Abstract
Survivors of early-onset colorectal cancer (EOCRC, i.e., diagnosed before age 50) are likely to experience recurrence after completing treatment. In this international, multi-centric, phase I-II-III EDRN biomarker study, we identified a panel of tumor-derived biomarkers of EOCRC recurrence. We then trained and independently validated a machine learning model (XGBoost) to predict 5-year recurrence-free and overall survival (RFS and OS) of patients with stage I-III EOCRC. Patients with “low-risk” EOCRC demonstrated statistically higher rates of 2-, 5-, and 10 year RFS in both the training cohort (51.0 vs. 92.4%; 34.4% vs. 92.4%; 25.8% vs. 92.4%, respectively; p < 0.0001) and the validation cohort (78.9% vs. 100.0%; 75.0% vs. 100.0%; 75.0% vs. 100.0%, respectively; p = 0.0019). We also report a significant reduction in both over-treatment and missed recurrences compared to current clinically available options. This tissue-based, machine learning-powered assay was prognostic of long-term RFS and OS outcomes after curative-intent treatment of EOCRC (ENCORE was first registered on ClinicalTrial.gov [ID: NCT06271980] on February 15th, 2024).

Similar content being viewed by others
Introduction
Colorectal cancer (CRC) once predominantly affected older individuals but, in recent years, has witnessed a progressive increase in incidence among young adults1. Once rare, early-onset CRC (EOCRC) now constitutes 15−20% of all newly diagnosed CRC cases and stands as the first cause of cancer-related death in young men in the US and the second for young women2,3. In the wake of the increasing incidence, the growing population of EOCRC survivors introduces distinctive clinical challenges4,5.
EOCRC survivors face significant risk of recurrence after primary treatment6,7,8,9,10,11, a risk that is elevated compared to late-onset CRC12,13,14,15,16,17 and can manifest years after initial therapy15,18,19,20. These considerations have prompted a trend toward offering more aggressive therapy or surveillance, a practice not yet substantiated by evidence15,21,22,23. Clinical guidelines recognize this elevated risk but also acknowledge that intensified surveillance might constitute overtreatment24,25,26. To address these gaps in knowledge, we developed a predictive model of post-resection tumor recurrence for the growing population of EOCRC survivors, leveraging state-of-the-art machine learning (ML) driven by biological and clinical data.
ML involves fitting predictive models to data for pattern recognition27. Ensemble classifiers, like random forests, combine multiple weak learners (decision trees) to boost accuracy27. However, the boosting mechanism may reduce interpretability, a crucial aspect for assessing predictions, detecting bias, extracting knowledge, validating models, and generating hypotheses28. XGBoost (eXtreme Gradient Boosting) is a tree-based and boosted ensemble classifier that overcomes these limitations, allows the evaluation of the importance of each predictor, and has a reduced sensitivity to feature scaling and normalization28,29. MiRNAs, non-coding single-stranded RNAs regulating gene expression and various cellular processes, influence CRC growth, progression, and treatment response and have proven promising as cancer biomarkers30,31,32,33. Notably, our group has reported key genetic and epigenetic alterations in EOCRC34,35,36,37,38,39,40,41, including a recent 4-miRNA liquid biopsy assay for its early detection42.
In this research effort, we leveraged XGBoost to predict recurrence-free and overall survival (RFS and OS, respectively) in two independent EOCRC cohorts followed up for over 1000 person-years. We first identified a panel of candidate biomarkers with RNA sequencing. Thereafter, we developed, trained, and independently validated a tissue-based RT-qPCR assay to predict RFS and OS outcomes up to 10 years after treatment.
Results
Cohort characteristics
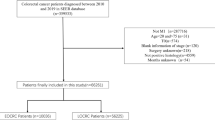
This study enrolled 177 survivors of stage I-III EOCRC from five medical centers who were followed-up for 1084.9 person-years (Supplementary Table 1). Forty-two patients developed a CRC recurrence during follow-up, while 135 remained recurrence-free. The biomarker discovery cohort comprised 20 patients with stage II-III EOCRC. The remaining patients were assigned to the training cohort (Clinic cohort 1, all from Hospital Clinic Barcelona, Spain; n = 88) or the external and independent validation cohort (Clinic cohort 2, from Kumamoto University, Tokyo Medical and Dental University (TMDU), and University of Tokyo, all in Japan; n = 69). Patients received a median follow-up of 85.4 months (95% confidence interval [CI95%]: 68.9–104.8).
