Abstract
Prostate cancer (PCa) is a leading cause of cancer-related mortality in men, with Gleason grading critical for prognosis and treatment decisions. Machine learning (ML) models offer potential for automated grading but are limited by dataset biases, staining variability, and data scarcity, reducing their generalizability. This study employs generative adversarial networks (GANs) to generate high-quality synthetic histopathological images to address these challenges. A conditional GAN (dcGAN) was developed and validated using expert pathologist review and Spatial Heterogeneous Recurrence Quantification Analysis (SHRQA), achieving 80% diagnostic quality approval. A convolutional neural network (EfficientNet) was trained on original and synthetic images and validated across TCGA, PANDA Challenge, and MAST trial datasets. Integrating synthetic images improved classification accuracy for Gleason 3 (26%, p = 0.0010), Gleason 4 (15%, p = 0.0274), and Gleason 5 (32%, p < 0.0001), with sensitivity and specificity reaching 81% and 92%, respectively. This study demonstrates that synthetic data significantly enhances ML-based Gleason grading accuracy and improves reproducibility, providing a scalable AI-driven solution for precision oncology.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the third most commonly diagnosed cancer globally and a leading cause of cancer-related deaths among males1,2,3,4,5,6,7. The diagnostic pathway typically starts with measuring prostate-specific antigen (PSA) levels8,9,10,11,12,13,14,15,16,17,18,19. If PSA levels are elevated, clinicians may proceed with additional assays, such as 4Kscore20,21,22,23,24,25,26, PCA327,28,29,30,31,32,33, or phi34,35,36,37,38,39, followed by imaging studies like MRI to identify potential PCa lesions40,41,42,43,44,45. In cases where abnormalities are detected, biopsies are often performed for histopathological and genomic evaluation. This diagnostic approach is critical for determining PCa prognosis, especially in men under active surveillance, which has significantly increased in the past decade46,47,48,49. However, overdiagnosis remains a significant concern, contributing to adverse physical and psychological health outcomes50. Therefore, improving diagnostic accuracy is essential to enhance patient outcomes.
Machine learning (ML) has gained traction in recent years for automating various stages of PCa diagnosis and prognosis51. ML algorithms, trained on data from tissue samples, medical images, and clinical information, can detect patterns associated with the disease51,52,53,54,55,56,57. Particularly effective in the automated analysis of MRI scans and biopsy slides, ML models are being used to identify suspicious prostate areas with greater precision than traditional manual methods58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78. Technologies such as PathAI, PaigeAI, and Deciplex convert digital histology into data that can differentiate between cancerous and non-cancerous areas79,80,81,82,83.
Despite these advances, current technologies face limitations, primarily due to the reliance on clinical trial datasets for training84,85,86,87. Clinical trial data often represent specific patient groups and disease stages, limiting the generalizability of models to the broader population81,88,89,90,91. Additionally, inconsistencies in staining, slide preparation, and imaging resolution introduce variability that can negatively impact ML model performance92,93. Moreover, ethical concerns arise from the use of sensitive patient data, which must be handled in accordance with strict privacy guidelines.
To address these challenges, synthetic data generation has emerged as a complementary strategy to expand training datasets while preserving patient privacy94. Our previous study have demonstrated that GANs and other generative models can create histopathological images that capture critical morphological features for training AI models94. While prior research has successfully used synthetic data augmentation in medical imaging and general histopathology, its application in Gleason grading for prostate cancer remains underexplored95. Furthermore, the integration of synthetic data into AI-driven pathology models has largely focused on image augmentation rather than standalone training and validation strategies95,96, highlighting the need for further investigation.
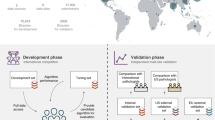
To address these challenges, we propose reducing reliance on clinical trial data by generating synthetic data. This study presents a novel approach using customized generative adversarial networks (GANs) to create synthetic histopathological images from a small set of original radical prostatectomy and needle biopsy images. The synthetic images were analyzed using spatial heterogeneous recurrence quantification analysis (SHRQA) modules to assess granularity compared to real images. We then trained a convolutional neural network (CNN, EfficientNet) on these synthetic image patches and evaluated its grading performance against a CNN trained on real image patches. For validation, we utilized histology images from various sources, including the TCGA, PANDA Challenge83 and MAST trial.
The CNN model trained on synthetic data significantly improved grading accuracy compared to models trained solely on original data. These findings demonstrate the potential of this method to overcome data scarcity in AI model training, offering a promising solution for PCa diagnostics in clinical settings.
Results
Network architecture and model selection
We evaluated four CNN models using 20 randomly selected histology images from the TCGA Prostate Adenocarcinoma dataset. AlexNet was the first CNN to be defined, and it consists of a series of convolutional pooling layers and an interconnected output layer97, achieved 55% accuracy, matching pathologists’ assessments and TCGA scoring. ResNet, which incorporates residual networks based on skip connections within the network, significantly improving AlexNet98, also achieved 55% accuracy on the same images. Xception model, which is based on the Inception model, improves traditional network architectures such as AlexNet and separates space and depth into different dimensions in its convolutional layers99, improved accuracy to 60%. EfficientNet, which utilizes a novel scaling method that employs a compound coefficient to scale up a CNN model in a more structured manner100,101, outperformed all models with 65% accuracy. Based on these results, EfficientNet was selected for this study (Supplementary Fig. 1, Supplementary Table 1).
Image preprocessing
To address variability in staining protocols, tissue quality, and section thickness102, pre-processing normalization was applied to the datasets. For the TCGA dataset (500 images), 21 outliers with mean RGB intensity beyond two standard deviations were excluded. Similarly, one outlier was removed from 32 radical prostatectomy section images from the University of Miami. For 3949 needle biopsy slides from the PANDA challenge83, area evaluation and color normalization excluded 257 images. These steps minimized variability and ensured a more consistent dataset for AI model predictions (Table 1).
Quality-controlled annotation and patch generation
RP sections from TCGA and UM were divided into pseudo-cohorts based on Gleason patterns and analyzed with HistoQC103 to exclude low-quality samples. Of 143 randomly reviewed sections, only 33 with agreement between two pathologists and TCGA scoring were selected for gold standard training data. Using PyHIST, 96 × 96 and 256 × 256 pixel patches were generated, each containing at least 75% tissue and no more than 25% whitespace. The minimum patch size was set at 96 pixels, matching the smallest overlapping annotation. From 33 images, 219 patches were created and augmented to 2082 patches (Fig. 1A).
A Illustration showing pipeline used in generating synthetic images from prostate cancer digital histology. Images were preprocessed by PyHist and HistoQC. Those that passed QC were then given to pathologist for scoring, and then cut into small patches for modeling. B Original and Synthetic images were generated for each primary Gleason pattern 3, 4, and 5, respectively.
GAN model selection
GANs are a class of deep learning models used to generate synthetic images by learning patterns from real data through an adversarial training process. A GAN consists of two neural networks: a generator, which produces synthetic images, and a discriminator, which differentiates between real and generated images. The networks are trained iteratively, with the generator improving its ability to create realistic images while the discriminator refines its capacity to distinguish them. Various GAN architectures have been developed to optimize image synthesis for different applications. In this study, three GAN variants—conditional GAN (cGAN), StyleGAN, and deep convolutional GAN (dcGAN)104,105,106 were evaluated. cGAN incorporates labeled input data to guide the generation process, allowing for controlled image synthesis based on predefined categories. StyleGAN introduces a style-based generator that enhances image quality and provides greater control over fine-grained image features. dcGAN, a simpler and computationally efficient architecture, utilizes convolutional layers to improve image synthesis while maintaining relatively low training complexity.
