Abstract
Pancreatic ductal adenocarcinoma (PDAC) is notably resistant to conventional chemotherapy and radiation treatment. However, clinical trials indicate that carbon ion radiotherapy (CIRT) with concurrent gemcitabine is effective for unresectable locally advanced PDAC. This study aimed to identify patient characteristics predictive of CIRT response. We utilized a panel of human PDAC cell lines with diverse genetic profiles to determine their sensitivity to CIRT compared to γ-rays, assessing relative biological effectiveness (RBE) at 10% survival, which ranged from 1.96 to 3.04. Increased radiosensitivity was linked to impaired DNA double-strand break (DSB) repair, particularly in cell lines with deficiencies in the homologous recombination (HR) repair pathway and/or elevated genomic instability from replication stress. Furthermore, pretreatment with the HR inhibitor B02 significantly enhanced CIRT sensitivity in a radioresistant PDAC cell line when irradiated in the spread-out Bragg peak but not at the entry position of the beam. These findings suggest that PDAC tumors with HR pathway mutations or high replication stress are more likely to benefit from CIRT while minimizing normal tissue toxicity.
Similar content being viewed by others
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in both men and women in the United States (U.S.)1. The primary curative treatment is surgery for resectable disease, however, only 15–20% of PDAC patients are eligible for this treatment option due to the location of the tumor and disease stage2. Gemcitabine (GEM) is the only active single-agent chemotherapeutic that improves survival, however, combination chemotherapy with either folinic acid, fluorouracil, irinotecan, oxaliplatin (FOLFIRINOX), or albumin-bound paclitaxel (nab-PTX)/GEM resulted in greater survival impact for localized and metastatic disease PDAC3,4,5. Thus, these combination chemotherapies are now considered standard of care. Clinical studies have shown that a combination of chemotherapy and radiation can transition an unresectable PDAC to a resectable state in roughly 20% of cases. However, the role of radiotherapy in improving survival in localized disease remains to be demonstrated, likely due to inherent tumor radioresistance, lack of biological triage, and/or an insufficient total dose of ionizing radiation (IR)6. Despite improvements in treatment options, the 5-year overall survival remains low at just over 10%7. Thus, there is a great need for the development and validation of additional innovative, potent, and targeted/selective treatment approaches.
Heavy ion therapy, such as carbon ion radiotherapy (CIRT), could be a game changer for PDAC. Particle radiotherapy in general, and CIRT in particular encompass numerous physical and biological therapeutic advantages when compared to conventional radiotherapy and some chemotherapeutics. The physical advantages include the generation of a spread-out Bragg peak (SOBP) to focus the most damaging portion of a particle track inside the tumor, an enhanced dose distribution that more effectively spares nearby, at-risk structures, lateral beam focusing to improve field homogeneity, dose verification allowing for real-time treatment modification, an increased linear energy transfer (LET) that causes more damage to the tumor cells, and finally the capability to magnetically steer the ion beam leading to a more precise dose delivery8,9. The biological advantages of CIRT include a higher relative biological effectiveness (RBE) damaging cells more effectively per unit of physical dose as compared to photons or protons, a reduced dependency on molecular oxygen, which results in a lower oxygen enhancement ratio (OER) thus creating more damage in a hypoxic tumor, and the generation of complex DNA damage which leads to more persistent stress and cell death8,9,10. Furthermore, a carbon ion beam offers an ideal energy distribution that induces a maximum ionizing effect at the site of the tumor and less damage to the surrounding normal tissue, leading to the prediction that CIRT will result in better tumor control with fewer side effects than conventional radiotherapy.
Clinical data has been encouraging; as a Phase I dose escalation study in unresectable PDAC by the Japanese Working Group for Pancreatic Cancer showed a 2-year survival rate of 48% for patients treated with 45.6–55.2 GyE (Gray equivalent) and concurrent gemcitabine11. A follow-up single-institution study demonstrated a 53% 2-year survival for patients that received 55.2 GyE12. Local control in this latter group was 82%, suggesting that CIRT is improving local control of later-stage disease that is not observed with conventional or hypofractionated X-ray radiotherapy6,12,13.
Due to the concentrated energy deposition within a confined volume, carbon ions cause DNA damage of greater complexity. A special feature of this densely ionizing radiation is the induction of clustered DNA lesions, which is defined as two or more DNA lesions, such as DNA double-strand breaks (DSBs), single-strand breaks, or base damage, within one or two helical turns of the DNA14,15. Multiple studies have revealed that as LET increases, DNA repair slows as complex DNA lesions are more difficult to repair16,17,18. In response to clustered DNA damage, cells activate multiple DNA damage response (DDR) pathways. The most toxic of the CIRT-induced DNA lesions are DSBs, which if left unrepaired or misrepaired can result in genomic instability or cell death by a number of mechanisms including apoptosis, mitotic catastrophe, or senescence. DSBs are repaired by four pathways: homologous recombination (HR), non-homologous end joining (NHEJ), alternative end joining (alt-EJ), and single-strand annealing (SSA)19. Multiple studies have addressed the different contributions of the NHEJ and HR pathways to DSB repair according to the complexity of the DSBs generated, with HR being more important for the repair of high-LET than low-LET generated DNA lesions20,21,22. However, the contributions of the NHEJ and HR pathways to the repair of clinical carbon ion beam-induced DSBs have not been clarified in human PDAC cell lines.
Sequencing and chromosomal copy number variation analyses of PDAC revealed a complex genomic landscape23,24. Activating mutations of KRAS are near ubiquitous and inactivation of TP53, SMAD4, and CDKN2A occurs at rates of >50%. Genomic classification via patterns of variation in chromosome structure identified four subtypes of PDAC that were termed (i) stable, (ii) locally arranged, (iii) scattered, and (iv) unstable24. The unstable subtype accounts for 15–20% of human PDACs and the majority of these tumors harbor a mutation(s) in a gene required for the DDR, including BRCA1, BRCA2, PALB2, and ATM. Similarly, mutations in DDR genes are commonly found in inherited forms of PDAC25,26. DDR deficiency renders some tumors preferentially sensitive to DNA-damaging agents such as platinum agents and PARP inhibitors27,28,29. Additionally, DDR deficiency has been observed to be predictive of FOLFIRINOX (platinum-containing regimen) efficacy in PDAC30. Unfortunately, FOLFIRINOX is suitable only for patients with good performance status given the drug combination’s toxicity. Moreover, the burden of morbidity, and even mortality, associated with platinum chemotherapy is a major challenge complicating the use of this chemotherapeutic regimen30. Therefore, it is hypothesized that a less toxic, tailored, and targeted therapy for PDACs with either somatic or germline mutations in DDR genes could be leveraged to improve treatment outcomes and the overall toxicity profile.