Discovery of the 10-microRNA panel
Conducting genome-wide, high-throughput small RNA-sequencing on FFPE-derived RNA, we identified 35 differentially expressed miRNA candidates (18 up-regulated and 17 down-regulated, Fig. 1A). After univariate LASSO-based Cox regression and AUC-based ranking, 10 best-performing candidates were selected: hsa-miR-365a-3p, hsa-miR-410-3p, hsa-miR-654-3p, hsa-miR-125b-5p, hsa-miR-125b-2-3p, and hsa-miR-99a-5p were up-regulated in cases, while hsa-let-7g-5p, hsa-miR-142-3p, hsa-miR-15b-3p, and hsa-miR-30e-5p were down-regulated (Fig. 1B). Applying the unweighted pair-group centroid method for unsupervised clustering, only the development of recurrence co-segregated with unsupervised clustering, while other clinical characteristics did not (Fig. 1C). Finally, the hazard ratio (HR) of recurrence for each miRNA was increased for six candidates and reduced for four (Fig. 1D). Interestingly, we also observed that these microRNAs collectively regulated several shared target genes as a network (Supplementary Fig. 1A). In fact, functional enrichment analysis further demonstrated that these microRNAs were involved in multiple cancer-related pathways, including neo-angiogenesis, NFκB, TGFβR, VEGF, and other inflammatory pathways (Supplementary Fig. 1B).

A The volcano plot displays the differential expression of microRNAs between cases that experienced recurrence and those that did not. MicroRNAs are color-coded based on their level of significance. B The ridgeline plot visualizes the distribution of expression levels for the 10 best-performing candidates. Each ridgeline represents the expression density of a specific microRNA across the patient samples. C The heatmap displays the expression levels of the top 10 prognostic microRNAs across the patient samples. Unsupervised clustering was applied to group patients based on their microRNA expression profiles. The annotation bar above the heatmap indicates the recurrence status (recurrent vs. non-recurrent) and other clinical characteristics of the patients. D This Forrest plot shows the hazard ratio of recurrence for each of the 10 best-performing microRNA candidates. CI Confidence intervals, FDR False-discovery rate, MSI Microsatellite instability.
The discovery phase identified a panel of 10 candidate miRNAs associated with EOCRC recurrence using RNA sequencing.
Development of the ENCORE assay
Upon transitioning our sequencing efforts into an RT-qPCR-based assay, we fit a XGBoost ML model on the miRNA expression levels from the first clinic cohort. The resulting tissue-based assay, “ENCORE,” relied primarily on the differential expression of hsa-let-7g-5p, hsa-miR-365a-3p, and hsa-miR-410-3p and excluded one candidate (hsa-miR-99a-5p, Fig. 2A). Because gain and cover values (Supplementary Table 2) are aggregated measures, we explored ENCORE inner workings with patient-specific SHAP values. SHAP values for hsa-let-7g-5p consistently deviated from 0 for almost all patients. SHAP values for hsa-miR-365a-3p and hsa-miR-410-3p mostly separated from the 0-value line, but their impact was not as pronounced in the positive direction, which implies that their contribution was selective, affecting most patients but not all (Fig. 2B). This tissue-based assay demonstrated high accuracy in predicting 5 year RFS with an AUROC value of 90.1% (CI95% = 83–97%), 84.0% sensitivity (CI95% = 68-96%), 81.0% specificity (CI95% = 70–91%), and 81.8% accuracy (CI95% = 76-93%) (Fig. 2C). Interestingly, patients with RFS < 5 years had higher ENCORE values (Fig. 2D).

A Simplified decision tree of the ENCORE decision forest. This panel presents the ensemble of trees that constitute the ENCORE forest model. This simplified view illustrates the hierarchical decision-making process of the algorithm, showing how different microRNA expression levels (and potentially combinations thereof) lead to risk stratification. The nodes represent decision points based on microRNA levels, and the branches represent the possible outcomes, ultimately leading to a predicted risk score or risk group. B This beeswarm plot visualizes the SHAP values of each microRNA included in the ENCORE model. SHAP values quantify the contribution of each microRNA to the model’s prediction for individual patients. Points further from zero on the x-axis indicate a greater impact on the prediction (either increasing or decreasing the risk score). The color gradient of the points corresponds to the measured expression level of the corresponding microRNA, providing insight into how the expression level influences the prediction. C AUROC of ENCORE. D The raincloud plots with superimposed box and whisker plots provide a comprehensive visualization of the distribution of ENCORE-derived risk scores in two groups of EOCRC survivors: those who experienced recurrence and those who remained recurrence-free survivors. AUROC Area under the receiver-operating characteristic curve, CI Confidence intervals, ENCORE Early Onset Colorectal Cancer Recurrence, SHAP SHapley Additive exPlanations.
