncRNAs associated with malignant tissue using spatial transcriptomics. We have demonstrated the use of GPT-4 for solving challenges to cell-type deconvolution for spatial data from archival tissues. We identified 301 lncRNAs from our spatial transcriptomics data that were specifically upregulated in the malignant epithelium and cross-validated with a publicly available single cell and spatial dataset and the TCGA-COAD cohort. We prioritised 3 lncRNAs from a pool of 120, which were expressed in two or more of the datasets, and used HCR-FISH to show that LINC01978, PLAC4 and LINC01303 are observable in stage II disease and have high expression in metastatic tissues, indicating their potential as prognostic markers. This work establishes spatial transcriptomics as a powerful method to identify relevant cancer-associated lncRNAs and presents three lncRNAs which have the potential to be clinically relevant predictors of metastatic disease.
Discussion
In this paper we have demonstrated for the first time the potential of using spatial methods to identify lncRNAs with potential clinical relevance in CRC. We used the Visium spatial gene expression platform to characterise patient CRC tissues, and annotated tumours based on both histo-pathology and transcriptomic signature. We identified 301 lncRNAs specifically upregulated in malignant regions and cross-validated their expression in publicly available datasets. We further examined the expression patterns of four prioritised lncRNAs with specific expression in malignant tissue across a larger cohort of patient tissues using HCR–FISH.
Our spatial analysis showed good correspondence between pathologist-annotated regions of interest and transcriptome-characterised malignant regions. Use of spatial transcriptomics instead of single cell or bulk sequencing ensured that we were able to select specifically which tumour regions to characterise, and focus on lncRNA expression in malignant epithelium, while also being able to capture information about the tissue architecture as a whole.
A limitation of the spatial platform used here is the spot resolution currently available. We observed that in CRC tumours, there can be more cells per spot than the 1–10 cells per spot generally expected. However, using GPT-4 to provide a reference-free estimation of cell types beneath spots, we were able to reveal a layer of depth which is not delivered by histology alone. This tool was particularly useful in resolving challenges posed by the use of archival tissues, where the gradual loss of transcriptomic complexity over time may result in the loss of canonical markers. This would usually result in a loss of information if traditional cell-type classification methods were used or biases if manual classification was used. GPT-4 was able to predict cell types based on both canonical and non-canonical markers in an unbiased fashion. We acknowledge that despite the accuracy described by Hou and Ji62, the use of GPT-4 for cell type prediction is still in its infancy, and therefore highly recommend validating predictions against published literature.
We compared lncRNA expression lists between another single cell and spatial dataset and the TCGA-COAD cohort and found only seven lncRNAs common between all three datasets. While the single cell and spatial datasets showed reasonable comparability, there was less overlap with the TCGA-COAD cohort, and only seven lncRNAs common between all three datasets. While this is a very small percentage of the total TCGA-COAD differentially upregulated lncRNAs, this discrepancy may have arisen from differences in the chemistry of bulk sequencing compared to single and spatial methods or due to the much larger nature of the cohort. However, bulk RNA-seq served as a valuable additional validation of lncRNAs identified in single-cell and spatial data.
This study used patient tissues from a New Zealand CRC cohort. To increase the power of the study. we used expression data (including lncRNAs) from the TCGA-COAD cohort61 and a second single-cell and spatial CRC dataset60, from which we extracted SCLs. The TCGA-COAD cohort originates from the United States, while the SCLs dataset contains patient data of Chinese origin. Having datasets from multiple global regions increases the likelihood of the findings being generalisable to a larger cohort of patients, and reduces the likelihood of systematic error originating from use of a biased cohort, however does introduce the possibility of collecting ethnicity specific expression profiles. Another limitation to be aware of is the potential for confounding in the use of CRC samples. While still classified as a single instance of disease, colon and rectal cancers can have differences in molecular carcinogenesis and treatment, which may impact the generalisability of findings to one instance or another6,77.