In this study, molecular mechanisms of CIRT were identified that impact the therapeutic response in PDAC cell lines. We investigated the contributions of the NHEJ and HR pathways to the repair of carbon ion-generated DNA lesions with the goal of identifying genomic features that can guide the treatment of pancreatic cancer and distinguish patients who would benefit the most from CIRT. We demonstrate that PDAC cell lines are more sensitive to CIRT than γ-rays and that this correlates with the burden of unrepaired DSBs. Furthermore, we found that the most sensitive cell lines are those with defects in the DDR. If we target a radioresistant PDAC cell line with the HR inhibitor, B02, there is an increased sensitivity to CIRT at the SOBP but not at the entry region of the depth-dose distribution in vitro, indicating an effect potentially driven by LET. Collectively, our data support the notion that PDAC patients or tumors with defects in DDR can benefit the greatest from CIRT. However, if a DDR defect is not identified then the HR pathway becomes a critical target to enhance response without complication of adverse normal tissue events.
Results
Differential response of PDAC cell lines to γ-ray and 12C ion irradiation
PDAC has been shown to be effectively targeted by CIRT clinically6,11,13; however, it is unknown whether genetics affect the response to this treatment modality. To assess the impact of genetic variability, we examined the radiation response of a panel of seven human tumor-derived PDAC cell lines with varied genetic backgrounds to both γ-rays and CIRT. Each cell line contained a KRAS G12 mutation and additional mutation or deletion events in at least two of the other three commonly mutated genes (TP53, CDKN2A, and SMAD4) found in PDAC (Supplementary Fig. 1A). Furthermore, we noted various mutations of known or unknown significance in DNA damage response (DDR) genes and other putative oncogenic mutations (Supplementary Fig. 1B). Clonogenic survival curves were generated using 137Cs for γ-ray (low LET) irradiation at UT Southwestern Medical Center (UTSW) in Dallas, TX and carbon ions (high LET) irradiation at the National Center for Oncological Radiotherapy (CNAO) in Pavia, Italy. As demonstrated in Fig. 1A, B, all cell lines were significantly more sensitive to carbon ions compared to γ-rays per unit physical dose (Gray/Gy), consistent with previous studies31,32. The surviving fraction at 2 Gy (SF2) to γ-rays ranged from 0.51 to 0.78 and at 3.5 Gy (SF3.5) from 0.20 to 0.51, indicating a broad range of inherent radioresponses. Alternatively, the SF2 values for carbon ions ranged from 0.09-0.23 and SF3.5 from 0.01 to 0.05, indicating much less cell-specific variability. Relative biological effectiveness (RBE) using 10% SF was estimated to be 1.96–3.04 and 2.38–3.53 when using the Mean Inactivation Dose (MID/\(\bar{D}\))33,34,35. The most radioresistant cell lines to CIRT were PANC.03.27 and PANC-1 and the most radiosensitive cell lines were CAPAN-1 and PANC.04.03 (Fig. 1A, B).
A Clonogenic survival assays were performed to compare the radiation sensitivities of seven PDAC cell lines. Cells were irradiated at the indicated doses of γ-rays (solid) or carbon ions (dashed) and plated for analysis of survival and colony-forming ability. B Survival fraction (SF) at 2 Gy and 3.5 Gy in response to γ-rays and carbon ions. Relative biological effectiveness (RBE) is calculated using multiple methods. RBESF10%, RBE calculated using 10% survival and RBEDAUC, RBE calculated using mean inactivation dose derived from Reimann sum. Statistical comparisons (p values) of clonogenic survival curves of cells treated with γ-rays vs γ-rays were conducted using a student’s t-test comparing the MID for each irradiation treatment. C Immunostaining of γH2AX foci in PANC03.27, MIA PaCA-2, and CAPAN-1 cells after exposure to 1 Gy of γ-rays (solid) or carbon ions (crosshatch). Cells were fixed at 0.5 h, 8 h, and 24 h after IR and immunostained for γH2AX foci. γH2AX foci were counted for each cell and averaged. Student’s t-test (two-sided) was performed to assess statistical significance (****p < 0.0001).
The radioresponse to γ-ray and CIRT correlated with unrepaired DNA double-strand breaks (DSBs), as γ-H2AX (surrogate marker from DSBs) focus resolution was attenuated in a higher proportion in response to 12C ions than γ-rays in the representative cell lines PANC.03.27, MIA-PaCa-2, and CAPAN-1 at 8- and 24-h post-IR (Fig. 1C). Moreover, the number of CIRT-induced γ-H2AX foci was significantly higher at 24 h in the known homologous recombination (HR)-defective cell line, CAPAN-136, compared to PANC.03.27 and MIA-PaCA-2 cells. The PANC.04.03 cell line had the highest RBE among the lines (Fig. 1A, B) likely due to its relative radioresistance to γ-rays, suggesting that this radioresistance was diminished when exposed to CIRT. Evaluation of the cBioPortal and COSMIC databases failed to identify a pathogenic mutation or deletion in a DDR gene in the PANC.04.03 cell line, indicating an unknown factor/phenotype is driving the increased sensitivity to CIRT. Clonogenic survival assays found that, unlike CAPAN-1, PANC.04.03 was not sensitive to treatment with the PARP inhibitor (PARPi) olaparib, indicating that this cell line is likely not HR-deficient (Fig. 2A)36. High γH2AX focus formation in G1 cells without exogenous stress indicates increased intrinsic replication stress37; therefore, we assessed if this was increased in PANC.04.03 cells. We found that PANC.04.03 has increased γH2AX foci in G1 cells without exogenous stress, suggesting that this cell line has elevated replication stress (Fig. 2B). Moreover, increased DNA damage in the PANC.04.03 cell line in the absence of treatment with an exogenous DNA damaging agent revealed that DNA-PKcs is autophosphorylated at serine 2056 and KAP1 and CHK2 are phosphorylated at serine 824 and threonine 68, respectively, in untreated cells (Fig. 2C, compare UT lanes). Finally, cells with increased replication stress are sensitive to ATR inhibition27. We found that PANC.04.03 cells are more sensitive to ATRi treatment than PANC.03.27 and MIA-PaCa-2, which further supports the notion that PANC.04.03 has elevated replication stress (Fig. 2D).