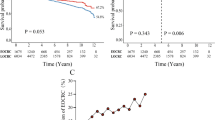
We then evaluated the survival characteristics beyond the 5 year RFS. Over 20+ years of follow-up, there was a single case of disease recurrence among patients with “low-risk” tumors. In fact, “low-risk” survivors had significantly higher OS probabilities than “high-risk” survivors (OS differences at 2, 5, and 10 years: +18.6% [CI95% = 6.6–35.4%], +33.8% [CI95% = 13.1–57.9%], and +45.8% [CI95% = 21.0–69.5%], respectively). In fact, patients classified as “high-risk” had higher rates of recurrence (2 year RFS: 51.0 vs. 92.4%, p < 0.0001; 10 year RFS: 25.8% vs. 92.4%, p < 0.0001, Fig. 3A) and death (2 year OS = 81.4% vs. 100%, p = 0.0012; 5 years OS = 64.1% vs. 97.9%, p = 0.0002; 10 year OS = 52.1% vs. 97.9%, p < 0.0001, Fig. 3B).

Kaplan Meier recurrence-free (A) and overall (B) survival curves stratified by ENCORE status: low-risk (green) vs. high-risk (purple). The statistical significance of the difference between the two survival curves is assessed using a log-rank test. The number of patients at risk in each group at various time points is below the graph. CI Confidence intervals, ENCORE Early Onset Colorectal Cancer Recurrence, OS Overall survival, RFS Recurrence-free survival.
Independent and external validation of the ENCORE assay
Transitioning the assay to the clinical cohort 2 for independent and external validation (Supplementary Table 3), the assay showed a sustained sensitivity for both recurrence and mortality, with no statistically significant decline in performance. Recurrent cases demonstrated higher ENCORE values than non-recurrent cases (p = 0.008, Supplementary Fig. 2A, B). Importantly, ENCORE could stratify survivors with statistically significant differences both in the short and long term (at 2 years, +21.1% [CI95%: 5.9; 40.3%]; and 10 years, +25% [CI95%: 4.9; 50.3], respectively) (Fig. 4A). We also observed a statistically significant difference in OS stratification at 5 and 10 years ( + 16% [CI95%: 0.2% to 37.2%], and +23% [CI95%: 4.0 to 46.0%], respectively), although not yet at two (7.4% [CI95%: -3.8 to +23.0%], Fig. 4B and Table 1).

A, B Kaplan-Meier recurrence-free (A) and overall (B) survival curves in the independent cohort, stratified by ENCORE status: high-risk (red) vs. low-risk (blue). The statistical significance of the difference between the two survival curves is assessed using a log-rank test. The number of patients at risk in each group at various time points is below the graph. CI Confidence intervals, ENCORE Early Onset Colorectal Cancer Recurrence, OS Overall survival, RFS Recurrence-free survival.
We finally investigated whether additional clinical or pathological factors could further improve the performance of this tissue-based test. We performed univariate and multivariate Cox proportional hazard analysis in each cohort separately and observed that patients with a “low-risk” ENCORE status had a reduced risk of recurrence at both univariate (HR 0.072 [CI95% = 0.02–0.21]) and multivariate analysis (HR 0.06 [CI95% = 0.01–0.34], p < 0.001, Supplementary Table 4).
Decision curve and net benefit analysis
The ENCORE assay demonstrated higher net benefit values than other surveillance strategies based on clinical characteristics. At the 25% high-risk threshold used to dichotomize ENCORE assay results, the ENCORE-based approach maintained a more favorable net benefit than all other strategies (Fig. 5A). At the same 25% risk threshold, 37% of the EOCRC survivors would be considered “high-risk” (CI95% = 33.0–44.3%), with a low false-positive rate (19%, CI95% = 11-30%, Fig. 5B). We finally evaluated the recurrence risk among the ENCORE “low-risk” survivors. At the same 25% risk threshold, 63% of survivors would be classified as “low-risk”. For these “low-risk” survivors the recurrence risk was significantly lower than that of all EOCRC survivors, even among stage I/II EOCRC survivors, whose recurrence risk is already low (Fig. 5C). Thus, this assay may complement clinical-feature-based strategies to assess the risk of recurrence after EOCRC treatment.

A Unstandardized net benefit of a surveillance strategy based on ENCORE vs. clinical characteristics. The y-axis represents the net benefit of each recurrence prediction model. The x-axis represents the probability threshold for considering a patient high-risk and intervening. The plot compares the net benefit of using a strategy based on the ENCORE assay against strategies based on clinical characteristics alone and the default strategies of considering all or no patients at risk of recurrence. The model with the highest net benefit across a clinically relevant range of probability thresholds has the greatest clinical utility. The net benefit is calculated by weighing the benefits of true positives against the harms of false positives. B Clinical Impact of a strategy based on ENCORE (circles represent the 25% high-risk threshold). The x-axis represents the probability threshold used to classify patients as high-risk. The curve shows the number of events (recurrences) captured by the strategy (true positives) and the number of patients unnecessarily classified as high-risk (false positives) at different risk thresholds. C Recurrence risks in survivors classified as “low-risk” compared to all EOCRC survivors. CI Confidence intervals, ENCORE Early Onset Colorectal Cancer Recurrence, EOCRC Early-onset colorectal cancer.