To select the optimal GAN architecture, 2082 RP image patches were divided by Gleason patterns (GS3, GS4, GS5) and tested using each GAN model. Each GAN generated 1,000 synthetic images, which were classified using a CNN. StyleGAN and dcGAN showed similar accuracies (0.65 and 0.64, respectively), but dcGAN was significantly faster (901 min vs. 2372 min for 1000 images). Based on these results, we selected dcGAN to generate synthetic images for training. The Adam optimization algorithm determined the best iteration value at 14,000 iterations. We generated synthetic images in 128 × 128 and 256 × 256 pixel sizes and performed a manual quality control (QC) assessment on a randomized set of images by board-certified pathologists, who approved 80% of the images.
For needle biopsy analysis, 300 annotated biopsies produced 1712 cancer-specific (752 for GS3, 592 for GS4, and 368 for GS5, respectively) and 539 benign tissue patches (256 × 256 pixels). Various patch sizes (from 512 × 512 to 32 × 32) were tested, with CNN accuracy dropping significantly below 64 × 64 pixels. We selected 64 × 64 patches, upscaled to 256 × 256, for optimal performance. GAN training on a single NVIDIA T1000 GPU generated 1000 synthetic images in 2.5 h, with loss functions balancing at approximately 50,000 epochs (Supplementary Fig. 2). A repository of 2000 patches was created for tumor and normal samples (Fig. 2). Pathologists manually assessed these images, with an 80% approval rate for adequacy in patch size.
Benchmarking
To determine the optimal number of synthetic images for EfficientNet training, we fine-tuned the model with synthetic data generated by dcGAN and assessed overfitting using the similarity index and Frechet Inception Distance (FID)107. We generated synthetic images based on Gleason patterns in batches of 10 K, 50 K, and 100 K. FID scores for RP images were 25.1 (10 K), 18.8 (50 K), and 36.2 (100 K). The 50 K batch produced the best results, with accuracy increasing from 33% to 70% for GS3, 35% to 65% for GS4, and 41% to 71% for GS5, stabilizing beyond 50 K images. Thus, we integrated a maximum of 50 K synthetic images into the model.
To further avoid overfitting, we applied 10-fold cross-validation, randomly organizing all real and synthetic image tiles. Accuracy remained consistent, ranging from 0.68 to 0.71 for GS3, 0.63 to 0.70 for GS4, and 0.66 to 0.73 for GS5, with similarity index values between 0.8 to 1.0 (Fig. 2) (Table 1). This process confirmed the quality and variability of synthetic images.
For needle biopsies, FID scores were 21.2 (10 K), 20.2 (50 K), and 33.7 (100 K). Cross-validation of 1000 samples yielded stable accuracy across Gleason scores (GS3: 0.67–0.71, GS4: 0.63–0.70, GS5: 0.65–0.73) with similarity index values ranging from 0.92 to 1.0 (Table 1), confirming the quality of synthetic needle biopsy images (Supplementary Fig. 2).
Quality evaluation of synthetic images
Next, we sought to address the extent of technical variations that exist between the synthetically generated images and the original images. This examination is crucial because each Gleason pattern possesses specific morphological characteristics108,109 that synthetically generated images should ideally replicate. For this, we utilized SHRQA, a robust technique capable of measuring complex microstructures based on spatial patterns110,111,112. The SHRQA process, as shown and detailed in Supplementary Fig. 3, involves six key steps111,112,113,114,115,116,117,118. It begins with the application of the 2D-Discrete Wavelet Transform (2D-DWT) using the Haar wavelet to reveal patterns that are not apparent in the original image111,112,113,114,115,116. Subsequently, each image is converted into an attribute vector using the Space-Filling Curve (SFC), which plays a crucial role in maintaining the spatial proximity between pixels within the vector. This step is essential for representing the image’s geometric recurrence in vector form. The attribute vector is then projected into state space, forming a trajectory that emphasizes the geometric structure of the image. To further analyze spatial transition patterns, Quadtree segmentation is employed to divide the state space into distinct subregions117,118. Following this, an Iterated Function System projection is applied, transforming each attribute vector into a fractal plot that captures recurrence within the fractal topology. Finally, the fractal structures are quantified to provide insights into the geometric properties of the image. In one of our earlier studies, we used the SHRQA method to quantify synthetically generated image patches from eight different genitourinary organs, including the testis, kidney, prostate, bladder, vagina, cervix, ovary, and uterus94. Following a similar approach, the SHRQA method was applied to examine the spatial recurrence properties of real and synthetic image patches across four Gleason patterns (GS0/3/4/5) in the RP section and needle biopsies.
For RP, our sample set included an equal number of patches from real and synthetic sources, with a balanced representation of each Gleason pattern. We analyzed 4000 image patches, each 256 × 256 pixels, evenly split between real and synthetic. A one-layer 2D-DWT with Haar wavelet decomposed each image into four sub-images, revealing fine details. SHRQA quantitatively outlined each patch’s microstructures. From an initial extraction of 1997 spatial recurrence features per patch, LASSO119 selected 1819 features as significant to the Gleason pattern. Hotelling’s T-squared test, compared the spatial recurrence attributes. The resulting p-values of 0.8991 signified no significant differences in spatial recurrence properties between real and synthetic patches, as confirmed by the T-squared tests’ p-values for each Gleason pattern (refer to Table 2, left). We also employed PCA on the spatial recurrence properties120,121,122,123,124,125,126, visualized using radar charts, revealing that the six principal components capture 82% of the variability. This allowed us to map the distributions of spatial properties for real and synthetic images across Gleason patterns, as depicted in Fig. 3. Notably, while distributions for the same Gleason pattern aligned closely between real and synthetic images, significant differences were evident across different patterns.
A The distributions of spatial recurrence properties (in the first 6 Principal Components (PCs), which contain 82% of data variability) underlying different Gleason scores for both real and synthetic patches on Radical Prostatectomy. Note that the purple lines indicate the mean values of each feature, and the gray area shows the 95% confidence interval. Our results indicate that while the distributions of spatial properties are closely aligned between real and synthetic images under the same Gleason Score, they markedly differ when comparing different Gleason Scores. B The comparison of spatial recurrence properties between real and synthetic on the first six PCs (contain 82% of data variability). The distributions of this four PCs are similar between real and synthetic.
A similar analysis was conducted for the needle Biopsies section (Fig. 4), examining 1600 patches with an equal split between real and synthetic images distributed evenly across GS0, GS3, 4, and 5. SHRQA extracted 2585 initial features, with LASSO identifying 1578 as significant. Hotelling’s T-squared tests in the NB section corroborated the RP section’s results (refer to Table 2, right), demonstrating that synthetic images reliably replicate the geometric nuances of real images for each Gleason pattern. These findings across both RP and NB sections validate the model’s efficiency in capturing the geometric intricacies consistent with real images.
A The distributions of spatial recurrence properties (in the first 16 Principal Components (PCs), which contain 80% of data variability) underlying different Gleason scores for both real and synthetic patches on Needle Biopsy. Note that the purple lines indicate the mean values of each feature, and the gray area shows the 95% confidence interval. Our results indicate that while the distributions of spatial properties are closely aligned between real and synthetic images under the same Gleason Score, they markedly differ when comparing different Gleason Scores. B The comparison of spatial recurrence properties between real and synthetic on the first eight PCs (contain 70% of data variability). The distributions of this four PCs are similar between real and synthetic.