A Clonogenic survival assays were performed to compare the sensitivities of PANC.03.27, PANC.04.03, and CAPAN-1 to increase doses of the PARP inhibitor olaparib. B Immunostaining of γH2AX foci in PANC03.27, MIA-PaCA-2, and PANC.04.03 in G1 cells in the absence of exogenous DNA damage. γH2AX foci were counted for each cell and averaged. Student’s t-test (two-sided) was performed to assess statistical significance (****p < 0.0001). C PANC03.27, MIA-PaCA-2, and PANC.04.03 were irradiated using a dose of 0 (Untreated, UT) or 1 Gy carbon ions and then allowed to recover at times indicated in the figure. Phosphorylation of DNA-PKcs at S2056, KAP1 at S824, and CHK2 at T68 were assessed via immunoblotting. Immunoblotting for Ku70 was used as a loading control. D Clonogenic survival assays were performed to compare the sensitivities of PANC03.27, MIA-PaCA-2, and PANC.04.03 to increasing doses of the ATR inhibitor AZD6738.
Non-homologous end joining and homologous recombination mediate the repair of 12C ion-induced DSBs
The pathway or pathways integral to the repair of DSBs induced by therapy-relevant carbon ions in PDAC cell lines is still not clear. Thus, we aimed to determine if a specific DSB repair pathway is required for the repair of γ-ray and carbon ion-induced damage in PDAC cell lines. Specifically, we pretreated cells with either the DNA-PKcs inhibitor NU7441, RAD51 inhibitor B02, or ATR inhibitor AZD6738 to inhibit the NHEJ, HR, and ATR-CHK1 pathways, respectively. Clonogenic survival assays show that inhibiting the NHEJ pathway resulted in a marked sensitivity to γ-rays in the PANC.03.27 (Fig. 3A, B) and MIA-PaCa2 (Fig. 3D, E) cells. No significant increase in radiosensitivity to γ-rays was observed when cells were pretreated with the RAD51 and ATR inhibitors (Fig. 3B, E). This observation correlated with unrepaired DNA double-strand breaks (DSBs), as treatment with the DNA-PKcs inhibitor resulted in a significant increase in unrepaired DSBs as monitored by γH2AX foci resolution at 8- and 24-h post-IR (Fig. 3C, F). Next, we assessed clonogenic survival in response to CIRT with the three inhibitors. As shown in Fig. 3A, C, treatment with either the DNA-PKcs, RAD51, or ATR inhibitor resulted in increased radiosensitivity to 12C ions, with significant cell killing in response to pretreatment with the DNA-PKcs and RAD51 inhibitors (Fig. 3B, E). Similar to the γ-ray data, the increased radiosensitivity correlated with unrepaired DSBs (Fig. 3C, F). Moreover, we assessed if ATM inhibition affected the response to CIRT. We found that pretreatment with the ATM inhibitor KU55933 resulted in a significant radiosensitization in PANC.03.27, MIA-PaCa-2, and PANC.04.03 (Supplementary Fig. 3A–C). We also examined if alt-EJ played a role in response to CIRT-induced DNA damage by examining if pretreatment with an inhibitor to PARP (olaparib) affects radiosensitization to CIRT in the PANC.03.27 cell line. We observed that treatment with olaparib did not result in increased radiosensitization to carbon ions (Supplementary Fig. 4A). Moreover, we found that inhibition of the protein kinases CDK4/6, which are required for the transition from G1 to S phase, did not significantly affect the cellular response to CIRT (Supplementary Fig. 4B). Finally, sensitization enhancement ratios (SER) using the MID support significantly increased radiosensitization by DNA-PKi, RAD51i, and ATMi when cells are treated with CIRT compared to the DMSO control-treated cells (Fig. 3B, E and Supplementary Fig. 3)33,34,35. Together, the data show that multiple DDR proteins and pathways are required for the repair of carbon ion-generated DSBs, including NHEJ, HR, and ATM.
Clonogenic survival assays were performed to compare the radiation sensitivities of PANC03.27 (A) and MIA-PaCa-2 (D) cells in the presence of DNA damage response inhibitors. Cells were pretreated with DMSO (Control), 3 μM NU7441 (DNA-PKi), 20 μM B02 (RAD51i), or 100 nM AZD6738 (ATRi) and then irradiated at the indicated doses of γ-rays (solid) or carbon ions (dashed) and plated for analysis of survival and colony-forming ability. B, E Sensitization enhancement ratio (SER) was calculated using the mean inactivation dose (MID) for each clonogenic survival (γ-rays or carbon ions) and calculating the ratio of the control (DMSO) vs each inhibitor individually. Variance (VAR), standard deviation (StDEV), and p values as generated by a student’s t-test are also shown. Immunostaining of γH2AX foci in PANC03.27 (C) and MIA PaCA-2 (F) cells after pretreatment with DMSO (control), 3 μM NU7441 (DNA-PKi), 20 μM B02 (RAD51i), or 100 nM AZD6738 (ATRi), and then exposed to 1 Gy of γ-rays (solid) or carbon ions (crosshatch). Cells were fixed at 0.5 h, 8 h, and 24 h after IR and immunostained for γH2AX foci. γH2AX foci were counted for each cell and averaged. Student’s t-test (two-sided) was performed to assess statistical significance (****p < 0.0001).
Inhibition of HR is a more viable option as a radiosensitizer compared to NHEJ inhibition
Our data indicate that treatment with DNA-PKcs and RAD51 inhibitors mediates increased radiosensitivity to CIRT, suggesting that each of these could be used as tumor radiosensitizers. However, a common issue with radiosensitizers is that they not only sensitize the tumor, but also the normal tissue in the radiation field, which can result in adverse tissue events38. Given the lack of radiosensitization seen for γ-rays when combined with some of these inhibitors, the cellular response to these inhibitors was tested in the low LET entry region of the carbon ion beam to determine if there may be a LET-driven component to the cellular response of the combination of inhibitor and carbon ions. Another consideration to this examination of response based upon beam profile position is that the entry region of the beam is where the carbon ion beam would be expected to predominantly traverse normal tissue in a clinical setting. The LET at the entrance position (15 mm) correlates with a LET of 13.0–16.4 keV/µm (Fig. 4A). Survival assays found that the radiation response at the entrance position of the carbon ion beam was similar to that found for γ-rays, with the RBESF10% being 1.08 (Fig. 4B, C). And like the γ-ray data, pretreatment with either the RAD51 or the ATR inhibitor did not result in a significant increase in radiosensitivity as assessed by MID and SER at the entrance position of the carbon ion beam, whereas the addition of the DNA-PKcs inhibitor resulted in a significant increase in radiosensitivity (Fig. 4D).
