Abstract
Intratumoral microbiota are implicated in colorectal cancer liver metastasis (CRLM), but their relationships with clinical characteristics and prognosis remain to be explored. Using 5R 16S rRNA gene sequencing method, we analyzed the intratumoral microbiota of patients with colorectal cancer with and without liver metastasis. CRLM patients were classified into three intratumoral microbial community subtypes (IMCSs). These included IMCS1 (sugar metabolism-related liver metastasis with T cell activation, moderate proliferation and invasion, and median disease-free survival (mDFS) of 22 months), IMCS2 (protein metabolism-related liver metastasis with natural killer cell activation, high proliferation and invasion, and mDFS of 12 months), and IMCS3 (lipid metabolism-related liver metastasis with a pauci-immune phenotype, the highest level of proliferation and invasion, and mDFS of 10 months). Our study establishes intratumoral microbiome subtypes as a novel risk stratification strategy for CRLM, facilitating microbiota-directed therapeutic strategies with potential survival benefits.

Similar content being viewed by others
Introduction
The incidence of colorectal cancer (CRC) has been increasing annually, making it the third most common cancer worldwide and the second leading cause of cancer-related mortality, thereby posing a significant socioeconomic burden1,2. Distant metastasis is a major factor contributing to the high mortality rate of colorectal cancer, with liver metastasis being the most frequent and strongly associated with poor prognosis and a low five-year survival rate3,4. Therefore, early identification of colorectal cancer liver metastasis is crucial. However, current methods for early detection of colorectal cancer liver metastasis are limited by insufficient sensitivity and specificity, and the direct monitoring of liver metastasis is challenging due to the difficulty in obtaining continuous liver tissue samples. Consequently, there is a pressing need to investigate novel strategies for the early detection of colorectal cancer liver metastasis.
Numerous studies have demonstrated that dysbiosis of the gut microbiota influences the occurrence and development of colorectal cancer. Specific oncogenic bacteria, such as Fusobacterium nucleatum, Bacteroides fragilis, Parvimonas micra, and Helicobacter pylori, have shown significantly altered abundances in the gut of colorectal cancer patients5,6. The traditional view considered tumors to be sterile environments. However, the concept of intratumoral microbiota has since challenged this perspective. Research has revealed that various cancer tissues harbor microorganisms whose abundance in colorectal cancer changes along the adenoma-carcinoma sequence7. The distribution of microorganisms within colorectal cancer is not random but highly organized within specific ecological niches, where they can promote cancer progression by modulating immune and epithelial cell functions8. Furthermore, studies have shown that Fusobacterium nucleatum is strongly present in the superficial regions of stage III/IV colorectal cancer, and its presence is also increased in the deep areas of adjacent normal mucosa9. These findings provide additional insights into the role of intratumoral microbiota in the occurrence and development of colorectal cancer. Intratumoral microbiota in colorectal cancer can originate from microorganisms crossing the mucosal barrier, microorganisms present in normal adjacent tissues, and hematogenous spread. Intratumoral microbiota can contribute to tumor initiation and progression by inducing genomic instability and mutations, influencing epigenetic modifications, promoting inflammatory responses, evading immune surveillance, modulating metabolic processes, and activating invasion and metastasis10,11. Moreover, intratumoral microbiota can affect the spatial and cellular heterogeneity of colorectal cancer liver metastasis by stimulating the production of specific metabolites, disrupting the integrity of the intestinal vascular barrier, and remodeling the microenvironment of distant organs, thereby indirectly promoting the distant metastasis of colorectal cancer12,13.