Discussion
In this translational study, the ENCORE assay was prognostic of RFS and OS outcomes of EOCRC survivors in two independent cohorts. This tissue-based assay is powered by machine learning and, to the best of our knowledge, the longest follow-up of EOCRC survivors to date ( > 1000 person-years). It was developed and validated independently in two international, multi-centric, prospective cohorts to identify the risk of recurrence and death.
The current guidelines exhibit a divided perspective on the recurrence risk in EOCRC, posing a challenge for survivors’ management24,25,26. EOCRC survivors face both a higher risk than stage-matched LOCRC survivors and longer lifespan after treatment15,18,19,20, leading to an accumulation of risk over time. Diverse investigations into post-surgical surveillance strategies have yielded varied and sometimes conflicting outcomes. A US study identified the recurrence risk peaking at 24 months21. Yet, this mono-centric study had a median follow-up of only 48.1 months21. A recent UK study, with a 10.1 year follow-up, found that most metachronous CRCs occurred after over 3 years20. Moreover, an IDEA meta-analysis from six randomized controlled trials revealed higher 3 year recurrence and 5 year mortality rates among patients with EOCRC compared to stage-matched late-onset CRC patients. This trend persisted despite their receipt of more intensive treatment regimens (which led to increased gastrointestinal toxicity with no survival benefit)15. In our study most recurrence events occurred in the initial 24 months, but some occurred at later time points. Our findings underscore the importance of extended timeframes and caution against designating a patient as “cured” after the first years, because metachronous CRC may develop several years after primary EOCRC treatment.
Our study, conducted within the EDRN framework, is subject to the inherent limitations of observational studies, including potential biases related to patient selection, data quality, and treatment heterogeneity within our retrospective cohorts. Furthermore, we acknowledge a lack of detailed clinicopathologic and treatment data limited our ability to assess interactions between ENCORE results and treatment outcomes. The extended follow-up represents both a strength of our collaborative effort and a limitation, because treatment paradigms evolved over this study’s follow-up period. This temporal variability may limit the generalizability of our findings, particularly in the context of minimal residual disease assessment with contemporary approaches43. The temporal variability in treatment may confound the association between our biomarker signature and recurrence risk. Additionally, despite multi-institutional collaboration, the relatively modest sample size, especially in the second cohort, may influence the statistical power of our analyses. Moving forward, it will be crucial to validate the prognostic and predictive value of ENCORE in prospective clinical trials, including interventional studies, to determine whether ENCORE status, CMS status, other tools (for example, ColoPrint or Oncotype DX), or combinations thereof predict any benefit from specific treatments, thereby further enhancing its clinical utility32,44,45,46,47. On the other hand, it should be noted that while the ENCORE study population may have sample size limitations, it represents the largest cohort to date focused on EOCRC recurrence patterns and it comprises individuals with EOCRC and a long follow-up period who were enrolled during a time of lower EOCRC prevalence and incidence. Therefore, even with its limits, this cohort represents a unique and valuable resource to study EOCRC recurrence patterns. Moreover, the nature of our cohorts, encompassing both rectal and colon cancers, warrants consideration: while rectal and colon cancers have distinct management strategies, our findings appear to be applicable to both. Finally, given the predominantly distal tumor presentation in EOCRC in general and in our study population specifically, the extrapolation of our results to patients with proximal EOCRC requires caution.
We would like to highlight a few unique strengths of this study. The ENCORE assay was prognostic of long-term outcomes for both RFS and OS endpoints. The second cohort reaffirmed the replicability of our findings48. Furthermore, both cohorts underwent extensive follow-up, providing sufficient time to detect statistically significant differences in RFS and OS outcomes over a 10 year period. Moreover, the utilization of an explainable machine learning model allowed for a comprehensive understanding, highlighting the pivotal factors influencing the model—three main candidates for coarse computations and six for fine-tuning, all collectively regulating gene networks related to colorectal carcinogenesis and some previously associated with EOCRC specifically17. Lastly, although we acknowledge the potential for RNA degradation in FFPE and the value of future validation on alternative tissue types, the use of FFPE tissues makes this approach intrinsically scalable, potentially benefiting a sizable number of EOCRC survivors.
In conclusion, this study represents the first effort to tailor a surveillance strategy for the unique and expanding population of EOCRC survivors. Leveraging biological data and machine learning, we investigated the risk of recurrence and death following curative-intent resection in a large, multi-centric, international cohort of patients with sporadic EOCRC, independently and externally validating our findings. This assay may not only heighten recurrence risk awareness in those identified as having “high-risk” EOCRC but also mitigate concerns, anxiety, and financial burdens in survivors with “low-risk” EOCRC.