A Schematic showing the placement of the cells at the entry and spread-out Bragg peak (SOBP). B Clonogenic survival assays were performed to compare the radiation sensitivities of PANC03.27 and MIA-PaCa-2 cells in the presence of DNA damage response inhibitors when exposed to carbon ions at the beam entry. Cells were pretreated with DMSO (Control), 3 μM NU7441 (DNA-PKi), 20 μM B02 (RAD51i), or 100 nM AZD6738 (ATRi) and then irradiated at the indicated doses of γ-rays (solid) or carbon ions (dashed) and plated for analysis of survival and colony-forming ability. C Survival fraction (SF) at 2 and 3.5 Gy in response to γ-rays and 12C ions. Relative biological effectiveness (RBE) is calculated using multiple methods. RBESF10%, RBE calculated using 10% survival and RBEDAUC, RBE calculated using mean inactivation dose derived from Reimann sum. D Sensitization enhancement ratio (SER) was calculated using the Mean Inactivation Dose (MID) for each clonogenic survival (carbon ions at the entry) and calculating the ratio of the control (DMSO) vs each inhibitor individually. Variance (VAR), standard deviation (StDEV), and p values as generated by a student’s t-test are also shown.
Treatment with carbon ions results in a differential expression of genes as compared to γ-rays
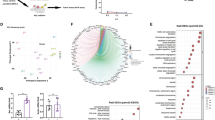
The transcriptomic response of PDAC cells with carbon ions compared to that of γ-rays has not been fully examined. To assess this, we treated five PDAC cell lines (MIA-PaCa-2, PANC.03.27, CAPAN-1, PANC.04.03, and PANC.02.03) with carbon ions or γ-rays and performed expression analysis 4-, 12-, and 24-h post-treatment against untreated cells at the same times post-irradiation. Per unit dose, transcriptome analysis found that CIRT induces a greater transcriptomic change in pancreatic cancer cell lines than γ-rays. This is demonstrated by the larger number of genes differentially expressed in response to CIRT (557 genes) compared to γ-rays (89 genes) (Fig. 5A) and greater variance in principal components for carbon-irradiated cells than the γ-ray-irradiated samples when both are compared against the unirradiated samples using supervised principal component analysis (PCA) (Fig. 5B). The signaling pathways most highly enriched with differentially expressed genes in response to radiation were similar between the carbon and γ-ray groups, as was their activation/suppression status. However, carbon particle irradiation induced the expression of several signaling pathways that were not identified in the γ-ray response based on z-score determination. These include the upregulated pathways, “Role of CHK Proteins in Cell Cycle Checkpoint Control”, “p53 Signaling”, “Role of BRCA1 in DNA Damage Response”, and “Ferroptosis Signaling Pathway”, and the downregulated pathway “SPINK1 General Cancer Pathway” (Fig. 5C). These pathways illustrate an increased DDR, with the activation of pathways that promote HR more prominent in the carbon ion irradiated cell lines per unit dose.
A Venn-diagram showing numbers of differentially expressed genes after γ-ray or carbon ion irradiations in comparison to sham-irradiated cell lines (FDR < 0.1). B Plot of cell line samples with the top 3 principal components after supervised PCA analysis using 745 differentially expressed genes. C Pathway analysis of differentially expressed genes in response to radiations showed significantly changed signaling pathways (FDR < 0.1 in at least one condition) with activation z-scores. A positive z-score indicated up-regulation and a negative z-score indicated down-regulation of the pathway.
Discussion
The proposed benefits from carbon ions in the treatment of malignant tumors in general, and PDAC in particular, is mostly motivated by the physical properties of high-LET radiation beams allowing for the delivery of a more conformal dose distribution to the target which limits the extent to which normal tissue is irradiated, as well as a greater biological effect per unit dose, which is described by the RBE. The RBE is strongly influenced by both physical and biological factors and the endpoint chosen. In this study, the RBE of carbon ions was primarily determined by in vitro clonogenic cell survival experiments where established cell lines were exposed to various doses of high LET carbon ions as expected in a typical Bragg peak, as well as the entry region of the carbon ion beam. Here, we observe that the RBE range is 1.96–3.04 (SF10%) and 2.38–3.53 (\(\bar{D}\)) for PDAC cell lines irradiated in the SOBP of the clinical beam at CNAO.
Currently, there is little opportunity to optimize CIRT or CIRT plus targeted agents for a given individual based on their known or determined radiosensitivity and/or targeted vulnerabilities as identified by genetics/genomics. We believe that the genetic makeup of a tumor plays a significant role, as defects in the DNA damage response impact the tumor response to CIRT. Our data shows that PDAC cell lines with defects in HR and increased replication stress are more sensitive to CIRT than those without. If possible, the genetics/genomics of the patient’s tumor should be taken into account when choosing between therapeutic options.
We hypothesized that there would be a difference in the DNA repair pathway(s) required for the response to clinical carbon ions in human PDAC cell lines when compared to γ-rays which are predominantly reliant upon the DNA-PKcs-dependent NHEJ pathway. For carbon ions, we observed the greatest reduction in DSB resolution was with the combination therapy of DNA-PKcs-dependent NHEJ pathway and RAD51 HR pathway inhibitors given the reduction in DNA DSB foci resolution and increased unresolved chromosomal aberrations. These results are similar to those obtained with NHEJ and HR-defective Chinese hamster ovary (CHO) cell lines21 and human skin fibroblasts treated with a DNA-PKi39. Furthermore, our results indicate that the ATM protein kinase is involved in the repair of DSBs induced by clinical carbon ion beams. In addition, we found that PARP inhibition does not affect cell killing by CIRT in our PDAC panel of cell lines. Previous work showed that combining CIRT with the PARPi olaparib enhanced radiosensitivity in the BRCA1-mutated triple-negative breast cancer cell line HCC1937 but had no effect on the BRCA wild-type cell line MDA-MB-23140. We postulate that PARP activity is not required for the response to CIRT-induced DNA damage unless the cell line is HR-defective.