The biological implications of intratumoral microbiota on colorectal cancer liver metastasis remain largely unknown. Previous large-scale cohort studies have focused on individual pathogens, and our understanding of intratumoral microbial community subtypes (IMCSs) remains limited. Therefore, we explored the changes in intratumoral microbiota in patients with colorectal cancer with and without liver metastasis. This is the first study to identify the intratumoral microbiota in patients with colorectal cancer liver metastasis. By integrating variations in intratumoral microbiota with host metabolic and immune status, we aimed to elucidate the mechanisms driving liver metastasis and identify intratumoral microorganisms that may serve as predictors of liver metastasis. By doing so, we identified three IMCSs associated with colorectal cancer liver metastasis, namely IMCS1, IMCS2, and IMCS3. We investigated the metabolic and immunological implications of these IMCSs and highlighted the associations of the OCS group with the risk stratification and prognosis of colorectal cancer. This detailed subtyping of intratumoral microbiota provides a novel approach for risk stratification of colorectal cancer liver metastasis. This approach may facilitate the development of microbiota-targeted cancer therapies and inform precise clinical decision-making for patients with colorectal cancer liver metastasis.
Results
Baseline characteristics of patients with colorectal cancer
Based on the inclusion and exclusion criteria, we initially enrolled a total of 256 patients in the study. Following a rigorous screening process, the final study cohort consisted of 44 patients in the colorectal cancer with liver metastasis (metastasis) group and 85 patients in the colorectal cancer without liver metastasis (non-liver metastasis) group. We retrospectively collected data on tumor markers, immunohistochemical indicators, maximum tumor diameter, number of metastatic lymph nodes, and lymphocytes in serum, as well as follow-up data. We analyzed the colorectal cancer tissue samples from these patients for intratumoral microbiota (Table 1). Comparative analysis of the clinical characteristics between the two groups revealed significantly higher levels of carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA19-9) in the metastasis group compared with the non-metastasis group. The metastasis group also exhibited enhanced proliferation, invasion, and malignancy, as evidenced by the significantly increased expression of Ki-67 and p53 in the tumor tissue (p < 0.05). Analysis of the serum metabolic markers related to liver and renal function showed that the metastasis group had significantly higher levels of fasting blood glucose, albumin, triglyceride, and creatinine compared with the non-metastasis group, suggesting active metabolism in colorectal cancer with liver metastasis. Additionally, immune cell profiling in patient sera showed increased proportions of T cells and natural killer (NK) cells, indicating co-activation of the metabolic and immune systems in colorectal cancer with liver metastasis.
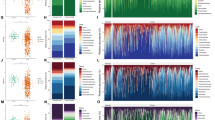
As microorganisms play a crucial role in the onset and progression of colorectal cancer, being closely associated with host immunity and metabolism, we measured the intratumoral microbiota of the patients. Alpha diversity analysis indicated that the colorectal cancer patients with and without liver metastasis had significantly different intratumoral microbial compositions, specifically significantly different Chao1 and Shannon indices (p < 0.05; Fig. 1A–C). Beta diversity analysis using nonmetric multidimensional scaling and principal coordinate analysis did not reveal any significant differences between samples (Fig. 1D, E), likely due to the similarity in intratumoral microbiota across colorectal cancer tissue samples. These results suggest that the sources of intratumoral microbiota are generally consistent among patients with colorectal cancer. Nevertheless, the significant differences in intratumoral microbiota and survival outcomes between the metastasis and non-metastasis groups indicate that variations in intratumoral microbiota may play an important role, warranting further investigation. Despite the differences, the small number of microbial communities that are similar among the different samples can be further analyzed.
A–C Analysis of α-diversity between colorectal cancer liver metastasis and non liver metastasis groups (Chao1, Shannon, and Simpson indices). D, E Analysis of β-diversity between colorectal cancer liver metastasis and non liver metastasis groups (NMDS and PCoA analysis). (*p < 0.05, **p ≤ 0.01, ***p < 0.001, ****p ≤ 0.0001, indicating statistical significance).