Methods
Study design
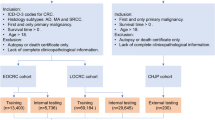
The ENCORE (Early oNset COlorectal cancer REcurrence prediction) study was an international, multi-centric, multi-phase, REMARK- and CONSORT-compliant (Supplementary Tables 5 and 6) biomarker study covering EDRN-defined phases I, II, and III (Supplementary Fig. 3). Briefly, EDRN phase I is aimed at the discovery of a panel of biomarkers that are associated with the outcome of interest (EOCRC recurrence, in this case); EDRN phase II uses these biomarkers to develop a clinical-level test, the performance of which is then confirmed during the EDRN phase III leveraging an independent and external cohort. There was no overlap of patients across study phases. The study was approved by the Institutional Review Board of each participating institution (IRB No. 23228), conducted in accordance with the Declaration of Helsinki, and it was registered and completed on clinicaltrials.gov (https://www.clinicaltrials.gov/study/NCT06271980, first registered on February 15th, 2024, where the protocol is available). All participants provided written informed consent.
Study population, settings, and specimens
This study involved adult participants who were diagnosed with stage I, II, or III CRC (TNM classification, 8th edition) diagnosed before age 50. They underwent standard diagnostic, staging, and therapeutic procedures per local guidelines, received stage-specific curative-intent resection (with or without systemic therapy, as appropriate), and were confirmed cancer-free survivors at the time of study inclusion. Exclusion criteria comprised hereditary CRC syndromes (identified through genetic testing) and inflammatory bowel diseases.
Patients were enrolled from five institutions in Europe and Asia (Supplementary Fig. 4), and, given the unpredictable nature of recurrence development, cases and controls were not initially matched, but subsequent analyses incorporated stage-specific stratification (as appropriate). Only the biomarker discovery phase was intentionally devised to have a 50:50 representation of recurrent and non-recurrent cases (age- and sex-matched, Supplementary Fig. 5).
The specimens were formalin-fixed and paraffin-embedded (FFPE) tissues. Initial pathological analyses, including tumor grading, lymphatic invasion, and vascular invasion, were conducted at the recruiting centers. Subsequently, all samples were sequentially collected at participating sites and gathered at the principal study site for centralized analyses.
Definitions and study endpoints
RFS was defined as the duration from curative-intent treatment to the first disease recurrence (locally assessed) or death from any cause. OS was defined as the time from curative-intent treatment to death from any cause. Survival was censored at the last follow-up. Patients with RFS ≥ 60 months were dichotomously categorized as “non-recurrent” (or controls), while those with RFS < 60 months were considered “recurrent” (or cases). The co-primary endpoints of this study were RFS and OS survival differences by virtue of our assay, with statistically significant differences at 2, 5, and 10 years. Independent associations with RFS were assessed using univariate and multivariate Cox proportional hazards analysis.
Laboratory procedures
For the biomarker discovery phase, total RNA was isolated from 10-μm-thick FFPE specimens by microdissection from cancer cell−rich areas ( ≥ 75% of tumor cells) and using AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany). Construction of next-generation sequencing libraries for miRNAs from tissue was performed using a modified protocol for the Truseq Small RNA Kit (Illumina) with 200 ng of total RNA input. The quality of individual libraries was assessed using a High Sensitivity DNA Kit (Agilent). Libraries were size selected individually ( ~ 148 nt) by gel electrophoresis using a Pippin HT instrument (Sage Science). The efficiency of size selection was assessed using a High Sensitivity DNA Kit. Libraries were equimolar-pooled; pooled libraries were quantitated via qPCR using a KAPA Library Quantification Kit, Universal (KAPA Biosystems) prior to sequencing on an Illumina HighSeq 2500 with single-end 35-base read lengths at an average of 10 million reads per sample. Illumina small RNA-seq 3’ adapters were trimmed (cutadapt), and all retained sequences contained high-quality scores peaking at 22 nt. Preprocessed reads were aligned to the human genome build 38 and annotated using GENCODE. Candidate miRNAs were selected based on differential gene expression ( | Log2(Fold-Change)| >0.5 and p-values < 0.05) using DESeq2, Cox-LASSO regression, and AUC-based ranking.
For the two clinical cohorts, complementary DNA was synthesized from tissue-isolated total RNA using the TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA). In both cohorts, miRNA expression was assessed by RT-qPCR on a StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA) using the TaqMan miRNA probes (ThermoFisher Scientific, Supplementary Table 7). Expression levels were evaluated using Applied Biosystems’ Real-Time PCR System Software, and the relative abundance normalized to the expression levels of U6b as an internal control using the by 2-ΔCt method and a log10 transformation. ΔCt refers to the difference of Ct values between the transcript of interest and the normalizer.