Previous studies have implicated DNA polymerase Θ (POLQ) and SSA in promoting the repair of complex DNA damage after exposure to high LET radiations. POLQ knockout in the human cell line U2-OS was found to sensitize cells to both carbon ions and low LET X-rays41. In vitro data shows that POLQ efficiently anneals and extends substrates mimicking complex DSBs41. Loss of SSA significantly increased radiation sensitivity following X-ray or high LET alpha particle irradiation in CHO cells during the S phase of the cell cycle42. Further, it was found in CHO cells that increased DSB complexity correlates with increased structural chromosomal abnormalities (SCAs) and that this high level of SCAs is promoted by SSA or alt-EJ43. In both cases described above, knockout of either POLQ or use of an SSA-deficient derivative of CHO AA8 cells, sensitized cells to both low LET X-rays, as well as high LET carbon ions or α-particles suggesting that there is little benefit to the clinical use of inhibitors of these pathways as the differential response based upon position in the depth dose profile for carbon ions would be at best modest or non-existent like was observed here with the DNA-PKcs inhibitor. However, while we did not examine if SSA mediates the repair of carbon ion-induced DSBs in our panel of human PDAC cell lines, treatment with γ-rays in combination with the RAD52 inhibitor D-I03 did not result in increased radiation-induced cell killing in the HR-defective cell line CAPAN-1 with or without PARP inhibition, suggesting that SSA does not mediate repair of DSBs after γ-ray exposure in the absence of HR at least in this cell line (Supplementary Fig. 5). We believe that future experiments are required to fully assess the requirement of SSA and/or POLQ in the cellular response to carbon ions and if there may be an advantageous differential response to carbon ions based upon the position of cells in the carbon ion depth dose profile as was seen here with the HR inhibitor.
Finally, our data indicates that HR-defective cancer cells can be effectively targeted by CIRT. This is of importance as approximately 10% of PDAC tumors are HR-defective and approximately 20% have a defect in the DNA damage response24,27,44. Our preclinical data extends this to the use of inhibitors of the HR pathway to potentiate CIRT, where pretreatment with RAD51i, B02, results in increased CIRT-induced cell killing in vitro. A similar effect was seen for proton exposures of lung cancer cells defective in HR45, although the RBE difference was not as great as that seen with CIRT. Additionally, B02 treatment did not increase cell killing at the entry of the depth-dose distribution, implying that normal tissue will not be greatly affected by the combination of CIRT and the inhibition of HR. This statement is not without caveats. For example, for tumors that do not carry germline or somatic mutations in the HR pathway, this statement is applicable as there is an LET advantage to the response of normal tissues in the entry region of the beam. This positional advantage is also likely the case when a tumor bears somatic mutations in HR, that is, no HR mutations would be seen in the normal tissues. However, it is sometimes the case where DDR mutations are germline and the positional LET advantage may be limited or non-existent. This issue can be resolved by simple blood cell analysis for such germline mutations. Future studies should be directed to fully examine the response of normal cells with or without mutations in the HR pathway, as well as other DDR pathways, to understand the radioresponse based on beam profile position.
In conclusion, we evaluated CIRT on a cohort of PDAC cell lines and found that the cell lines with defects in HR and those that have increased replication stress are more sensitive to CIRT than γ-rays. Further, we elucidated the molecular mechanisms underlying the effects of CIRT on PDAC cell lines and found that NHEJ and HR, but not alt-EJ, are required for the repair of carbon ion-induced DSBs. Furthermore, we found that targeting a radioresistant PDAC cell line with the HR inhibitor B02 results in markedly increased sensitivity to CIRT at the SOBP but not as effectively at the entry of the depth-dose distribution in vitro. These results support the notion that further experiments should be performed that combine HR inhibitors with CIRT in vivo to further test this hypothesis, especially where no DDR defect has been identified. Lastly, while DNA-PKcs, ATM, and ATR inhibitors are in clinical trials, no HR inhibitor has yet entered the clinic or been approved by the FDA46. Our data suggest that there is a real need for the effective clinical application of an HR inhibitor with favorable properties.
Methods
Cell culture
The cell lines PANC02.03, PANC03.27, PANC04.03, PANC10.05, MIA-PaCa-2, PANC-1, and CAPAN-1 were purchased from American Type Culture Collection (ATCC). PANC02.03, PANC03.27, PANC04.03, and PANC10.05 were cultured in RPMI-1640 medium supplemented with 10 units/mL human recombinant insulin and 15% fetal bovine serum (FBS). MIA-PaCa-2 and PANC-1 were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. CAPAN-1 was cultured in Iscove’s Modified Dulbecco’s Medium supplemented with 20% FBS. The cells were grown in an atmosphere of 5% CO2 at 37 °C. To inhibit the activity of DNA-PKcs, ATR, Rad51 nucleofilament formation, ATM, PARP1, or CDK4/6, cells were incubated for 2 h before the experiment with 3 μM NU7441 (SelleckChem), 100 nM AZD6738 (SelleckChem), 20 μM B02, (SelleckChem), 3 μM KU55933 (SelleckChem), 1 μM olaparib (SelleckChem), or 1 μM palbociclib (SelleckChem), respectively. To inhibit PARP and RAD52 alone or in combination, cells were incubated for 2 h before the experiment with 10 nm olaparib (SelleckChem) or 15 μM D-I03 (SelleckChem), respectively.
Photon irradiations
Photon irradiation was conducted at the University of Texas Southwestern Medical Center (UTSW) using a J. L. Shepherd sealed horizontal 137Cs-sourced irradiator. Dosimetry for these sealed source irradiators is validated on an annual basis. Briefly, cells in culture were placed on a 360° platform revolving at 13 RPM, irradiated, removed from the irradiator, and immediately returned to the incubator. At the time of treatment, the irradiator had a dose rate of ~3.25 Gy/min.