Intratumoral microbiota composition in colorectal cancer patients with liver metastasis and non-liver metastasis
To analyze the differences of intratumoral microbiota composition between colorectal cancer patients with liver metastasis and those without liver metastasis, we performed differential abundance analysis of microbial taxa. We focused on the top ten microbial taxa with significant relative abundance (Fig. 2A–F). As expected, we observed significant changes in the phylum-level abundance between tumor and normal states. Specifically, in the liver metastasis group, the relative abundance of Actinobacteria, Thermi, and Firmicutes increased, while Fusobacteria, Proteobacteria, and Bacteroidetes decreased. At the genus level, Deinococcus, Bacillus, and Corynebacterium showed increased abundance in the metastasis group, whereas Pseudomonas, Burkholderia, and Enterobacter were less abundant. At the species level, in the non-liver metastasis group, the relative abundance of Isoptericola variabilis, Bacillus flexus, Pseudomonas aeruginosa, and Enterobacter cancerogenus increased, whereas in the liver metastasis group, Enterococcus casseliflavus, Bacillus niabensis, and Deinococcus murrayi exhibited higher abundance, indicating some structural differences. We further explored differential bacteria between the liver metastasis and non-liver metastasis groups. Using library size-normalized counts and non-parametric significance tests, we identified bacterial taxa that differed significantly between groups. At the genus level, Odoribacter, Leptothrix, Clavibacter, and Caulobacter were significantly more abundant in the liver metastasis group (p < 0.05), with Odoribacter and Leptothrix showing the most significant differences. In contrast, Agrobacterium, Fusobacterium, Methylobacterium, and Faecalibacterium were more abundant in the non-liver metastasis group (p < 0.05), with Fusobacterium showing the most notable decrease (Fig. 2G). At the species level, common bacteria such as Faecalibacterium prausnitzii were significantly more abundant in the non-liver metastasis group, while in the liver metastasis group, it was significantly reduced (p < 0.05) (Fig. 2H). These differences provide important evidence for further investigation into the role of intratumoral microbiota in the development of liver metastasis in colorectal cancer.
A–F The proportion of dominant intratumoral microbiota communities at the phylum, class, order, family, genus, and species between colorectal cancer liver metastasis and non-liver metastasis groups. G, H Differentiating the genus and species of intratumoral microbiota between colorectal cancer liver metastasis and non-liver metastasis groups. (*p < 0.05, **p ≤ 0.01, ***p < 0.001, ****p ≤ 0.0001, indicating statistical significance).
Functional prediction analysis of intratumoral microbiota in colorectal cancer patients with liver metastasis and non-liver metastasis
We used PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2) to further analyze sequencing data and predict the functional potential of the microbiota. By associating the microbiota with functional information, we can better understand the functional composition of the microbiome, elucidating the potential mechanisms of interaction between the microbiota, host, and environment. This approach provides valuable insights for subsequent, deeper investigations. Using gene function annotation from databases such as COG, KO, PFAM, and TIGRFAM, we conducted differential analysis with the STAMP (STatistical Analysis of Metagenomic Profiles) software to identify the top 10 significantly different gene functions between the liver metastasis and non-liver metastasis groups. KO analysis revealed differences between the two groups in pathways related to Fatty Acid Biosynthesis, Secondary Bile Acid Biosynthesis, and other carbohydrate synthesis metabolic pathways, with statistically significant differences (Fig. 3A). In COG database analysis, we identified significant differences in enzymes related to pyruvate kinase, alpha-amylase/alpha-mannosidase, and amino acid synthesis between the two groups (Fig. 3B). Additionally, we predicted protein families and domains in the liver metastasis group (Fig. 3C, D). In the PFAM database, we found that Oxaloacetate Decarboxylase and Immunity Protein 17 were significantly more abundant in the liver metastasis group compared to the non-metastasis group, suggesting their roles as important proteases. These enzymes are potentially linked to the tricarboxylic acid cycle and immune responses. In the TIGRFAM database, we observed an increase in Pyruvate Kinase in the liver metastasis group, which may be related to carbohydrate metabolism. In summary, by performing functional predictions on the intratumoral microbiota in both liver metastasis and non-liver metastasis colorectal cancer groups, we found that the microbiota is associated with the host’s metabolic and immune systems, showing significant statistical differences. These findings provide important direction for further research and lay the groundwork for future studies.