Statistical analyses
All statistical analyses were computed in R. The diagnostic assay, designated ‘ENCORE,’ leverages XGBoost, a prominent ensemble learning algorithm. XGBoost employs the gradient boosting framework and iteratively combines a series of weak decision tree learners to construct a more robust predictive model. This approach is particularly well-suited for high-dimensional and complex datasets, owing to its efficient computational performance, extensive hyperparameter optimization capabilities, and robust handling of missing data. Our ML model was therefore trained for a maximum of 5000 rounds. To mitigate overfitting during training, we implemented several key strategies: restricting the maximum tree depth to three splits to prevent over-complex trees, imposing a 75% subsample parameter to train each tree on a random subset of the data and promote generalization, and employing a high-pruning strategy (γ = 8) to penalize the addition of new nodes and prune less informative splits. This gamma parameter specifies the minimum loss reduction required for XGBoost to make a further split, leading the algorithm to make splits up to a maximum depth of three and then prune the tree backward by removing any splits that do not significantly improve the model’s performance. Furthermore, we monitored the performance of the training data using cross-validation and would have implemented early stopping if performance began to degrade before reaching the maximum number of rounds. This performance was measured by multi-class logarithmic loss, where lower values indicate better performance. After training in the clinical cohort 1, the ENCORE assay was fully locked to prevent modifications and applied to the second cohort for external and independent validation. Cohorts were stratified into “high-risk” and “low-risk” (Youden’s index). RFS and OS differences at 2, 5, and 10 years were evaluated with Kaplan-Meier curves, with censoring at the last follow-up, and statistical significance evaluated with the log-rank test. To assess independent associations with RFS, univariate and multivariate Cox proportional hazard regressions were performed. 95% confidence intervals for proportions were computed with the Wilson method, while confidence intervals for the ROC curves were estimates with 2000 stratified bootstrap replicates. Statistical significance was defined at p < 0.05 for all analyses and was tested by the paired Wilcoxon method for pairwise comparisons, by the student t-test for two groups (paired or unpaired, as appropriate), by ANOVA for multiple groups, and by log-rank test for survival analyses.
Patient and public involvement
The patients and the public were not involved in formulating the research question(s), designing the study, conducting the experiments, evaluating the outcome measures, or in the decision to publish. However, the authors wish to express their appreciation to the study participants and their families.
Data availability
Data collected for the study, including de-identified participant data and the code, will be made available to others at publication via a signed data access agreement and at the discretion of the investigators’ approval of the proposed use of such data.
Code availability
The code generated for the study will be made available to others at publication via a signed code access agreement and at the discretion of the investigators’ approval of the proposed use of the code.
Abbreviations
- AUC :
-
Area under the curve
- AUROC :
-
Area under the receiver-operating characteristic curve
- CI :
-
Confidence intervals
- CRC :
-
Colorectal cancer
- EDRN :
-
Early detection research network
- ENCORE :
-
Early Onset Colorectal Cancer Recurrence
- EOCRC :
-
Early-onset colorectal cancer
- FFPE :
-
Formalin-fixed and paraffin-embedded
- HR :
-
Hazard ratio
- miRNA :
-
MicroRNA
- ML :
-
Machine learning
- OS :
-
Overall survival
- REMARK :
-
Reporting Recommendations for Tumor Marker Prognostic Studies
- RFS :
-
Recurrence-free survival
- RNA :
-
Ribonucleic acid
- ROC :
-
Receiver-operating characteristic curve
- RT-qPCR :
-
Reverse-transcription quantitative polymerase chain reaction
- SHAP :
-
SHapley Additive exPlanations
- XGB :
-
eXtreme Gradient Boosting
References
Sinicrope, F. A. Increasing incidence of early-onset colorectal cancer. N. Engl. J. Med. 386, 1547–1558 (2022).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49 (2024).
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin.73, 233–254 (2023).
Eng, C. et al. A comprehensive framework for early-onset colorectal cancer research. Lancet Oncol. 23, e116–e128 (2022).
Patel, S. G., Karlitz, J. J., Yen, T., Lieu, C. H. & Boland, C. R. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol. Hepatol. 7, 262–274 (2022).
Collaborative, R. et al. Characteristics of early-onset vs late-onset colorectal cancer: a review. JAMA Surg. 156, 865–874 (2021).
Gausman, V. et al. Risk factors associated with early-onset colorectal cancer. Clin. Gastroenterol. Hepatol. 18, 2752–2759.e2 (2020).