12C ion irradiations
All 12C ion irradiations were performed at the Centro Nazionale di Adroterapia Oncologica (CNAO) facility in Pavia, Italy utilizing established methods47. Specifically, cells were irradiated in T12.5 cm flasks while immersed in a water bath at 37 °C using CNAO’s clinical pencil beam scanning 12C-ion beam. A spread-out Bragg peak (SOBP) was created to deliver a homogenous ( ± 2.5%) physical dose across the target volume. The beam quality has been previously characterized48 and adheres to the recommendations of an NCI special panel on particle beam characterization49. The dimensions of the SOBP were 17 cm in width, 7 cm in height, and 2 cm in depth (Supplementary Fig. 2B). Cells were centered in the SOBP using a leucite holder with the adherent cells aligned in their flasks back-to-back such that the depth of the cells in the upstream flask was 80.0 mm water equivalent depth (WED) while the position of cells in the downstream flask was 84.0 mm of WED (Supplementary Fig. 2A). LETs at the aligned positions were 74.1 keV/µm and 89.3 keV/µm, respectively at a typical dose rate of 0.60 Gy/min (no difference in biological response was observed based upon position.). The entrance LET was 13.0-16.4 keV/µm at a depth of 15 mm.
Clonogenic cell survival assays
Cells undergoing log phase growth at roughly 70–80% maximum cell culture density were trypsinized and then seeded into T-12.5 flasks at low density in complete growth medium 8 h prior to irradiation. If using an inhibitor, the regular media was changed and supplemented with the specific inhibitor and allowed to incubate for 2 h. Five minutes prior to irradiation with either γ-rays or carbon ions, cell culture flasks were filled to the neck with complete growth media containing 2% FBS with or without inhibitors. Cells were irradiated with doses of 1 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy of γ-rays, or 0.5 Gy, 1 Gy, 2 Gy, and 4 Gy, 5 Gy, and/or 6 Gy carbon ions. Following irradiation, growth medium containing 2% FBS was immediately aspirated and replaced with growth medium containing 10% FBS, and dishes were allowed to incubate for ~10 population doublings based on cell-specific doubling times. Media with 2% FBS was used during the irradiations to limit the overall volume of FBS needed to completely fill the T12.5 flasks for only a few minutes as was necessary at CNAO. Using 2% FBs for such a limited time had no effect on cell growth or radioresponse. Following incubation, the media was removed and the cultures were incubated in 100% ethanol solution containing 0.1% crystal violet in order to fix and stain the colonies. After drying, colonies were counted to determine the number of surviving cells following irradiation. Only colonies identified as having more than 50 cells per colony were scored as surviving, and the surviving fraction was determined by dividing the number of colonies by the product of the plating efficiency of the cell line multiplied by the number of cells seeded.
Survival curve fits
Survival curves were fitted based upon the repairable conditionally repairable (RCR) model as described in Eq. 1 where d is the dose per fraction and a, b, and c are parameters determined using a curve fitting algorithm50.
The γ-ray survival assays were performed at least twice for each cell line. If the coefficient of variation at 2 Gy was greater than 25%, they were repeated.
RBE calculations
Because all experiments were performed as single exposure irradiations the RBE values were calculated by using the simplest determination of RBE, i.e., by comparing a radiosensitivity value from 137Cs exposures (reference) to that same radiosensitivity value determined from 12C exposures (test) as in Eq. 2.
Radiosensitivity parameters included:
Dose at 10% survival
The dose at SF10% was calculated using values generated with the RCR model as described in Eq. 1.
\(\bar{D}\): \(\bar{D}\) was defined by Kelleher and Hug to represent cell survival across the entire dose response34,51. \(\bar{D}\) is calculated by \(\bar{D}={\int }_{0}^{\infty }D\,{\cdot}\,s\left(D\right){dD}\) where s(D) = −dS(D)/dD and S(D) is the survival function representing the fraction of cells surviving a radiation dose D. S(D) is the probability that a dose greater than D is required to kill a randomly selected cell. s(D) = −dS(D)/dD defines the corresponding probability density. \(\bar{{{D}}}\) can be represented by the area under the survival curve which in this case was calculated using a trapezoidal method of the area under the curve for each survival assay. \(\bar{D}\) is equal to D0 for exponential survival curves.
Determination of sensitization ratio (SER) using the mean inactivation dose (MID)
The sensitization enhancement ratio (SER) for each radiation type and inhibitor was calculated using a ratio of the mean inactivation dose (MID, Dparm) originally theorized to be a comprehensive parameter to accurately account for variations in clonogenic survival curve shapes determined by estimating the area under the curve (AUC) and recommended by the International Committee on Radiation Units and Measurements (ICRU) Report 3033,34,35.
Statistical comparison of radiation survival curves
Statistical comparisons of all radiation clonogenic survival curves were conducted using a two-tailed student’s t-test comparing the MID for each irradiation and treatment condition.
γH2AX foci kinetics
IR-induced γH2AX kinetics were determined as previously outlined with modifications52,53. Cells were grown on coverslips one day before the experiment and on the day of the experiment, cells were exposed to 1 Gy of γ-rays or 12C ion using the protocols outlined above. At 0 h, 0.5 h, 8 h, or 24 h time points after IR, cells were washed twice with ice-cold 1 × phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (in 1 × PBS) for 20 min at RT, washed 5 times with 1 × PBS, and incubated in 0.5% Triton X-100 on ice for 10 min. Cells were washed 5 times with 1 × PBS and incubated in a blocking solution (5% goat serum (Jackson Immuno Research) in 1 × PBS) overnight. The blocking solution was then replaced with the γH2AX (Cell Signaling Technology, 7631) and/or 53BP1 (ab175933, Abcam) primary antibody diluted in 5% goat serum in 1 × PBS and the cells were incubated for 2 h. Cells were then washed five times with wash buffer (1% BSA in 1 × PBS). Cells were incubated with the Alexa Fluor 488 (Molecular Probes) and/or anti-mouse IgG conjugated with Texas Red (Molecular Probes) (1:1000 dilution for both antibodies) secondary antibodies in 1% BSA, 2.5% goat serum in 1 × PBS for 1 h in the dark, followed by five washes. After the last wash, cells were mounted in a VectaShield mounting medium containing 4’6-diamidino-2-phenylindole (DAPI). The images were acquired using a Zeiss Axio Imager fluorescence microscope utilizing a 63× oil objective, ≥50 cells were analyzed for each time point.
Immunoblotting
Immunoblotting for protein expression and phosphorylation levels was performed as previously described54. The following antibodies were used in this study: antibodies from anti-DNA-PKcs phospho-S2056 (Abcam, ab124918), anti-Ku70 (Santa Cruz, sc-515736), anti-CHK2 phospho-T68 (Cell Signaling, 2661), and anti-KAP1 phospho-S824 (Bethyl Laboratories, A300-767A). Secondary antibodies anti-mouse IgG (HRP-linked) (7076) and anti-rabbit IgG (HRP-linked) (7074) were purchased from Cell Signaling.