Our HCR-FISH results showed that LINC01978, LINC01303 and PLAC4 were upregulated in metastatic tissue while also being detectable in early-stage disease. Because CRCs are highly heterogeneous, and this expression pattern is observable across three candidate lncRNAs and seven patients, we hypothesise that it may be of significance. Previous work has shown quantitative evidence that CRC tumours often seed early82. We hypothesise that these three lncRNAs may be involved in early seeding and metastatic disease establishment, which would explain their raised levels in stage II and metastatic tissues, but not stage IV tissues. Validation across a larger patient cohort, paired with mechanistic studies, would need to be conducted to confirm this hypothesis. In addition, further work is required to assess the sensitivity and specificity of these lncRNAs. This will determine whether they can be classified as clinically useful biomarkers for risk of disease progression. Our careful selection process for lncRNA candidates (using spatial transcriptomics to selectively characterise malignant epithelium) is likely to result in good statistical sensitivity metrics (the ability to detect a disease in patients in whom the disease is truly present), however, statistical specificity (the ability to rule out the disease in patients in whom the disease is truly absent) is equally as important83, and will need assessment. Expression of these candidate lncRNA markers should be analysed in stage II patient tissues that did not have disease recurrence or progression to assess whether the three lncRNAs are specific to increased risk of recurrence and progression.
LINC01978 has been identified in a list of differentially expressed, glycolysis-related lncRNAs in pancreatic ductal adenocarcinoma84. Reprogramming of energy metabolism occurs when cancer cells exchange aerobic metabolism for anaerobic, glycolytic metabolism, even in the presence of oxygen85. The possibility that LINC01978 plays a role in the glycolytic fuelling of the tumour fits with the survival data observed in the TCGA-COAD cohort, as the glycolytic fuelling process benefits the tumour as it grows and develops a hypoxic core (as reviewed in ref.86).
Prior to an update of Ensembl annotations in 2017 which classified it as a lncRNA, PLAC4 was described as a placenta-specific mRNA87. Cancer cells can often show embryonic-like plastic, multipotent phenotypes contributing to invasiveness and tumorigenicity (as reviewed in ref.88). Its high expression in placental tissue as well as CRC tumours suggests that PLAC4 may be involved in the de-differentiation and reprogramming of tumour cells.
LINC01303 has been described by several papers as acting as a sponge for microRNAs (miRNAs). However, there is conflicting information about which miRNAs it binds, and what effect this may have89,90,91. Further characterisation of the mechanism of action for LINC01303 is warranted.
In the landscape of current testing, lncRNA biomarkers have the distinct advantage of being easily implementable with current standard-of-care clinical tests. Due to their rapid and easy targeting with specific probes and primers, lncRNA biomarkers can be detected both in patient tissue concurrently with immunohistochemistry testing for mismatch repair (MMR) protein status, or during PCR testing concurrently with microsatellite instability tests. LINC01978, PLAC4 and LINC01303 are able to be specifically targeted and, due to their similar expression patterns across disease stages described above, could, therefore, be implemented as a panel for early triaging of metastatic disease. The benefit of this approach is that the lncRNA expression levels are traceable over time, as opposed to a binary MSI-status test result.
The future of testing for lncRNA biomarkers in plasma is also promising, with evidence that lncRNAs can be exported out of the cell into extracellular vesicles92. There is potential for their detection in patient plasma, as demonstrated in pancreatic adenocarcinoma by Yu et al.93. Future studies should investigate whether LINC01978, PLAC4 and LINC01303 are detectable in patient plasma, with their cytoplasmic localisation indicating that they could potentially be exported. In addition, in this study, we investigated lncRNAs expressed in malignant epithelium, and future studies will focus on identifying and characterising lncRNAs in the tumour microenvironment. Specifically, lncRNAs in tumour-associated immune cells might serve as promising new biomarkers and/or potential new drug targets and should be explored further in the context of immune-modulatory treatments such as checkpoint inhibitors94,95,96. Finally, lncRNA expression can be heterogeneous across subpopulations of malignant cells as the tumour evolves, therefore, a more in-depth characterisation of epithelial clonality using single cell sequencing could be performed.
Due to the current lack of predictive and prognostic markers for CRC, further investigation of LINC01978, LINC01303 and PLAC4 as potential biomarkers is justified in order to meet clinical need, and could have impact on outcomes for patients if further studies confirm their ability to predict risk of metastatic disease.
Methods
Sample Identification
Research on human samples in this study was performed in accordance with the Declaration of Helsinki. This study was approved by the New Zealand Health and Disability Ethics Committee (HEDC; reference number: 14/NTA/33/AM07, study title: Dunedin Colorectal Cohort). All participants provided informed consent.