Low, E. E. et al. Risk factors for early-onset colorectal cancer. Gastroenterology 159, 492–501.e7 (2020).
Chen, H. et al. Metabolic syndrome, metabolic comorbid conditions and risk of early-onset colorectal cancer. Gut 70, 1147–1154 (2021).
Yan, H. H. N. et al. Organoid cultures of early-onset colorectal cancers reveal distinct and rare genetic profiles. Gut 69, 2165–2179 (2020).
Kong, C. et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut 72, 1129–1142 (2023).
Kim, T. J., Kim, E. R., Hong, S. N., Chang, D. K. & Kim, Y. H. Long-term outcome and prognostic factors of sporadic colorectal cancer in young patients: a large institutional-based retrospective study. Med. (Baltim.) 95, e3641 (2016).
You, Y. N. et al. Young-onset rectal cancer: presentation, pattern of care and long-term oncologic outcomes compared to a matched older-onset cohort. Ann. Surg. Oncol. 18, 2469–2476 (2011).
Foppa, C. et al. Early age of onset is an independent predictor for worse disease-free survival in sporadic rectal cancer patients. A comparative analysis of 980 consecutive patients. Eur. J. Surg. Oncol. 48, 857–863 (2022).
Fontana, E. et al. Early-onset colorectal adenocarcinoma in the IDEA database: treatment adherence, toxicities, and outcomes with 3 and 6 months of adjuvant fluoropyrimidine and oxaliplatin. J. Clin. Oncol. 39, 4009–4019 (2021).
Di Leo, M. et al. Risk factors and clinical characteristics of early-onset colorectal cancer vs. late-onset colorectal cancer: a case-case study. Eur. J. Gastroenterol. Hepatol. 33, 1153–1160 (2021).
Cavestro, G. M. et al. Early onset sporadic colorectal cancer: Worrisome trends and oncogenic features. Dig. Liver Dis. 50, 521–532 (2018).
Kim, S. B. et al. Comparison of colonoscopy surveillance outcomes between young and older colorectal cancer patients. J. Cancer Prev. 22, 159–165 (2017).
Chen, F. W., Sundaram, V., Chew, T. A. & Ladabaum, U. Advanced-stage colorectal cancer in persons younger than 50 Fs not associated with longer duration of symptoms or time to diagnosis. Clin. Gastroenterol. Hepatol. 15, 728–737.e3 (2017).
Al Maliki, H. & Monahan, K. J. The diagnostic yield of colonoscopic surveillance following resection of early age onset colorectal cancer. U. Eur. Gastroenterol. J. 12, 469–476 (2024).
Peacock, O. et al. Clinically significant metachronous colorectal pathology detected among young-onset colorectal cancer survivors: implications for post-resection surveillance guidelines. Gastroenterology 163, 1682–1684.e2 (2022).
Zaborowski, A. M. et al. Clinicopathological features and oncological outcomes of patients with young-onset rectal cancer. Br. J. Surg. 107, 606–612 (2020).
Bouvier, A. M. et al. The lifelong risk of metachronous colorectal cancer justifies long-term colonoscopic follow-up. Eur. J. Cancer 44, 522–527 (2008).
Monahan, K. J. et al. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG). Gut 69, 411–444 (2020).
Rutter, M. D. et al. British Society of Gastroenterology/Association of Coloproctology of Great Britain and Ireland/Public Health England post-polypectomy and post-colorectal cancer resection surveillance guidelines. Gut 69, 201–223 (2020).
Cavestro, G. M. et al. Delphi initiative for early-onset colorectal cancer (DIRECt) international management guidelines. Clin. Gastroenterol. Hepatol. 21, 581–603.e33 (2023).
Greener, J. G., Kandathil, S. M., Moffat, L. & Jones, D. T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55 (2022).
Rodriguez-Perez, R. & Bajorath, J. Interpretation of machine learning models using shapley values: application to compound potency and multi-target activity predictions. J. Comput. Aided Mol. Des. 34, 1013–1026 (2020).
Wichmann, R. M., Fernandes, F. T., Chiavegatto Filho, A. D. P. & Network, I.-B. Improving the performance of machine learning algorithms for health outcomes predictions in multicentric cohorts. Sci. Rep. 13, 1022 (2023).
Jung, G., Hernandez-Illan, E., Moreira, L., Balaguer, F. & Goel, A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 17, 111–130 (2020).
Okugawa, Y., Grady, W. M. & Goel, A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149, 1204–1225.e12 (2015).
Xu, C. et al. An exosome-based liquid biopsy predicts depth of response and survival outcomes to cetuximab and panitumumab in metastatic colorectal cancer: the exonerate study. Clin. Cancer Res. 31, 1002–1015 (2025).