A Differential analysis of functional prediction between colorectal cancer liver metastasis group and non-liver metastasis group using KEGG(Kyoto Encyclopedia of Genes and Genomes) database. B CGO(Clusters of Orthologous Groups of proteins) database predicts protein function between two groups. C PFAM (Protein Families Database) database predicts protein families between two groups. D TIGRFAM(TIGR defined protein families) database predicts protein families between two groups. (*p < 0.05, **p ≤ 0.01, ***p < 0.001, ****p ≤ 0.0001, indicating statistical significance).
Identification of intratumoral microbiota with predictive value in colorectal cancer patients with liver metastasis and non-liver metastasis
We further explored the intratumoral microbiota in colorectal cancer patients with liver metastasis and non-liver metastasis to identify microbiota with predictive potential. Through clustering and correlation analysis of the microbiota between the two groups (Fig. 4A), we identified several genera with significant correlations. Methylobacterium, Agrobacterium, Faecalibacterium, and Fusobacterium were more highly expressed in the non-metastasis group and showed lower expression in the metastasis group. Conversely, Caulobacter, Odoribacter, Leptothrix, and Clavibacter exhibited the opposite pattern, with higher expression in the metastasis group and lower expression in the non-metastasis group. At the species level (Fig. 4B), the clustering of microbial populations was not as pronounced. The Faecalibacterium prausnitzii cluster was less prominent, and the 16S RNA analysis at the genus level was more precise. Therefore, we chose to focus on genus-level analysis for further investigation. We then analyzed the diagnostic potential of the two intratumoral microbiota clusters using ROC (Receiver Operating Characteristic) curve analysis. When analyzing the four genera (Caulobacter, Odoribacter, Leptothrix, and Clavibacter) with high expression in colorectal cancer, individual ROC analysis showed that the diagnostic ability of these four bacteria for liver metastasis vs. non-liver metastasis was poor (p > 0.05) (Fig. 4C and Table 2), with AUC values all below 0.7. Moreover, when performing a combined diagnostic ROC analysis of these four clustered bacteria (Fig. 4D), we found the AUC to be 0.67 (p = 0.05263), with a sensitivity of 52.27% and specificity of 83.53%. However, this combination of microbiota was not an ideal diagnostic model for distinguishing between liver metastasis and non-liver metastasis.
A, B Cluster analysis of intratumoral microbiota in two groups at the genus and species levels (C) Individual ROC survival curve analysis of Odorobacter, Leptothrix, Clavibacter, and Clavibacter in colorectal cancer liver metastasis and non-liver metastasis groups. D Combined ROC survival curve analysis of Odorobacter, Leptothrix, Clavibacter, and Clavibacter between two groups. E Individual ROC survival curve analysis of Agrobacterium, Fusobacterium, Methylobacterium, and Faecalibacterium in colorectal cancer liver metastasis and non-liver metastasis groups. F Combined ROC survival curve analysis of Agrobacterium, Fusobacterium, Methylobacterium, and Faecalibacterium between two groups. G Correlation analysis between Agrobacterium, Fusobacterium, Methylobacterium, and Faecalibacterium. (*p < 0.05, **p ≤ 0.01, ***p < 0.001, ****p ≤ 0.0001, indicating statistical significance).
On the other hand, when analyzing the combined genera Methylobacterium, Agrobacterium, Faecalibacterium, and Fusobacterium, we found a significant correlation, with these genera being less expressed in the liver metastasis group. Individual ROC analysis of each genus showed a better diagnostic ability in the liver metastasis group compared to the non-metastasis group (p < 0.05). Specifically, Methylobacterium (AUC = 0.63, sensitivity 81.82%, specificity 44.71%), Agrobacterium (AUC = 0.63, sensitivity 72.73%, specificity 55.29%), Faecalibacterium (AUC = 0.61, sensitivity 52.27%, specificity 64.71%), and Fusobacterium (AUC = 0.62, sensitivity 93.18%, specificity 31.76%) (Fig. 4E and Table 3). Furthermore, when we performed a combined diagnostic ROC analysis for these four genera, the AUC increased to 0.78 (p < 0.0001), with sensitivity of 68.18% and specificity of 74.12% (Fig. 4F). Additionally, correlation analysis of Methylobacterium, Agrobacterium, Faecalibacterium, and Fusobacterium (Fig. 4G) revealed significant positive correlations between these genera (p < 0.05), suggesting that these four intratumoral microbiota have a strong positive correlation. Therefore, these genera could be used as a panel for predicting liver metastasis in colorectal cancer.