Patients from the DNCRC with Stage II disease, or Stage IV disease with distant metastasis and matched normal colon were selected. Archival fresh-frozen tissue samples from two DNCRC patients were retrieved as described in Table 1 and embedded in Tissue-Tek O.C.T. Compound (Sakura, 4583).
Visium sample preparation, optimisation and imaging
In total, 10 µm sections were taken from OCT-embedded tissues using a cryostat set at −22 °C and placed on the Tissue Optimisation slide (10× Genomics, 1000193) following the manufacturer’s instructions. Tissue optimisation was performed, and the slide was imaged as described in the manufacturer’s instructions (Technical note CG000241). Briefly, imaging was performed on a Nikon Eclipse Ti2 microscope using the 10× magnification at 0.73 µm/pixel capture resolution, with a 200 ms exposure time. The TRITC filter cube was used with 75% Sola pad. A permeabilisation time of 12 min was selected. Next, 10 µm sections were taken from OCT-embedded tissues using a cryostat set at −22 °C and placed on the Visium Spatial Gene Expression slide (10× Genomics, 1000187). The tissue sections were stained using Mayer’s Hematoxylin—Lillie’s Modification (Agilent, S30930-2), and Eosin Y solution (Agilent, CS70130-2) following manufacturer’s instructions (User Guide CG000160). Each capture area was imaged at 20× magnification using Brightfield settings on a Nikon Eclipse Ti2 microscope, with a 2.18 µm/pixel capture resolution, an exposure time of 10 ms, and with the stitch function enabled. Images were annotated by CR and TH, and highly malignant regions were identified.
Visium spatial RNA-seq
Spatial gene expression libraries were constructed following the manufacturer’s instructions (User Guide CG000239). Briefly, the tissue was permeabilised for 12 min, followed by reverse transcription, second strand synthesis and denaturation steps to convert the RNA into cDNA. Next, the cDNA was amplified using PCR (Cq = 16 cycles). The number of cycles to amplify for was determined using qPCR, selecting the Cq value at 25% of peak fluorescence per manufacturer recommendations. Adaptors were ligated, and index IDs for each tissue were assigned using the Dual Index Kit TT Set A (10× Genomics, 1000215). Sequencing was performed on a NovaSeq 6000D× with an SP 200 flowcell following 10× instructions for read depth per library (50,000 read-pairs/spot). Raw data from 11,950 spots across two patients and eight tissue sections was processed using 10× Genomics SpaceRanger 1.2.2. following the Spatial GEX pipeline for fresh frozen tissue. Expression data was visualised primarily using 10× Genomics Loupe Browser 6.3.0, followed by an analysis of spatial expression patterns using Seurat 4.097. Briefly, low quality spots were filtered out (spots with a mitochondrial content of >20%, and spots with <200 reads, with the exception of P1N, which filtered out spots with <100 reads due to the lower complexity of the sample). Normalisation and scaling were performed using the SCTransform function, with regression of spots with a high mitochondrial content. Dimensionality reduction was performed, and the first 10 principal components were selected for all samples except P1N, for which the first 20 components were selected. These principal components were selected as they captured the majority of the variation within the data. Spatial clustering was performed on each tissue dataset individually using the FindNeighbours and FindClusters tools. Differentially expressed genes were identified for each cluster using the FindAllMarkers tool. Next, due to the number of cells per spot in our tissues, we used GPT-4 to predict multiple tissue or cell types per spot based on their gene expression profiles, as described by Hou and Ji62, and guided by pathologist annotations (by co-authors CR and TH). Briefly, we accessed GPT-4 through a premium subscription to OpenAI at chat.openai.com (now chatgpt.com). We used the prompt “From this list of markers only, identify possible cell types present in human colorectal tissue. Do not make predictions if the marker list is inconclusive.” In this way, we were able to select spot clusters containing malignant cells based on their transcriptome profile, paired with annotations of highly malignant regions as described above. The GENCODE v4198 lncRNA subset annotation file was used to identify lncRNAs which were specifically expressed in the malignant regions, and a list was compiled of upregulated lncRNAs specific to these clusters (>1.5 fold upregulation, p < 0.05). Where additional sample aggregation was performed, the SpaceRanger aggr pipeline was used, and the same analysis was performed on the aggregated data.