Furthermore, we subjected a randomized number of synthetic image patches allocated into Gleason 3, 4, and 5 via SHRQA quantification models to cross-validation for characterization by pathologists. Specific features associated with each phenotype, such as uniform glandular arrangement, moderate differentiation, fibrous stroma, mild nuclear atypia, clear lamina, and minimal desmoplasia associated with Gleason 3; Moderate to severe nuclear atypia, poor differentiation, irregular and cribriform patterns, inflammatory infiltrate associated with Gleason 4; and Sheet of cells, scant stromal tissue, undifferentiated tumor cells, sever nuclear atypia, complete loss of glandular structure associated with Gleason 5 were accurately identified by the SHRQA models. These findings across both RP and NB sections validate the model’s efficiency in capturing the geometric intricacies consistent with real images (Fig. 5A–C)
Validation of CNN performance post-training with enhanced synthetic data
Next, to determine whether synthetic images could substitute original images for training a CNN model, we trained the CNN model with two sources of image patches. The first source was the patches derived from original RP sections from TCGA and inhouse images, which were classified according to the Gleason pattern (normal (n = 175), GS3 (n = 726), GS4 (n = 1029), and GS5 (n = 152)). The second source of patches was derived from a combination of original and synthetic images, with 5000 image patches used for each Gleason pattern. The grading capabilities of this CNN model were then compared by allowing it to assign Gleason scoring to the RP images in the TCGA (n = 475). The results demonstrated that the CNN’s accuracy significantly improved in GS3 from 0.53 to 0.67 (p = 0.0010), in GS4 from 0.55 to 0.63 (p = 0.0274), and in GS5 from 0.57 to 0.75 (p < 0.0001) when trained with the combination of original and synthetic image patches compared to just being trained with original image patches. Moreover, the comparative analysis revealed a notable enhancement in accuracy and the receiver operating characteristic curve (ROC) for the combined (original and synthetic) dataset relative to the original (p = 0.0381). Additionally, the in-house RP images (n = 24) were subjected to grading using CNN models described above. Results demonstrated a significant improvement of the CNN model to accurately assign the grade.
Furthermore, we extended the validation of the CNN model using needle biopsies. Similar to the RP sections, we trained the CNN model with two sources of image patches. The first source was the patches derived from original needle biopsy sections, classified according to the Gleason pattern (normal (n = 539), GS3 (n = 610), GS4 (n = 890), and GS5 (n = 212)). The second source was derived from a combination of original and synthetic images, classified according to the Gleason pattern, with a total of 2000 image patches used for each Gleason pattern. The grading capabilities of this CNN model were then compared by allowing it to assign Gleason grading to the needle biopsy images (n = 3649). The comparative analysis revealed a significant enhancement in accuracy and the ROC for the amalgamated dataset relative to the original (Fig. 6). Specifically, the enhancement was consistent across both benign and malignant samples, with an overall accuracy increase from 91% when using the original training images to an accuracy of 95% when using original and synthetic images combined (p = 0.0402). The original and synthetic combined training database yielded a sensitivity of 0.81, and a specificity of 0.92.
Moreover, to confirm the efficiency of the validated AI model, we subjected it to evaluate the images derived from MAST trial samples. While the MAST trial analysis focused on baseline data, it provides a unique real-world validation setting for AI applications in active surveillance workflows. The outcome of the AI model evaluation was compared to the pathologist grading. Results showed an overall accuracy of 87% for Gleason grading, with sensitivity and specificity of 81% and 92%, respectively. Together, cross-validation of the final model yielded consistent performance, with mean accuracy across folds varying by less than 1.5 percentage points (95% CI, ±1.5%). The improved accuracy across datasets highlights the potential of synthetic data for enhancing model robustness and reproducibility (Fig. 4).
Discussion
The integration of AI-based tools into cancer diagnostics, particularly for prostate cancer (PCa), holds significant promise. Over the past decade, numerous image analysis algorithms have been developed to assist with tumor grading, classification, and metastasis detection127,128,129,130,131,132,133,134,135,136,137. Despite their potential, many AI tools face substantial barriers to widespread clinical adoption138. These challenges stem primarily from biases in training data, the inability to generalize across diverse populations, and technological constraints such as model drift139,140. This study sought to address these limitations by employing GANs to generate synthetic histopathological data, supplementing real-world datasets, and enhancing the accuracy and robustness of convolutional neural network (CNN) models for PCa Gleason grading81,141,142.
AI models trained on datasets with narrow demographic representation often fail to generalize effectively to diverse clinical environments, risking misdiagnosis and suboptimal performance when applied to unseen populations. This study addressed this limitation by integrating data from three diverse datasets: TCGA, PANDA Challenge, and the MAST trial. While these datasets provided robust foundations for model training and validation, they are not fully representative of global patient populations. Future research should prioritize external validation across broader ethnic, geographic, and demographic cohorts. Doing so would ensure that AI models trained for Gleason grading are equitable and clinically reliable in a wide range of settings.
GANs were leveraged to generate synthetic histopathological images, providing a scalable solution to address data scarcity and augment training datasets. Unlike prior approaches where synthetic data was used primarily for general augmentation, this study validated synthetic images against pathologist-annotated real-world images using SHRQA, FID, and Hotelling’s T-squared test. These quantitative validation methods confirmed that GAN-generated images preserved key histological structures, including cribriform patterns and glandular arrangements, with no significant differences in spatial recurrence properties. However, while synthetic data significantly improved model performance, real-world validation remains critical, as synthetic images may not fully capture the biological variability inherent in tissue samples. Future studies should explore the optimal balance between synthetic and real-world data to maximize both accuracy and generalizability in AI-driven diagnostic models.
Given the growing prominence of diffusion models for synthetic data generation, their potential role in histopathology warrants discussion. While diffusion models offer superior image fidelity143,144,145, they require significantly greater computational resources and iterative refinement steps, making them less practical for large-scale AI training146,147. In contrast, dcGAN was chosen for its computational efficiency, rapid convergence, and ability to generate clinically relevant images validated through SHRQA and FID metrics. While diffusion models remain a promising alternative, their added complexity did not provide a clear advantage for this study’s objectives. However, future work should investigate hybrid models that integrate the strengths of both GANs and diffusion-based approaches for optimizing synthetic histopathological image generation.
A crucial consideration in this study was dataset heterogeneity, particularly in combining RP and needle biopsy datasets. While merging these datasets into a single training framework was considered, it posed technical and biological challenges. Technically, needle biopsy images were processed at 64 × 64 and upscaled to 256 × 256, whereas RP sections were directly processed at 256 × 256. Combining these datasets without accounting for resolution differences could have led to inconsistencies in feature extraction and CNN performance. Biologically, RP specimens provide larger, whole-mount tissue architecture, while needle biopsies capture smaller, fragmented tumor regions, leading to distinct histological patterns relevant for Gleason grading148,149. Training separate CNN models for RP and needle biopsies ensured that tissue-specific morphological patterns were preserved, improving the robustness and clinical applicability of the AI-based grading system.
The impact of patch size and resolution variability on CNN performance was also systematically evaluated as it influences feature extraction, model convergence, and classification accuracy150,151,152. Ablation studies demonstrated that patches smaller than 64 × 64 resulted in decreased accuracy, likely due to the loss of critical histological details, while patches larger than 512 × 512 increased computational cost without proportional accuracy gains. These findings align with prior studies in digital pathology (Litjens et al. 2017; Campanella et al. 2019) and emphasize the importance of optimizing input resolution to balance accuracy and efficiency. Additionally, image standardization techniques, including color normalization and histogram-based intensity adjustments, were employed to minimize artifacts and domain shift across datasets. A key challenge in deep learning models for histopathology is interpretability, which is essential for clinical adoption. While this study prioritized quantitative validation techniques (SHRQA, FID, Hotelling’s T-squared test, PCA), which provide structured insights into synthetic data fidelity, we acknowledge the importance of explicit interpretability techniques such as Grad-CAM or SHAP. Future work will explore attention-based visualization methods to further enhance clinician trust in AI-driven Gleason grading.
Performance comparison with existing AI-based Gleason grading models highlights the effectiveness of incorporating synthetic histopathological images into CNN training pipelines. The proposed model achieved 95% accuracy when trained on a combined dataset of real and synthetic images, surpassing the 91% accuracy of models trained on real data alone (p = 0.0402). Independent validation on the MAST trial dataset demonstrated 87% accuracy, reinforcing the robustness and generalizability of the approach in real-world clinical settings. These findings align with existing AI-based models such as PathAI and PaigeAI, which, despite high diagnostic accuracy, face dataset bias and generalizability issues153. Notably, the inclusion of GAN-generated synthetic data improved classification performance across all Gleason patterns. Unlike prior AI approaches, which rely solely on real-world datasets, this study demonstrates that synthetic data can be strategically integrated to enhance AI-based pathology models.