Isolation of RNA from pancreatic cancer cell lines
Pancreatic cancer cell lines were irradiated with 1 Gy of γ-rays (UTSW) or carbon particles (CNAO). Total RNA was isolated at 4-, 12-, and 24-h post-irradiation. Total RNA from sham-irradiated cell lines was isolated at the same time as the irradiated samples. The sham-irradiated cells were also collected right before irradiation and used as baseline (0 h) references. RNA extractions were performed using miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Once isolated, RNA concentration was determined using a Nanodrop 2000 Spectrophotometer (Thermo Scientific) and quality was assessed using an Experion Electrophoresis system (Bio-Rad).
Transcriptomic analysis of PDAC cell lines following γ-ray and 12C ion irradiations
The sequencing library was prepared using the Illumina (San Diego, CA) Truseq Stranded Total RNA Prep Kit. Paired end next-generation sequencing was performed by DNAlink (San Diego, CA) using an Illumina Novaseq 6000 sequencer. Sequencing reads were trimmed to remove adapter sequences and were aligned with human genome GRCh38 using STAR with the 2-pass option. The aligned reads were quantified using the RSEM program. Gene counts were summarized using tximport55 R library and subsequently normalized with the TMM algorithm using edgeR. Differential expression analysis was performed using log2 counts per million (cpm) values and the limma package56. Genes with low read counts were removed by filterByExpr function within the edgeR package using default parameters57. Differential expression analysis was performed by fitting the log2 cpm values using the limma-trend approach described in the R limma package. Differentially expressed genes in irradiated cells at any of the 3 time points were identified by performing a moderated F-test using FDR < 0.1 as a statistical cutoff. Prior to principal component analysis (PCA), the cell line-specific variances were removed using the limma function and PCA was performed using the R prcomp function. Pathway analysis of differentially expressed genes was performed using the Ingenuity Pathway Analysis software (Qiagen).
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files). The RNA sequencing data can be accessed under BioProject accession number PRJNA1208067 via NCBI.
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Kolbeinsson, H. M., Chandana, S., Wright, G. P. & Chung, M. Pancreatic cancer: a review of current treatment and novel therapies. J. Invest Surg. 36, 2129884 (2023).
Conroy, T. et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825 (2011).
Von Hoff, D. D. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 369, 1691–1703 (2013).
Conroy, T. et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 379, 2395–2406 (2018).
Hammel, P. et al. Effect of chemoradiotherapy vs chemotherapy on survival in patients with locally advanced pancreatic cancer controlled after 4 months of gemcitabine with or without erlotinib: the LAP07 randomized clinical trial. JAMA 315, 1844–1853 (2016).
Ryan, D. P., Hong, T. S. & Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 371, 1039–1049 (2014).
Mohamad, O. et al. Carbon ion radiotherapy: a review of clinical experiences and preclinical research, with an emphasis on DNA damage/repair. Cancers https://doi.org/10.3390/cancers9060066 (2017).
Ahmad, R. et al. Particle beam radiobiology status and challenges: a PTCOG radiobiology subcommittee report. Int J. Part Ther. 13, 100626 (2024).
Durante, M., Cucinotta, F. A. & Loeffler, J. S. Editorial: charged particles in oncology. Front. Oncol. 7, 301 (2017).
Shinoto, M. et al. Carbon ion radiation therapy with concurrent gemcitabine for patients with locally advanced pancreatic cancer. Int J. Radiat. Oncol. Biol. Phys. 95, 498–504 (2016).
Shinoto, M. et al. A single institutional experience of combined carbon-ion radiotherapy and chemotherapy for unresectable locally advanced pancreatic cancer. Radiother. Oncol. 129, 333–339 (2018).
Zhu, X. et al. Patterns of local failure after stereotactic body radiation therapy and sequential chemotherapy as initial treatment for pancreatic cancer: implications of target volume design. Int. J. Radiat. Oncol. Biol. Phys. 104, 101–110 (2019).
Sutherland, B. M. et al. Clustered DNA damages induced by high and low LET radiation, including heavy ions. Phys. Med. 17, 202–204 (2001).
Hada, M. & Georgakilas, A. G. Formation of clustered DNA damage after high-LET irradiation: a review. J. Radiat. Res. 49, 203–210 (2008).
Sollazzo, A. et al. Live dynamics of 53BP1 foci following simultaneous induction of clustered and dispersed DNA damage in U2OS cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19020519 (2018).
Nikitaki, Z. et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 50, S64–S78 (2016).
Lorat, Y., Timm, S., Jakob, B., Taucher-Scholz, G. & Rube, C. E. Clustered double-strand breaks in heterochromatin perturb DNA repair after high linear energy transfer irradiation. Radiother. Oncol. 121, 154–161 (2016).
Sishc, B. J. & Davis, A. J. The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers https://doi.org/10.3390/cancers9070081 (2017).
Zafar, F., Seidler, S. B., Kronenberg, A., Schild, D. & Wiese, C. Homologous recombination contributes to the repair of DNA double-strand breaks induced by high-energy iron ions. Radiat. Res. 173, 27–39 (2010).
Gerelchuluun, A. et al. The major DNA repair pathway after both proton and carbon-ion radiation is NHEJ, but the HR pathway is more relevant in carbon ions. Radiat. Res. 183, 345–356 (2015).
Allen, C. P. et al. Low- and high-LET ionizing radiation induces delayed homologous recombination that persists for two weeks before resolving. Radiat. Res. 188, 82–93 (2017).
Li, Y. et al. Patterns of somatic structural variation in human cancer genomes. Nature 578, 112–121 (2020).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Hu, C. et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 319, 2401–2409 (2018).
Yurgelun, M. B. et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet. Med. 21, 213–223 (2019).
Dreyer, S. B. et al. Targeting DNA damage response and replication stress in pancreatic cancer. Gastroenterology 160, 362–377.e313 (2021).
Golan, T. et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019).
Basourakos, S. P. et al. Combination platinum-based and DNA damage response-targeting cancer therapy: evolution and future directions. Curr. Med. Chem. 24, 1586–1606 (2017).
Palacio, S. et al. DNA damage repair deficiency as a predictive biomarker for FOLFIRINOX efficacy in metastatic pancreatic cancer. J. Gastrointest. Oncol. 10, 1133–1139 (2019).