Analysis of publicly available data
Data from a Visium and single-cell RNA-seq experiment in CRC were analysed60. This dataset included transcriptomic information from a single-cell experiment (54,103 cells from 5 patients) and a spatial experiment (8705 spots from 4 patients). Significant differentially expressed genes from major and minor cell types were collected (log2FC > 2, padj < 0.05). LncRNAs were identified from the “malignant cells” cluster (Provided as “Supplementary data 3, Supplementary data 4”60) using the GENCODE v41 lncRNA subset annotation file. This list is referred to as Shanghai cohort lncRNAs (SCLs). Next, bulk gene expression count data from the TCGA-COAD cohort (n = 473) and matched normal tissue (n = 41 patients) were downloaded using the TCGAbiolinks package99. Differential gene expression analysis between tumour and matched normal tissue was performed using DESeq2100, and significantly upregulated lncRNAs (log2FC > 2, padj < 0.05) were identified. Significantly upregulated lncRNAs in the SCLs and TCGA-COAD datasets were compared with the DCLs list and common lncRNAs identified (common cohort lncRNAs, CCLs). All CCLs were assessed for expression in healthy tissues using GTEx (version 8), and any with expression >10 transcripts per million (TPM) were excluded from further analysis. The genomic position was investigated, and those whose genomic position overlapped completely with other mRNA or ncRNA genes were excluded, leaving only lncRNAs which were specifically targetable.
Cell culture
HCT116 (ras-mutant epithelial colorectal adenocarcinoma) (ATCC: CCL-247) and HT29 (ATCC: HTB-38) (epithelial colorectal adenocarcinoma) cells were cultured as follows. Chambered slides were coated in 15 µg/mL Poly-l-Lysine (Sigma, P9404-25MG). Twenty-thousand cells were then seeded into each chamber and grown in Dulbecco’s Modified Eagle Medium (DMEM, Thermofisher, 11995073) with 10% foetal bovine serum (FBS, Thermofisher, 10091148), and 1% penicillin and streptomycin (Thermofisher, 15140122). Cells were grown to 80% confluency at 37 °C in 95% Oxygen, and 5% CO2 before fixing with 4% formaldehyde (Sigma, 252549-100 ML). Fixed cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) depleted of Mg or Ca ions (Thermofisher, 14190144) and then dehydrated using an ethanol gradient (20%, 30%, 70%). Fixed cells were stored in 70% ethanol at −20 °C until used for HCR–FISH.
FFPE tissues
FFPE blocks were obtained from the DNCRC for patients with either stage II disease at first diagnosis with a later metachronous second primary cancer or stage IV disease with distant metastasis (n = 7). All primary tumours were treatment-naïve. Samples included normal colon, primary tumour, second metachronous primary tumour and metastasis tissues (n = 17) as described in Table 2. Tissue preparation for HCR–FISH, including deparaffinisation and pre-hybridisation steps, was carried out as described in ref.101.
HCR–FISH assay
HCR–FISH was carried out as described in detail in ref.101. Briefly, split initiator probe sets for PLAC4, LINC01978, LINC01303, and MALAT1 were ordered from Molecular Instruments (Supplementary Table 1). Following deparaffinisation and pre-hybridisation steps, we prepared two probe pools. Pool one comprised 8 pmol each of the PLAC4 and MALAT1 probe sets. Pool two comprised 8 pmol each of LINC01978, LINC01303 and MALAT1 probe sets. Probes were hybridised for 12–18 h in a humidified chamber at 37 °C. To fluorescently amplify the probe signal, excess probes were washed off using probe wash buffer, and two pools of hairpin components were prepared and snap cooled, with the h1 components in pool one and the h2 components in pool two. In each pool, 0.6 pmol of each of the hairpin components conjugated to Alexa 488, Alexa 546 and Alexa 647 were used. The snap-cooled hairpin components were then pooled and added to the tissue in an amplification buffer and incubated for 12–18 h at room temperature (21 °C). On each slide, the MALAT1 signal was amplified using Alexa 647 conjugated hairpins. PLAC4 and LINC01978 signal was amplified (on separate slides) using Alexa 488 conjugated hairpins, while LINC01303 signal was amplified using Alexa 546 conjugated hairpins. Following washing steps to remove excess hairpins, glass coverslips were mounted using Fluoromount-G with DAPI (Thermofisher, 00-4959-52) to stain for DNA.