While this study demonstrated significant technical advancements, transitioning from research to real-world clinical implementation requires addressing practical challenges. Integration into clinical workflows demands robust digital infrastructure, clinician training, and seamless interoperability with existing pathology systems. To facilitate adoption, future efforts should focus on developing intuitive user interfaces and decision-support systems that enhance, rather than disrupt, clinician workflows. Moreover, regulatory approvals and adherence to ethical guidelines will be pivotal in fostering trust and acceptance among healthcare providers and patients.
Although this study focused on PCa diagnosis, the methodologies demonstrated here—particularly the use of GAN-generated synthetic data—can be extended to other cancer types. The ability to generate high-fidelity synthetic data holds immense potential for augmenting AI models across a spectrum of oncological applications. Future research should refine these models for PCa-specific contexts, such as active surveillance and advanced disease stages, while also exploring their adaptability to other malignancies.
Together this study addresses key limitations in AI-driven cancer diagnostics by introducing synthetic data to overcome dataset scarcity, integrating diverse datasets to improve generalizability, and demonstrating the feasibility of using CNN models for Gleason grading. While challenges such as model drift, workflow integration, and demographic diversity remain, these findings lay a strong foundation for future longitudinal studies and clinical applications. As AI continues to evolve, its potential to transform cancer diagnostics and personalized medicine becomes increasingly evident, particularly when developed in collaboration with clinicians and validated in real-world settings.
Methods
Scanning and image annotation
Images are deconstructed by first taking individual areas in patchwise fashion using the software package PYHist. Briefly, the image is taken from raw.svs format and scanned for areas that are defined as tissue regions. This is done by color definition compared to whitespace background. Each predefined block is taken at a 512 × 512 pixel area which has been identified as the lowest amount of space that our pathologist graded any given image. These patches were then used as the training database from which other images are annotated. The image annotation for a given test image then is a whole image from which a sliding window approach is taken that moves through each of the given pixel windows and scored against the training database.
Cohorts used
Samples were taken from TCGA. Histology images from 500 individuals were considered. 32 local samples were taken from the University of Miami pathology core. The Institutional Review Board (IRB) protocol was approved by the University of Miami Miller School of Medicine, Miami, FL, to ensure that the research adhered to ethical guidelines and principles. Furthermore, the study included 3949 needle biopsies sourced from Radboud University Medical Center and Karolinska Institute (the PANDA challenge)83,154. The MAST Trial dataset comprised 141 patients stratified into NCCN risk groups: Very Low (44.2%), Low (38.1%), and Intermediate (17.3%). Mean patient age in the MAST dataset was 62.8 years (SD, 8.5 years; range, 43–85 years) (Table 3). Across datasets, pathologists annotated Gleason patterns, ensuring robust and consistent training data (Fig. 1).
Algorithm design training and testing procedure
We selected 3 initial algorithms from which to test out the convolutional network. These were defined as the major stepwise breakthroughs in AI with the AlexNET, ResNet, and Xception models. Initially, training images were taken from a single pathologist defined areas and 10 random images were used as test images. We used granular level annotation (detail in result section) to select the model with the highest accuracy. Post model selection, the hyperparameters were turned. Here a Tree-structured Parzen Estimator was implemented to complete a sequential model optimization. In addition, tree weights were investigated by a population based training approach. From this, the highest performing hyperparameters were used along with the appropriate tree weights that were used to define the network.
Calculation of performance
We defined accuracy in 2 different ways. First, in granular accuracy we started with the areas that were selected and annotated with the exact same Gleason pattern by all the 3 pathologists. Gold standard granular accuracy is when the convolutional network overlaps with the agreement between all pathologists. This is defined by the number of overlapping pixels in the defined area compared with the definition between the annotated areas of the pathologist pattern. Second, patient level accuracy is defined when the artificial intelligence pipeline has correctly identified a patient’s Gleason Score as defined by the TCGA pathology confirmed score.
Development and evaluation of a conditional Generative Adversarial Network
A preliminary conditional Generative Adversarial Network (cGAN) was designed and implemented to assess the performance accuracy of various GAN architectures. The cGAN was developed utilizing Python 3.7.3 and the Tensorflow Keras 2.7.0 package. The generator component of the cGAN comprises three input layers and a single output layer. In parallel, the discriminator component is configured with analogous input, hidden, and output layers. The cGAN’s total parameter count was 19.2 million for each of the evaluated Gleason patterns (Supplementary Fig. 1).
Implementation and adaptation of StyleGAN for tissue image analysis
StyleGAN, a progressive generative adversarial network architecture, serves as a baseline for comparison to the cGAN, featuring a distinct generator configuration. The architecture was adopted from the original StyleGAN publication with minimal alterations to the generator and discriminator networks. A notable modification involved substituting human face images in the StyleGAN with tissue images to create a tissue image GAN. The generator’s total parameter count amounted to 28.5 million, in contrast to 26.2 million in the original StyleGAN publication and 23.1 million in a conventional generator. Particular emphasis was placed on refining the GS4 and GS5 images to ensure adequate representation of tumor heterogeneity.
dcGAN
The dcGAN weights were initialized randomly from a normal distribution with mean = 0 and a standard deviation of 0.02. The generator neural network was constructed using 13 layers consisting of transpose function with batch normalization and ReLU functions. The walkthrough of the generator layers is as follows :
ConvTranspose2d -> BatchNorm2d -> ReLU -> ConvTranspose2d -> BatchNorm2d -> ReLU -> ConvTranspose2d -> BatchNorm2d -> ReLU -> ConvTranspose2d -> BatchNorm2d -> ReLU -> ConvTrnaspose2d -> Tanh.
The discriminator neural network was constructed using 12 layers of Conv2d, LeakyReLU functions with batch normalization. The walkthrough of the layers is as follows :
Conv2d -> LeakyReLU -> Conv2d -> BatchNorm2d -> LeakyReLU -> Con v2d -> BatchNorm2d -> LeakyReLU -> Conv2d -> BatchNorm2d -> LeakyReLU -> Conv2d -> Sigmoid.
Estimation of the number of parameters used for a single run ranged from 1.3 T to 1.4 T calculations per run to generate 1,000 synthetic images that were run on standard GPU chips (GET INFO). Time taken for a single run was dependent on the number of GPU processors available. For multi-threaded GPU the time taken ranged from 1.5 h to 5 h for a run while for a single GPU it ranged from 3 h to 12 h. Time was dependent on the level of resolution desired.
Finally, the BCE loss function was used and the Adam optimizer was implemented for both the generator and discriminator. Iteration level statistics were generated at the end of each run and saved for further analysis in matplot.
EfficientNET
The EfficientNET baseline model was setup with the B3 function within the TensorFlow python backend. Initial testing for the B6 function was found to be too restrictive and B1 was too simple to form complex patterns so B3 was selected as our model function. To form the model based on our images, a GlobalMaxPooling2D layer was added after the initial base as well as a Dropout layer which will help avoid overfitting. The dropout rate was set to 0.2 after initial estimations were too high. The number of classes for prediction layer was set to 4 which represented the normal tissue group as compared to the primary Gleason patterns GS3, GS4, and GS5. The EfficientNET model was setup using pre-trained weights from the “imageNet” to take advantage of transfer learning to reduce analysis time.
Image augmentation was completed using Keras ImageDataGenerator function. Image rotation range was set to 45, width_shift_range and height_shift_range was set to 0.2, and the horizontal flip was set to true for flipping the image. Fill mode was defaulting to “nearest”. Validation data was not augmented, but the images were put through a rescale function to ensure that every test image was uniform before annotation.
Statistical calculations
FID was implemented in custom scripts developed in house. The FID model was pre-trained using Inception V3 weights for transfer learning. In house code was centered around the FID model and inserted into the dcGAN to be run during each iteration. Stats were reported at intervals of 1000 and graphed with in house python scripts.