Okuno, K. et al. A microRNA signature for risk-stratification and response prediction to FOLFOX-based adjuvant therapy in stage II and III colorectal cancer. Mol. Cancer 22, 13 (2023).
Giraldez, M. D. et al. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin. Cancer Res. 16, 5402–5413 (2010).
Goel, A. et al. De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int J. Cancer 128, 869–878 (2011).
Antelo, M. et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS ONE7, e45357 (2012).
Perea Garcia, J. et al. Association of polyps with early-onset colorectal cancer and throughout surveillance: novel clinical and molecular implications. Cancers (Basel) 11, 1900 (2019).
Jansen, A. M. L. et al. Novel candidates in early-onset familial colorectal cancer. Fam. Cancer 19, 1–10 (2020).
Arriba, M. et al. Intermediate-onset colorectal cancer: a clinical and familial boundary between both early and late-onset colorectal cancer. PLoS One 14, e0216472 (2019).
Antelo, M. et al. Lynch-like syndrome is as frequent as Lynch syndrome in early-onset nonfamilial nonpolyposis colorectal cancer. Int J. Cancer 145, 705–713 (2019).
Perea, J. et al. A clinico-pathological and molecular analysis reveals differences between solitary (early and late-onset) and synchronous rectal cancer. Sci. Rep. 11, 2202 (2021).
Nakamura, K. et al. A liquid biopsy signature for the detection of patients with early-onset colorectal cancer. Gastroenterology 163, 1242–1251.e2 (2022).
Mannucci, A. & Goel, A. Stool and blood biomarkers for colorectal cancer management: an update on screening and disease monitoring. Mol. Cancer 23, 259 (2024).
Lenz, H. J. et al. Impact of consensus molecular subtype on survival in patients with metastatic colorectal cancer: results from CALGB/SWOG 80405 (Alliance). J. Clin. Oncol. 37, 1876–1885 (2019).
Tie, J. et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 26, 1715–1722 (2015).
Tan, I. B. & Tan, P. Genetics: an 18-gene signature (ColoPrint(R)) for colon cancer prognosis. Nat. Rev. Clin. Oncol. 8, 131–133 (2011).
Kelley, R. K., Van Bebber, S. L., Phillips, K. A. & Venook, A. P. Personalized medicine and oncology practice guidelines: a case study of contemporary biomarkers in colorectal cancer. J. Natl. Compr. Canc Netw. 9, 13–25 (2011).
Chen, Q. H., Wang, Q. B. & Zhang, B. Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: a HuGE meta-analysis. Tumour Biol. 35, 529–543 (2014).
Acknowledgements
This work was financially supported by CA72851, CA181572, CA184792, CA187956, CA202797, and CA227602 grants from the National Cancer Institute (National Institutes of Health) and by Fight Colorectal Cancer and the Collaborative Group of the Americas on Inherited Gastrointestinal Cancer. The funders had no role in the design, conduct, or analysis of the results or in the decisions to publish. The internship of Goretti Hernández was supported by Fundación MAPFRE Guanarteme, Las Palmas de Gran Canaria, Spain. This work was also supported by a grant from the Ministry of Science and Technology Bureau of Foreign Experts, China. (G2023016006L). This work has been partly supported by Zotti Prize, 3th Edition, on behalf of Surgical, Oncological and Gastroenterological Department, University of Padova, Padova- Italy, with the patronage of Kauri Holding Spa, Alumni and Amici Associations and Zotti Minici family. Finally, the authors appreciate the contributions made by the study participants, their families, and all those who assisted with the study. Additionally, the authors thank Drs. Caiming Xu, Katsutoshi Shoda, Silei Sui, Yoh Asahi, Takayuki Noma, and Yuan Li for their thoughtful discussions and advice during this project.
Author information
Authors and Affiliations
Contributions
A.M. and A.G. had direct and full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of data analysis. All authors had access to the data and accept responsibility to submit for publication. Conceptualization A.M., G.H., E.Q., A.G. Methodology A.M., G.H., A.G. Investigation A.M., G.H., H.U., Y.Y., F.B., H.B., C.R.B., T.C., E.Q., A.G. Visualization A.M., A.G. Funding acquisition A.M., A.G. Project administration C.R.B., E.Q., A.G. Supervision A.M., F.B., C.R.B., E.Q., A.G. Writing—original draft A.M., C.R.B., A.G. Writing—review/edits A.M., G.H., H.U., Y.Y., F.B., H.B., J.C., C.R.B., T.C., G.M.C., E.Q., A.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mannucci, A., Hernández, G., Uetake, H. et al. Prediction of long-term recurrence-free and overall survival in early-onset colorectal cancer: the ENCORE multi-centre study. npj Precis. Onc. 9, 202 (2025). https://doi.org/10.1038/s41698-025-00978-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00978-7