Imaging
Imaging was performed on an Andor Dragonfly spinning disk confocal attached to a Nikon Ti2 microscope. 10 µm z-stack images were taken at 60X magnification. DAPI signal was imaged at 405 nm, with an exposure time of 200 ms, and 2.5% laser intensity. PLAC4 and LINC01978 were imaged (on separate slides) at 488 nm with an exposure time of 200 ms and 5% laser intensity. LINC01303 was imaged at 561 nm with an exposure time of 200 ms and 5% laser intensity. MALAT1 was imaged at 637 nm with an exposure time of 50 ms and 2.5% laser intensity. All channels used a gain setting of 250 and a frame averaging of 2.
Image analysis
Image analysis was performed using the Nikon Elements General Analysis 3 software. A binary threshold was used to segment the DAPI signal in the 405 channel, and the nuclear area was recorded. Target lncRNA signal was detected in the 488 and 561 channels. We applied a binary threshold to manually segment and remove any autofluorescence observed in these channels and then performed 1 µm radium rolling ball averaging before using the FindBrightSpots tool to detect spots ≥2 µm in diameter. MALAT1 signal was detected in the 637 channel using the FindBrightSpots tool to detect spots ≥2 µm in diameter. Spots were normalised to the nuclear area and recorded as spots/10,000 µm2.
Data availability
The data generated in this study are not publicly available due to patient privacy requirements, but are available upon reasonable request from the corresponding author. The publicly available TCGA-COAD data analysed in this study were obtained from the TCGA data portal (https://portal.gdc.cancer.gov/projects/TCGA-COAD). The publicly available Visium and single cell data analysed in this study were obtained from https://doi.org/10.1038/s41467-022-29366-6, Supplementary Data 3.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. https://doi.org/10.3322/caac.21660 (2021).
Ferlay, J. et al. Global Cancer Observatory: Cancer Today (version 1.1). (International Agency for Research on Cancer, Lyon, France, 2024). https://gco.iarc.who.int/today [cited 19 Mar 2024].
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Ogino, S. & Goel, A. Molecular classification and correlates in colorectal cancer. J. Mol. Diagn. 10, 13–27 (2008).
Fearon, E. R. Molecular genetics of colorectal cancer. Annu Rev. Pathol. 6, 479–507 (2011).
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet 394, 1467–1480 (2019).
Parsons, R. et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75, 1227–1236 (1993).
Pino, M. S. & Chung, D. C. The chromosomal instability pathway in colon cancer. Gastroenterology 138, 2059–2072 (2010).
Bakhoum, S. F. & Cantley, L. C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 (2018).
Hinoue, T. et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 22, 271–282 (2012).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Berntsson, J., Nodin, B., Eberhard, J., Micke, P. & Jirström, K. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int. J. Cancer 139, 1129–1139 (2016).
Dagogo-Jack I. & Shaw A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. https://doi.org/10.1038/nrclinonc.2017.166 (2018).
Wang, J. et al. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: a population-based analysis. Cancer Med. 9, 361–373 (2020).
Chan, G. H. J. & Chee, C. E. Making sense of adjuvant chemotherapy in colorectal cancer. J. Gastrointest. Oncol. 10, 1183–1192 (2019).
Sargent, D. J. et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 28, 3219–3226 (2010).
Argilés, G. et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 31, 1291–1305 (2020).
National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology. Colon Cancer (Version 1.2024). https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf (2024). [cited 25 Mar 2024].
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Coan, M., Haefliger, S., Ounzain, S. & Johnson, R. Targeting and engineering long non-coding RNAs for cancer therapy. Nat. Rev. Genet. https://doi.org/10.1038/s41576-024-00693-2 (2024).
Arun, G., Diermeier, S. D. & Spector, D. L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 24, 257–277 (2018).
Mattick, J. S. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447 (2023).
Kung, J. T. Y., Colognori D. & Lee, J. T. Long noncoding RNAs: past, present, and future. Genetics https://doi.org/10.1534/genetics.112.146704 (2013).
Wutz, A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. https://doi.org/10.1038/nrg3035 (2011).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell https://doi.org/10.1016/j.cell.2007.05.022 (2007).
He, R.-Z., Luo, D.-X. & Mo, Y.-Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. https://doi.org/10.1016/j.gendis.2019.01.003 (2019).
Sun, M., Gadad, S. S., Kim, D.-S. & Lee Kraus, W. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol. Cell https://doi.org/10.1016/j.molcel.2015.06.023 (2015).
Diermeier, S. D. et al. Mammary tumor-associated RNAs impact tumor cell proliferation, invasion, and migration. Cell Rep. https://doi.org/10.1016/j.celrep.2016.08.081 (2016).