PCA analysis was performed by first transforming the images into numerical arrays. Images were separated into normal and synthetic batches and then distributed by primary Gleason score. Intensity was calculated (using the R package imgpalr and magick) as the average of the color of the entire image while keeping the matrix framework (i.e., positional arguments were retained). PCA was conducted using the general prcomp function in R and plotted results were displayed in ggplot2.
Data availability
Deidentified participant data will be made available when all primary and secondary endpoints have been met. Any requests for data and supporting material (data dictionary, protocol, and statistical analysis plan) will be reviewed by the data management group in the first instance. Only requests that have a methodologically sound proposal and whose proposed use of the data has been approved by the independent data steering committee will be considered. Proposals should be directed to the corresponding author in the first instance; to gain access, data requestors will need to sign a data access agreement.
References
Brawley, O. W. Prostate cancer epidemiology in the United States. World J. Urol. 30, 195–200 (2012).
Badalament, R. A. & Drago, J. R. Prostate cancer. Dis. Mon. 37, 199–268 (1991).
Carthon, B., Sibold, H. C., Blee, S. & D Pentz, R. Prostate Cancer: Community Education and Disparities in Diagnosis and Treatment. Oncologist 26, 537–548 (2021).
Cook, E. D. & Nelson, A. C. Prostate cancer screening. Curr. Oncol. Rep. 13, 57–62 (2011).
Litwin, M. S. & Tan, H. J. The Diagnosis and Treatment of Prostate Cancer: A Review. Jama 317, 2532–2542 (2017).
Moon, T. D. Prostate cancer. J. Am. Geriatr. Soc. 40, 622–627 (1992).
Schatten, H. Brief Overview of Prostate Cancer Statistics, Grading, Diagnosis and Treatment Strategies. Adv. Exp. Med Biol. 1095, 1–14 (2018).
Wozniak-Petrofsky, J. The significance of prostatic specific antigen in men with prostate disease. elevated PSA Lev. may Indic. prostate cancer– it may not. Geriatr. Nurs. 14, 150–151 (1993).
Aghdam, A. M. et al. MicroRNAs as Diagnostic, Prognostic, and Therapeutic Biomarkers in Prostate Cancer. Crit. Rev. Eukaryot. Gene Expr. 29, 127–139 (2019).
Albertsen, P. C. PSA testing, cancer treatment, and prostate cancer mortality reduction: What is the mechanism?. Urol. Oncol. 41, 78–81 (2023).
Barry, M. J. & Simmons, L. H. Prevention of Prostate Cancer Morbidity and Mortality: Primary Prevention and Early Detection. Med Clin. North Am. 101, 787–806 (2017).
Borley, N. & Feneley, M. R. Prostate cancer: diagnosis and staging. Asian J. Androl. 11, 74–80 (2009).
Jalloh, M. & Cooperberg, M. R. Implementation of PSA-based active surveillance in prostate cancer. Biomark. Med 8, 747–753 (2014).
Merriel, S. W. D., Funston, G. & Hamilton, W. Prostate Cancer in Primary Care. Adv. Ther. 35, 1285–1294 (2018).
Nguyen-Nielsen, M. & Borre, M. Diagnostic and Therapeutic Strategies for Prostate Cancer. Semin Nucl. Med 46, 484–490 (2016).
Pezaro, C., Woo, H. H. & Davis, I. D. Prostate cancer: measuring PSA. Intern Med J. 44, 433–440 (2014).
Sharma, S., Zapatero-Rodríguez, J. & O’Kennedy, R. Prostate cancer diagnostics: Clinical challenges and the ongoing need for disruptive and effective diagnostic tools. Biotechnol. Adv. 35, 135–149 (2017).
Troyer, D. A., Mubiru, J., Leach, R. J. & Naylor, S. L. Promise and challenge: Markers of prostate cancer detection, diagnosis and prognosis. Dis. Markers 20, 117–128 (2004).
Venderbos, L. D. & Roobol, M. J. PSA-based prostate cancer screening: the role of active surveillance and informed and shared decision making. Asian J. Androl. 13, 219–224 (2011).
Parekh, D. J. et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur. Urol. 68, 464–470 (2015).
Borque-Fernando, Á et al. Role of the 4Kscore test as a predictor of reclassification in prostate cancer active surveillance. Prostate Cancer Prostatic Dis. 22, 84–90 (2019).
Konety, B. et al. The 4Kscore® Test Reduces Prostate Biopsy Rates in Community and Academic Urology Practices. Rev. Urol. 17, 231–240 (2015).
Mi, C., Bai, L., Yang, Y., Duan, J. & Gao, L. 4Kscore diagnostic value in patients with high-grade prostate cancer using cutoff values of 7.5% to 10%: A meta-analysis. Urol. Oncol. 39, 366.e361–366.e310 (2021).
Punnen, S. et al. The 4Kscore Predicts the Grade and Stage of Prostate Cancer in the Radical Prostatectomy Specimen: Results from a Multi-institutional Prospective Trial. Eur. Urol. Focus 3, 94–99 (2017).
Scuderi, S. et al. Implementation of 4Kscore as a Secondary Test Before Prostate Biopsy: Impact on US Population Trends for Prostate Cancer. Eur. Urol. Open Sci. 52, 1–3 (2023).
Zappala, S. M. et al. The 4Kscore blood test accurately identifies men with aggressive prostate cancer prior to prostate biopsy with or without DRE information. Int. J. Clin. Pract. 71, https://doi.org/10.1111/ijcp.12943 (2017).
Hessels, D. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44, 8–15 (2003).
Day, J. R., Jost, M., Reynolds, M. A., Groskopf, J. & Rittenhouse, H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 301, 1–6 (2011).
de la Taille, A. Progensa PCA3 test for prostate cancer detection. Expert Rev. Mol. Diagn. 7, 491–497 (2007).
Filella, X. et al. PCA3 in the detection and management of early prostate cancer. Tumour Biol. 34, 1337–1347 (2013).
Gunelli, R., Fragalà, E. & Fiori, M. PCA3 in Prostate Cancer. Methods Mol. Biol. 2292, 105–113 (2021).
Hessels, D. & Schalken, J. A. The use of PCA3 in the diagnosis of prostate cancer. Nat. Rev. Urol. 6, 255–261 (2009).
Yang, Z., Yu, L. & Wang, Z. PCA3 and TMPRSS2-ERG gene fusions as diagnostic biomarkers for prostate cancer. Chin. J. Cancer Res 28, 65–71 (2016).
Kim, L. et al. Clinical utility and cost modelling of the phi test to triage referrals into image-based diagnostic services for suspected prostate cancer: the PRIM (Phi to RefIne Mri) study. BMC Med 18, 95 (2020).
Barisiene, M. et al. Prostate Health Index and Prostate Health Index Density as Diagnostic Tools for Improved Prostate Cancer Detection. Biomed. Res Int 2020, 9872146 (2020).
Tosoian, J. J. et al. Use of the Prostate Health Index for detection of prostate cancer: results from a large academic practice. Prostate Cancer Prostatic Dis. 20, 228–233 (2017).
Yan, J. Q. et al. Prostate Health Index (phi) and its derivatives predict Gleason score upgrading after radical prostatectomy among patients with low-risk prostate cancer. Asian J. Androl. 24, 406–410 (2022).
Ye, C. et al. The Prostate Health Index and multi-parametric MRI improve diagnostic accuracy of detecting prostate cancer in Asian populations. Investig. Clin. Urol. 63, 631–638 (2022).
Zhang, G. et al. Assessment on clinical value of prostate health index in the diagnosis of prostate cancer. Cancer Med 8, 5089–5096 (2019).
Kaplan, I. et al. Real time MRI-ultrasound image guided stereotactic prostate biopsy. Magn. Reson Imaging 20, 295–299 (2002).
Fernandes, M. C., Yildirim, O., Woo, S., Vargas, H. A. & Hricak, H. The role of MRI in prostate cancer: current and future directions. Magma 35, 503–521 (2022).