El Shafie, R. A. et al. In vitro evaluation of photon and raster-scanned carbon ion radiotherapy in combination with gemcitabine in pancreatic cancer cell lines. J. Radiat. Res. 54, i113–119 (2013).
Maalouf, M. et al. Different mechanisms of cell death in radiosensitive and radioresistant p53 mutated head and neck squamous cell carcinoma cell lines exposed to carbon ions and x-rays. Int. J. Radiat. Oncol. Biol. Phys. 74, 200–209 (2009).
Anon. Quantitative Concepts and Dosimetry in Radiobiology ICRU Report 30 (International Commission on Radiation Units and Measurements, 1987).
Fertil, B., Dertinger, H., Courdi, A. & Malaise, E. P. Mean inactivation dose: a useful concept for intercomparison of human cell survival curves. Radiat. Res. 99, 73–84 (1984).
Subiel, A., Ashmore, R. & Schettino, G. Standards and methodologies for characterizing radiobiological impact of high-Z nanoparticles. Theranostics 6, 1651–1671 (2016).
McCabe, N. et al. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol. Ther. 4, 934–936 (2005).
Saxena, S. & Zou, L. Hallmarks of DNA replication stress. Mol. Cell 82, 2298–2314 (2022).
Moding, E. J., Kastan, M. B. & Kirsch, D. G. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat. Rev. Drug Discov. 12, 526–542 (2013).
Hagiwara, Y. et al. Clustered DNA double-strand break formation and the repair pathway following heavy-ion irradiation. J. Radiat. Res. 60, 69–79 (2019).
Kawanishi, M., Fujita, M. & Karasawa, K. Combining carbon-ion irradiation and PARP inhibitor, olaparib efficiently kills BRCA1-mutated triple-negative breast cancer cells. Breast Cancer 16, 11782234221080553 (2022).
Yi, G. et al. DNA polymerase theta-mediated repair of high LET radiation-induced complex DNA double-strand breaks. Nucleic Acids Res. 51, 2257–2269 (2023).
Frankenberg-Schwager, M. et al. Single-strand annealing, conservative homologous recombination, nonhomologous DNA end joining, and the cell cycle-dependent repair of DNA double-strand breaks induced by sparsely or densely ionizing radiation. Radiat. Res. 171, 265–273 (2009).
Mladenova, V., Mladenov, E., Chaudhary, S., Stuschke, M. & Iliakis, G. The high toxicity of DSB-clusters modelling high-LET-DNA damage derives from inhibition of c-NHEJ and promotion of alt-EJ and SSA despite increases in HR. Front. Cell Dev. Biol. 10, 1016951 (2022).
Perkhofer, L. et al. DNA damage repair as a target in pancreatic cancer: state-of-the-art and future perspectives. Gut 70, 606–617 (2021).
Liu, Q. et al. Lung cancer cell line screen links fanconi anemia/BRCA pathway defects to increased relative biological effectiveness of proton radiation. Int. J. Radiat. Oncol. Biol. Phys. 91, 1081–1089 (2015).
Curtin, N. J. Targeting the DNA damage response for cancer therapy. Biochem. Soc. Trans. 51, 207–221 (2023).
Ding, L. et al. Evaluation of the response of HNSCC cell lines to gamma-rays and (12)C ions: Can radioresistant tumors be identified and selected for (12)C ion radiotherapy? Front Oncol. 12, 812961 (2022).
Mirandola, A. et al. Dosimetric commissioning and quality assurance of scanned ion beams at the Italian National Center for Oncological Hadrontherapy. Med. Phys. 42, 5287–5300 (2015).
Durante, M. et al. Report of a National Cancer Institute special panel: characterization of the physical parameters of particle beams for biological research. Med. Phys. 46, e37–e52 (2019).
Lind, B. K., Persson, L. M., Edgren, M. R., Hedlof, I. & Brahme, A. Repairable-conditionally repairable damage model based on dual Poisson processes. Radiat. Res. 160, 366–375 (2003).
Kellerer, A. M. & Hug, O. in Strahlenbiologie/Radiation Biology: Teil 3/Part 3 (eds Hug, O. & Zuppinger, A.) 1–42 (Springer Berlin Heidelberg, 1972).
Saha, J. et al. Ablating putative Ku70 phosphorylation sites results in defective DNA damage repair and spontaneous induction of hepatocellular carcinoma. Nucleic Acids Res. https://doi.org/10.1093/nar/gkab743 (2021).
Lee, K. J. et al. Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 44, 1732–1745 (2016).
Lu, H., Saha, J., Beckmann, P. J., Hendrickson, E. A. & Davis, A. J. DNA-PKcs promotes chromatin decondensation to facilitate initiation of the DNA damage response. Nucleic Acids Res. 47, 9467–9479 (2019).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Acknowledgements
The authors would like the Medical Physics team at CNAO for assistance with the irradiations. Funds were provided through the Department of Radiation Oncology Seed Grant Program to M.D.S. and by the National Institutes of Health to A.J.D. (CA162804 and CA233594). The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Author information
Authors and Affiliations
Contributions
B.J.S. conceived of and performed both the photon and CIRT experiments, interpreted the data, and wrote and edited sections of the manuscript. J.S. performed the immunofluorescence experiments and interpreted the data. E.M.A. plotted all data, calculated survival fits, and performed statistical analysis. L.D. was responsible for all omics analysis, and interpretation of that data, and wrote sections of the manuscript. H.L. performed all the immunoblotting experiments. S.-Y.W. performed the survival data to assess the sensitivity to ATR inhibition. K.L.S. participated intellectually and edited the manuscript. J.H.N. performed additional photon experiments. A.F. was the CNAO biology contact person. She helped facilitate experiments and helped with the design of the irradiation setup at CNAO. A.P. was responsible for radiation physics and helped develop the beam configuration. M.C. was responsible for 12C dosimetry, developing the SOBP for CNAO, and all physics aspects of 12C irradiation. T.A.A. participated intellectually and assisted in experimental design. M.D.S. conceived and performed experiments, edited the manuscript, and helped lead the research program. A.J.D. conceived and performed experiments, wrote the manuscript, and led the research program. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sishc, B.J., Saha, J., Alves, E.M. et al. Defective homologous recombination and genomic instability predict increased responsiveness to carbon ion radiotherapy in pancreatic cancer. npj Precis. Onc. 9, 20 (2025). https://doi.org/10.1038/s41698-025-00800-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-025-00800-4