Li, Y., Egranov, S. D., Yang, L. & Lin, C. Molecular mechanisms of long noncoding RNAs‐mediated cancer metastasis. Genes Chromosomes Cancer https://doi.org/10.1002/gcc.22691 (2019).
Shin, V. Y. et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. https://doi.org/10.1038/s41419-019-1513-5 (2019).
Arun, G. et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51 (2016).
Ling, H. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. https://doi.org/10.1101/gr.152942.112 (2013).
Ren, J. et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 8, 3932–3948 (2018).
Bian, Z. et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 6, 23892 (2016).
Han, P. et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 16, 9 (2017).
Xiang, J.-F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014).
O’Brien, S. J. et al. Long non-coding RNA ZFAS1 is a major regulator of epithelial-mesenchymal transition through miR-200/ZEB1/E-cadherin, vimentin signaling in colon adenocarcinoma. Cell Death Discov. https://doi.org/10.1038/s41420-021-00427-x (2021).
Liang, W.-C. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525 (2015).
Xu, C., Yang, M., Tian, J., Wang, X. & Li, Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 39, 169–175 (2011).
Kogo, R. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
de Goede, O. M. et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 184, 2633–2648.e19 (2021).
Groskopf, J. et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 52, 1089–1095 (2006).
de Kok, J. B. et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 62, 2695–2698 (2002).
Marks, L. S. et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 69, 532–535 (2007).
Haese, A. et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur. Urol. 54, 1081–1088 (2008).
Zhou, X., Yin, C., Dang, Y., Ye, F. & Zhang, G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 5, 1–10 (2015).
Tang, H., Wu, Z., Zhang, J. & Su, B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 7, 761–766 (2013).
Wang, X.-S. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 12, 4851–4858 (2006).
Matouk, I. J. et al. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol. 21, 688–692 (2009).
Chang, H. H., Hemberg, M., Barahona, M., Ingber, D. E. & Huang, S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature 453, 544–547 (2008).
Ji, P. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Brown, C. J. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991).
Cabili, M. N. et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015).
Gloss, B. S. & Dinger, M. E. The specificity of long noncoding RNA expression. Biochim. Biophys. Acta 1859, 16–22 (2016).
Asp, M., Bergenstråhle, J. & Lundeberg, J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays 42, e1900221 (2020).
Lewis, S. M. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021).
Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP fibroblasts and SPP1 macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Hou, W. & Ji, Z. Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis. Nat. Methods https://doi.org/10.1038/s41592-024-02235-4 (2024).
Pardo, A. & Selman, M. MMP-1: the elder of the family. Int J. Biochem. Cell Biol. 37, 283–288 (2005).
Sunami, E. et al. MMP-1 is a prognostic marker for hematogenous metastasis of colorectal cancer. Oncologist 5, 108–114 (2000).
O’Regan, A. & Berman, J. S. Osteopontin: a key cytokine in cell-mediated and granulomatous inflammation. Int. J. Exp. Pathol. 81, 373–390 (2000).
Zhao, H. et al. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 9, 356 (2018).
Matsubara, E. et al. SPP1 Derived from macrophages is associated with a worse clinical course and chemo-resistance in lung adenocarcinoma. Cancers https://doi.org/10.3390/cancers14184374 (2022).
Spencer, J. & Sollid, L. M. The human intestinal B-cell response. Mucosal Immunol. 9, 1113–1124 (2016).
Wang, W. et al. Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer. Clin. Transl. Med. 11, e253 (2021).
Zhang, Z., Wang, Y., Zhang, J., Zhong, J. & Yang, R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol. Med. Rep. 17, 5037–5042 (2018).
Zhang, D. et al. Polymeric immunoglobulin receptor suppresses colorectal cancer through the AKT-FOXO3/4 axis by downregulating LAMB3 expression. Front. Oncol. 12, 924988 (2022).
Kita, M. et al. Role of creatine shuttle in colorectal cancer cells. Oncotarget 14, 485–501 (2023).
Della Chiara, G. et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat. Commun. 12, 2340 (2021).
Xu, W.-W., Jin, J., Wu, X.-Y., Ren, Q.-L. & Farzaneh, M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 22, 126 (2022).