Mendhiratta, N., Taneja, S. S. & Rosenkrantz, A. B. The role of MRI in prostate cancer diagnosis and management. Future Oncol. 12, 2431–2443 (2016).
Stabile, A. et al. Multiparametric MRI for prostate cancer diagnosis: current status and future directions. Nat. Rev. Urol. 17, 41–61 (2020).
Stempel, C. V., Dickinson, L. & Pendsé, D. MRI in the Management of Prostate Cancer. Semin Ultrasound CT MR 41, 366–372 (2020).
Wibmer, A. G., Vargas, H. A. & Hricak, H. Role of MRI in the diagnosis and management of prostate cancer. Future Oncol. 11, 2757–2766 (2015).
Abdulmajed, M. I., Hughes, D. & Shergill, I. S. The role of transperineal template biopsies of the prostate in the diagnosis of prostate cancer: a review. Expert Rev. Med Devices 12, 175–182 (2015).
Ahdoot, M. et al. MRI-Targeted, Systematic, and Combined Biopsy for Prostate Cancer Diagnosis. N. Engl. J. Med 382, 917–928 (2020).
He, Y., Shen, Q., Fu, W., Wang, H. & Song, G. Optimized grade group for reporting prostate cancer grade in systematic and MRI-targeted biopsies. Prostate 82, 1125–1132 (2022).
Pinto, F. et al. Imaging in prostate cancer diagnosis: present role and future perspectives. Urol. Int 86, 373–382 (2011).
Resnick, M. J. et al. Repeat prostate biopsy and the incremental risk of clinically insignificant prostate cancer. Urology 77, 548–552 (2011).
Tataru, O. S. et al. Artificial Intelligence and Machine Learning in Prostate Cancer Patient Management-Current Trends and Future Perspectives. Diagnostics (Basel) 11, https://doi.org/10.3390/diagnostics11020354 (2021)
Choi, R. Y., Coyner, A. S., Kalpathy-Cramer, J., Chiang, M. F. & Campbell, J. P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 9, 14 (2020).
Deo, R. C. Machine Learning in Medicine. Circulation 132, 1920–1930 (2015).
Lo Vercio, L. et al. Supervised machine learning tools: a tutorial for clinicians. J. Neural Eng. 17, https://doi.org/10.1088/1741-2552/abbff2 (2020)
Villoutreix, P. What machine learning can do for developmental biology. Development 148, https://doi.org/10.1242/dev.188474 (2021)
Colling, R. et al. Artificial intelligence in digital pathology: a roadmap to routine use in clinical practice. J. Pathol. 249, 143–150 (2019).
Yousif, M. et al. Artificial intelligence applied to breast pathology. Virchows Arch. 480, 191–209 (2022).
Jovic, S., Miljkovic, M., Ivanovic, M., Saranovic, M. & Arsic, M. Prostate Cancer Probability Prediction By Machine Learning Technique. Cancer Invest 35, 647–651 (2017).
Arvaniti, E. et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 8, 12054 (2018).
Bhattacharya, I. et al. Bridging the gap between prostate radiology and pathology through machine learning. Med Phys. 49, 5160–5181 (2022).
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020).
Duenweg, S. R. et al. Comparison of a machine and deep learning model for automated tumor annotation on digitized whole slide prostate cancer histology. PLoS One 18, e0278084 (2023).
Le, M. H. et al. Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Phys. Med Biol. 62, 6497–6514 (2017).
Li, H. et al. Machine Learning in Prostate MRI for Prostate Cancer: Current Status and Future Opportunities. Diagnostics (Basel) 12, https://doi.org/10.3390/diagnostics12020289 (2022)
Mehralivand, S. et al. Deep learning-based artificial intelligence for prostate cancer detection at biparametric MRI. Abdom. Radio. (NY) 47, 1425–1434 (2022).
Xiang, J. et al. Automatic diagnosis and grading of Prostate Cancer with weakly supervised learning on whole slide images. Comput Biol. Med 152, 106340 (2023).
Bhargava, H. K. et al. Computationally Derived Image Signature of Stromal Morphology Is Prognostic of Prostate Cancer Recurrence Following Prostatectomy in African American Patients. Clin. Cancer Res 26, 1915–1923 (2020).
Safarpoor, A., Kalra, S. & Tizhoosh, H. R. Generative models in pathology: synthesis of diagnostic quality pathology images(†). J. Pathol. 253, 131–132 (2021).
Xu, I. R. L. et al. Generative Adversarial Networks Can Create High Quality Artificial Prostate Cancer Magnetic Resonance Images. J. Pers. Med. 13, https://doi.org/10.3390/jpm13030547 (2023).
Alfano, R. et al. Prostate cancer classification using radiomics and machine learning on mp-MRI validated using co-registered histology. Eur. J. Radio. 156, 110494 (2022).
Bertelli, E. et al. Machine and Deep Learning Prediction Of Prostate Cancer Aggressiveness Using Multiparametric MRI. Front Oncol. 11, 802964 (2021).
Bonekamp, D. & Schlemmer, H. P. Machine learning and multiparametric MRI for early diagnosis of prostate cancer]. Urol. A 60, 576–591 (2021).
Cuocolo, R. et al. Machine learning for the identification of clinically significant prostate cancer on MRI: a meta-analysis. Eur. Radio. 30, 6877–6887 (2020).
Cuocolo, R. et al. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radio. Exp. 3, 35 (2019).
Hosseinzadeh, M. et al. Deep learning-assisted prostate cancer detection on bi-parametric MRI: minimum training data size requirements and effect of prior knowledge. Eur. Radio. 32, 2224–2234 (2022).
Michaely, H. J., Aringhieri, G., Cioni, D. & Neri, E. Current Value of Biparametric Prostate MRI with Machine-Learning or Deep-Learning in the Detection, Grading, and Characterization of Prostate Cancer: A Systematic Review. Diagnostics (Basel) 12, https://doi.org/10.3390/diagnostics12040799 (2022).
Vente, C., Vos, P., Hosseinzadeh, M., Pluim, J. & Veta, M. Deep Learning Regression for Prostate Cancer Detection and Grading in Bi-Parametric MRI. IEEE Trans. Biomed. Eng. 68, 374–383 (2021).
Madabhushi, A. & Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 33, 170–175 (2016).
Freeman, K. et al. Use of artificial intelligence for image analysis in breast cancer screening programmes: systematic review of test accuracy. BMJ 374, n1872 (2021).
Twilt, J. J., van Leeuwen, K. G., Huisman, H. J., Futterer, J. J. & de Rooij, M. Artificial Intelligence Based Algorithms for Prostate Cancer Classification and Detection on Magnetic Resonance Imaging: A Narrative Review. Diagnostics (Basel) 11, https://doi.org/10.3390/diagnostics11060959 (2021).
Van Booven, D. J. et al. A Systematic Review of Artificial Intelligence in Prostate Cancer. Res Rep. Urol. 13, 31–39 (2021).
Bulten, W. et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod. Pathol. 34, 660–671 (2021).
Bulten, W. et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med 28, 154–163 (2022).
Shah, P. et al. Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med 2, 69 (2019).
Weissler, E. H. et al. The role of machine learning in clinical research: transforming the future of evidence generation. Trials 22, 537 (2021).
Banerjee, I. et al. Weakly supervised natural language processing for assessing patient-centered outcome following prostate cancer treatment. JAMIA Open 2, 150–159 (2019).
Yagi, Y. et al. Development of a database system and image viewer to assist in the correlation of histopathologic features and digital image analysis with clinical and molecular genetic information. Pathol. Int 66, 63–74 (2016).
Collet, J. P. Limitations of clinical trials. Rev. Prat. 50, 833–837 (2000).
Cheng, J. Y., Abel, J. T., Balis, U. G. J., McClintock, D. S. & Pantanowitz, L. Challenges in the Development, Deployment, and Regulation of Artificial Intelligence in Anatomic Pathology. Am. J. Pathol. 191, 1684–1692 (2021).
van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med 27, 775–784 (2021).
Krzyszczyk, P. et al. The growing role of precision and personalized medicine for cancer treatment. TECHNOLOGY 06, 79–100 (2018).
Zerbe, N., Hufnagl, P. & Schlüns, K. Distributed computing in image analysis using open source frameworks and application to image sharpness assessment of histological whole slide images. Diagn. Pathol. 6, S16 (2011).
Aeffner, F. et al. Digital Microscopy, Image Analysis, and Virtual Slide Repository. ILAR J. 59, 66–79 (2018).
Van Booven, D. J. et al. Synthetic Genitourinary Image Synthesis via Generative Adversarial Networks: Enhancing Artificial Intelligence Diagnostic Precision. J Pers Med. 14, https://doi.org/10.3390/jpm14070703 (2024)
Hu, X. et al. ProstateGAN: Mitigating Data Bias via Prostate Diffusion Imaging Synthesis with Generative Adversarial Networks. ArXiv abs/1811.05817 (2018).
Liu, Y., Wang, S., Sui, H. & Zhu, L. An ensemble learning method with GAN-based sampling and consistency check for anomaly detection of imbalanced data streams with concept drift. PLoS. One. 19, e0292140 (2024).
Ramya, M., Kirupa, G. & Rama, A. Brain tumor classification of magnetic resonance images using a novel CNN-based medical image analysis and detection network in comparison with AlexNet. J. Popul Ther. Clin. Pharm. 29, e97–e108 (2022).
Kumaraswamy, A. K. & Patil, C. M. Automatic prostate segmentation of magnetic resonance imaging using Res-Net. MAGMA 35, 621–630 (2022).
Yi, S., Wei, Y., Luo, X. & Chen, D. Diagnosis of rectal cancer based on the Xception-MS network. Phys. Med. Biol. 67, https://doi.org/10.1088/1361-6560/ac8f11 (2022)
Park, S. H. et al. Comparison between single and serial computed tomography images in classification of acute appendicitis, acute right-sided diverticulitis, and normal appendix using EfficientNet. PLoS One 18, e0281498 (2023).
Gao, X. Y., Yang, B. Y. & Zhang, C. X. Combine EfficientNet and CNN for 3D model classification. Math. Biosci. Eng. 20, 9062–9079 (2023).
Maguluri, G. et al. Core Needle Biopsy Guidance Based on Tissue Morphology Assessment with AI-OCT Imaging. Diagnostics (Basel) 13, https://doi.org/10.3390/diagnostics13132276 (2023).
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M. & Madabhushi, A. HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides. JCO Clin. Cancer Inf. 3, 1–7 (2019).
Li, X. et al. An artificial intelligence-driven agent for real-time head-and-neck IMRT plan generation using conditional generative adversarial network (cGAN). Med Phys. 48, 2714–2723 (2021).
Yang, S. et al. ShapeEditor: A StyleGAN Encoder for Stable and High Fidelity Face Swapping. Front Neurorobot 15, 785808 (2021).
Bian, Y., Wang, J., Jun, J. J. & Xie, X. Q. Deep Convolutional Generative Adversarial Network (dcGAN) Models for Screening and Design of Small Molecules Targeting Cannabinoid Receptors. Mol. Pharm. 16, 4451–4460 (2019).
Pagella, F. et al. FID Score: an effective tool in Hereditary Haemorrhagic Telangiectasia - related epistaxis. Rhinology 58, 516–521 (2020).
Suekane, S. et al. Percentages of positive cores, cancer length and Gleason grade 4/5 cancer in systematic sextant biopsy are all predictive of adverse pathology and biochemical failure after radical prostatectomy. Int J. Urol. 14, 713–718 (2007).
Lattouf, J. B. & Saad, F. Gleason score on biopsy: is it reliable for predicting the final grade on pathology?. BJU Int 90, 694–698 (2002). discussion 698-699.
Chen, C. B., Wang, Y., Fu, X. & Yang, H. Recurrence Network Analysis of Histopathological Images for the Detection of Invasive Ductal Carcinoma in Breast Cancer. IEEE/ACM Trans. Comput Biol. Bioinform 20, 3234–3244 (2023).
Chen, C. B., Yang, H. & Kumara, S. Recurrence network modeling and analysis of spatial data. Chaos 28, 085714 (2018).
Yang, H., Chen, C. B. & Kumara, S. Heterogeneous recurrence analysis of spatial data. Chaos 30, 013119 (2020).
Zhang, L. et al. (IEEE).
Raja, L., Merline, A. & Ganesan, R. International Journal of Advanced Information and Technology. 2 (2013).
Kumar, K., Naga Sai Ram, K. N., Kiranmai, K. S. S. & Harsha, S. Denoising of Iris Image Using Stationary Wavelet Transform. 2018 Second International Conference on Inventive Communication and Computational Technologies (ICICCT), 1232-1237 (2018).
Wang, Y. & Chen, C.-B. Recurrence Quantification Analysis for Spatial Data. IIE Annual Conference. Proceedings, 1-6 (2022).
Shukla, P., Verma, A., Abhishek, Verma, S. & Kumar, M. Interpreting SVM for medical images using Quadtree. Multimed. Tools Appl 79, 29353–29373 (2020).
Baia, J., Zhao, X. & Chenb, J. Z.
Loher, P. & Karathanasis, N. Machine Learning Approaches Identify Genes Containing Spatial Information From Single-Cell Transcriptomics Data. Front Genet 11, 612840 (2020).
Zou, H., Hastie, T. & Tibshirani, R. Sparse Principal Component Analysis. J. Computational Graph. Stat. 15, 265–286 (2006).
Song, F., Guo, Z. & Mei, D. (IEEE).
Lin, W., Pixu, S., Rui, F. & Hongzhe, L. Variable selection in regression with compositional covariates. Biometrika 101, https://doi.org/10.1093/biomet/asu031 (2014)
Zou, H. & Hastie, T. Regularization and Variable Selection Via the Elastic Net. J. R. Stat. Soc. Ser. B: Stat. Methodol. 67, 301–320 (2005).
Matsui, H. & Konishi, S. Variable selection for functional regression models via the L1 regularization. Comput. Stat. Data Anal. 55, 3304–3310 (2011).
Wolfe, P. J., Godsill, S. J. & Ng, W.-J. Bayesian Variable Selection and Regularization for Time–Frequency Surface Estimation. J. R. Stat. Soc. Ser. B: Stat. Methodol. 66, 575–589 (2004).
Al Sudani, Z. A. & Salem, G. S. A. Evaporation Rate Prediction Using Advanced Machine Learning Models: A Comparative Study. Advances in Meteorology, 1-13. https://doi.org/10.1155/2022/1433835 (2022)
Niazi, M. K. K. et al. Visually Meaningful Histopathological Features for Automatic Grading of Prostate Cancer. IEEE J. Biomed. Health Inf. 21, 1027–1038 (2017).
Erratum to “Visually Meaningful Histopathological Features for Automatic Grading of Prostate Cancer”. IEEE J Biomed Health Inform 21, 1473-1474 (2017).
Hou, L. et al. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. Proc. IEEE Comput Soc. Conf. Comput Vis. Pattern Recognit. 2016, 2424–2433 (2016).
Ren, J. et al. Computer aided analysis of prostate histopathology images Gleason grading especially for Gleason score 7. Annu Int Conf. IEEE Eng. Med Biol. Soc. 2015, 3013–3016 (2015).
Fauzi, M. F. et al. Classification of follicular lymphoma: the effect of computer aid on pathologists grading. BMC Med Inf. Decis. Mak. 15, 115 (2015).
Kothari, S., Phan, J. H., Young, A. N. & Wang, M. D. Histological image classification using biologically interpretable shape-based features. BMC Med Imaging 13, 9 (2013).
Kong, J. et al. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS One 8, e81049 (2013).
Dundar, M. M. et al. Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans. Biomed. Eng. 58, 1977–1984 (2011).