Sun, Y. & Ma, L. New insights into long non-coding RNA in cancer and metastasis. Cancers https://doi.org/10.3390/cancers11020216 (2019).
Zhang, M. et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 11, 113 (2018).
Wang, X. et al. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol. Cancer 17, 110 (2018).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 1–9 (2017).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Gavin, P. G. et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin. Cancer Res. 18, 6531–6541 (2012).
Boland, C. R. & Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 138, 2073–2087.e3 (2010).
Hu, Z. et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51, 1113–1122 (2019).
Ray, P., Le Manach, Y., Riou, B. & Houle, T. T. Statistical evaluation of a biomarker. Anesthesiology 112, 1023–1040 (2010).
Zhu, L.-L. et al. Deciphering the genomic and lncRNA landscapes of aerobic glycolysis identifies potential therapeutic targets in pancreatic cancer. Int. J. Biol. Sci. 17, 107–118 (2021).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Ferreira, L. M. R., Hebrant, A. & Dumont, J. E. Metabolic reprogramming of the tumor. Oncogene 31, 3999–4011 (2012).
Tsui, N. B. Y. et al. Synergy of total PLAC4 RNA concentration and measurement of the RNA single-nucleotide polymorphism allelic ratio for the noninvasive prenatal detection of trisomy 21. Clin. Chem. 56, 73–81 (2010).
Hendrix, M. J. C. et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer 7, 246–255 (2007).
Cao, C. et al. LINC01303 functions as a competing endogenous RNA to regulate EZH2 expression by sponging miR-101-3p in gastric cancer. J. Cell Mol. Med. 23, 7342–7348 (2019).
Xiao, D., Cui, X., Fang, N., Yu, S. & Wang, X. LINC01303 promotes the proliferation and migration of laryngeal carcinoma by regulating miR-200c/TIMP2 axis. Am. J. Transl. Res. 13, 1643–1656 (2021).
Sun, B., Zheng, X., Ye, W., Zhao, P. & Ma, G. LncRNA LINC01303 promotes the progression of oral squamous cell carcinomas via the miR-429/ZEB1/EMT Axis. J. Oncol. 2021, 7974012 (2021).
O’Grady, T. et al. Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 20, 72 (2022).
Yu, S. et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 69, 540–550 (2020).
Li, K., Xie, T., Li, Y. & Huang, X. LncRNAs act as modulators of macrophages within the tumor microenvironment. Carcinogenesis 45, 363–377 (2024).
Park, E.-G., Pyo, S.-J., Cui, Y., Yoon, S.-H. & Nam, J.-W. Tumor immune microenvironment lncRNAs. Brief Bioinform. https://doi.org/10.1093/bib/bbab504 (2022).
Gao, Y. et al. Identifying immune checkpoint-related lncRNA biomarkers for immunotherapy response and prognosis in cancers. Sci. Data 10, 663 (2023).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Aken, B. L. et al. The Ensembl gene annotation system. Database https://doi.org/10.1093/database/baw093 (2016).
Colaprico, A. et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acheampong, K. K. et al. Subcellular detection of SARS-CoV-2 RNA in human tissue reveals distinct localization in alveolar type 2 pneumocytes and alveolar macrophages. MBio 13, e0375121 (2022).
Acknowledgements
This study was funded by the Royal Society Te Apārangi (Rutherford Discovery Fellowship RDF-18-UOO-010 awarded to author SDD). The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this paper. The authors would like to acknowledge the work of DNCRC staff Sonya Fenton and Fran Munro for their assistance in cohort selection and sample coordination. We would also like to thank the patients who donated tissue to the DNCRC, making this work possible.
Author information
Authors and Affiliations
Contributions
H.R.P. contributed to project design, performed all computational and experimental work, interpreted the results and was a major contributor to writing the manuscript. C.R.R., T.O.H. and S.T.P. provided access to patient samples, performed histo-pathological analysis of the tissues, and contributed to study design. S.D.D. conceived the project, contributed to data interpretation, manuscript writing and editing, and secured funding. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
Author S.D.D. is a cofounder and consultant to, and holds shares in, Amaroq Therapeutics and RNAfold.AI, but declares no competing interests. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pinkney, H.R., Ross, C.R., Hodgson, T.O. et al. Discovery of prognostic lncRNAs in colorectal cancer using spatial transcriptomics. npj Precis. Onc. 8, 230 (2024). https://doi.org/10.1038/s41698-024-00728-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-024-00728-1