Sertel, O. et al. Computer-aided Prognosis of Neuroblastoma on Whole-slide Images: Classification of Stromal Development. Pattern Recognit. 42, 1093–1103 (2009).
Kong, J. et al. Computer-assisted grading of neuroblastic differentiation. Arch. Pathol. Lab Med 132, 903–904 (2008).
Koh, D. M. et al. Artificial intelligence and machine learning in cancer imaging. Commun. Med (Lond.) 2, 133 (2022).
Munjal, R., Arif, S., Wendler, F. & Kanoun, O. Comparative Study of Machine-Learning Frameworks for the Elaboration of Feed-Forward Neural Networks by Varying the Complexity of Impedimetric Datasets Synthesized Using Eddy Current Sensors for the Characterization of Bi-Metallic Coins. Sensors (Basel) 22, https://doi.org/10.3390/s22041312 (2022)
Celi, L. A. et al. Sources of bias in artificial intelligence that perpetuate healthcare disparities-A global review. PLOS Digit Health 1, e0000022 (2022).
Shreve, J. T., Khanani, S. A. & Haddad, T. C. Artificial Intelligence in Oncology: Current Capabilities, Future Opportunities, and Ethical Considerations. Am. Soc. Clin. Oncol. Educ. Book 42, 1–10 (2022).
Khan, B. et al. Drawbacks of Artificial Intelligence and Their Potential Solutions in the Healthcare Sector. Biomed Mater Devices, 1-8. https://doi.org/10.1007/s44174-023-00063-2 (2023)
Hodas, N. O. & Stinis, P. Doing the Impossible: Why Neural Networks Can Be Trained at All. Front Psychol. 9, 1185 (2018).
Pozzi, M. et al. Generating and evaluating synthetic data in digital pathology through diffusion models. Sci. Rep. 14, 28435 (2024).
Kazerouni, A. et al. Diffusion models in medical imaging: A comprehensive survey. Med Image Anal. 88, 102846 (2023).
Müller-Franzes, G. et al. A multimodal comparison of latent denoising diffusion probabilistic models and generative adversarial networks for medical image synthesis. Sci. Rep. 13, 12098 (2023).
Kadry, K., Gupta, S., Nezami, F. R. & Edelman, E. R. Probing the limits and capabilities of diffusion models for the anatomic editing of digital twins. NPJ Digit Med 7, 354 (2024).
Chen, M., Mei, S., Fan, J. & Wang, M. Opportunities and challenges of diffusion models for generative AI. Natl Sci. Rev. 11, nwae348 (2024).
Cookson, M. S., Fleshner, N. E., Soloway, S. M. & Fair, W. R. Correlation between Gleason score of needle biopsy and radical prostatectomy specimen: accuracy and clinical implications. J. Urol. 157, 559–562 (1997).
Loghin, A., Popelea, M. C., Nechifor-Boila, I. A. & Borda, A. Systematic Biopsy vs. Prostatectomy: Evaluating Correlations and Grading Discrepancies in Prostate Cancer. Cureus 16, e68075 (2024).
Quintana, G. I. et al. Exploiting Patch Sizes and Resolutions for Multi-Scale Deep Learning in Mammogram Image Classification. Bioengineering (Basel) 10, https://doi.org/10.3390/bioengineering10050534 (2023).
Shen, X., Lin, L., Xu, X. & Wu, S. Effects of Patchwise Sampling Strategy to Three-Dimensional Convolutional Neural Network-Based Alzheimer’s Disease Classification. Brain Sci 13. https://doi.org/10.3390/brainsci13020254 (2023)
Ashraf, M., Robles, W. R. Q., Kim, M., Ko, Y. S. & Yi, M. Y. A loss-based patch label denoising method for improving whole-slide image analysis using a convolutional neural network. Sci. Rep. 12, 1392 (2022).
Faryna, K. et al. Evaluation of Artificial Intelligence-Based Gleason Grading Algorithms “in the Wild. Mod. Pathol. 37, 100563 (2024).
Singhal, N. et al. A deep learning system for prostate cancer diagnosis and grading in whole slide images of core needle biopsies. Sci. Rep. 12, 3383 (2022).
Acknowledgements
This research was funded by the Scott R. MacKenzie Foundation (Grant No. AWD-009374). Additional financial support was provided by the University of Miami U-LINK (Grant No. PG013269) and the Provost Research Award (Grant Nos. PG012860 and PG015498). S.P. received additional funding from the National Institutes of Health/National Cancer Institute (NIH/NCI) (Grant Nos. U01CA239141, 1R01CA272766) and the Paps Corps Champions for Cancer Research. Primary financial support was provided by the Sylvester Cancer Center, the University of Miami, and the Desai and Sethi Institute of Urology. We extend our deepest gratitude to all patients whose willingness to participate made this study possible. We are also grateful to all principal investigators, sub-investigators, and local center staff for their dedication and commitment to patient recruitment. We acknowledge the members of the steering committee and the data monitoring and ethics committee for their valuable contributions. Special thanks to Dr. Dipen Parekh for providing critical insights from a patient perspective. The support of The Cancer Genome Atlas (TCGA) was instrumental to the successful execution of this study, and we appreciate the contributions of all its past and present staff. Finally, we extend our sincere appreciation to the laboratory team for their significant contributions to this research.
Author information
Authors and Affiliations
Contributions
H.A. was the chief investigator. D.V.B. and H.A. designed the study and developed the protocol. O.K., F.I., S.M., A.N., V.S., and A.N. carried out the pathological analysis plan. H.A. and D.V.B. coordinated the central modeling investigations. C.B.C. developed Spatial quantification methods. C.B.C. and D.V.B. validated the quantifications. H.A., O.K., and S.P. coordinated the data collection. D.V.B. and H.A. interpreted the data. H.A. and D.V.B. developed the first drafts of the manuscript. C.B.C., O.K., R.Q., S.P., S.M., V.S., A.N., F.I., A.B. and J.M.H. have accessed and verified all the data in the study. All authors had access to all the data reported in the study. All authors contributed to the review and amendments of the manuscript for important intellectual content and approved this final version for submission. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
Disclosure of Patent Information: The authors wish to inform that the technology presented in this study is part of a provisional patent application that has been filed with the United States Patent and Trademark Office (USPTO). The application has been assigned Serial No. 63/598,207 and was filed on November 13, 2023. The patent application is currently pending. Some of the authors of this paper are listed as inventors in the patent application. This patent filing may constitute a potential conflict of interest, and this statement serves to disclose this relationship in the interest of full transparency. Joshua M. Hare reports having a patent for cardiac cell-based therapy and holds equity in Vestion Inc., and maintains a professional relationship with Vestion Inc. as a consultant and member of the Board of Directors and Scientific Advisory Board. Vestion Inc. did not play a role in the design, conduct, or funding of the study. Dr. Joshua Hare is the Chief Scientific Officer, a compensated consultant, and a board member for Longeveron Inc. and holds equity in Longeveron. Dr. Hare is also the co-inventor of intellectual property licensed to Longeveron. Longeveron did not play a role in the design, conduct, or funding of the study. The University of Miami is an equity owner in Longeveron Inc., which has licensed intellectual property from the University of Miami.
Ethics statement
This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. Ethical approval was obtained from the Institutional Review Board (IRB) of the University of Miami Miller School of Medicine, Miami, FL (IRB Protocol Number: 20140372). The MAST Trial was registered on ClinicalTrials.gov (Identifier: NCT02242773). Informed consent was obtained from all participants prior to their inclusion in the study. Additionally, external datasets, including those from The Cancer Genome Atlas (TCGA), Radboud University Medical Center, and Karolinska Institute (PANDA challenge), were used in compliance with their respective data use agreements. All data were anonymized to ensure participant confidentiality and privacy.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Van Booven, D.J., Chen, CB., Kryvenko, O.N. et al. Mitigating bias in prostate cancer diagnosis using synthetic data for improved AI driven Gleason grading. npj Precis. Onc. 9, 151 (2025). https://doi.org/10.1038/s41698-025-00934-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00934-5