Identification of microbial community subtypes in colorectal cancer patients with liver metastasis
Next, we investigated the changes in intratumoral microbiota between colorectal cancer patients with liver metastasis and those without liver metastasis. By combining host and tumor microbiota data, we hypothesized whether the intratumoral microbiota composition in liver metastasis patients could be classified into distinct community subtypes. Based on previous analyses, we identified four microbiota most likely associated with liver metastasis: Methylobacterium, Agrobacterium, Faecalibacterium, and Fusobacterium. We analyzed the intratumoral microbiota expression profiles of each liver metastasis patient and performed dimensionality reduction clustering via Principal Component Analysis (PCA). As a result, colorectal cancer liver metastasis samples were broadly classified into three intratumoral microbial community subtypes (IMCSs), namely IMCS1, IMCS2, and IMCS3 (Fig. 5A, B).
IMCS1 subtype accounted for 57.0% of the total tumor samples. This subtype is characterized by either the absence of the four identified pathogens or the predominance of a single intratumoral pathogen. It includes samples where none of the four bacteria were detectable or where only Methylobacterium was present, labeled as “Methylobacterium+“ or “All negative.”
IMCS2 subtype represented 32.0% of the tumor samples. This subtype is defined by the presence of two intratumoral pathogens. It includes profiles such as Methylobacterium+ + Agrobacterium+, Methylobacterium+ + Fusobacterium+, and Methylobacterium+ + Faecalibacterium+.
IMCS3 subtype comprised 11.0% of the tumor samples. This subtype is characterized by the presence of three or all four of the identified intratumoral pathogens, including Methylobacterium+ + Fusobacterium+ + Faecalibacterium+, Methylobacterium+ + Agrobacterium+ + Fusobacterium+, and “All positive” (all four pathogens present).
Correlations between IMCSs and clinical characteristics in patients with colorectal cancer liver metastasis
We conducted a comprehensive retrospective analysis to further elucidate the associations between intratumoral microbiota and the clinicomolecular characteristics of patients with colorectal cancer liver metastasis. Building upon the previous functional enrichment analysis, which suggested the involvement of intratumoral microbiota in colorectal cancer liver metastasis, we collected the tumor markers, immunohistochemical indicators, and lymphocytes in serum as well as follow-up information from patients with IMCS1, IMCS2, and IMCS3 (Table 4). We focused on clinical indicators reflecting the metabolism of three major nutrients: sugars (fasting blood glucose), lipids (triglyceride, cholesterol, low-density lipoprotein, high-density lipoprotein), and proteins (albumin and globulin). Additionally, we examined the immunohistochemical markers Ki-67 and p53 associated with tumor proliferation and invasion and the relationships between intratumoral microbiota and host immune cells, including T cells, B cells, T helper cells, T suppressor cells, and NK cells.
To examine the relationships between the three IMCSs and metabolic indicators, we performed correlation analysis (Fig. 6A). After excluding patients with diabetes and potential measurement errors in the metastasis group, IMCS1 exhibited a strong positive correlation with fasting blood glucose (r = 0.85, p < 0.0001) and a negative correlation with albumin (r = −0.39, p < 0.05). IMCS2 demonstrated positive correlations with albumin (r = 0.70, p < 0.0001) and globulin (r = 0.61, p < 0.0001) but a negative correlation with fasting blood glucose (r = −0.64, p < 0.0001). IMCS3 showed positive correlations with triglyceride (r = 0.36, p < 0.05), cholesterol (r = 0.37, p < 0.05), and low-density lipoprotein (r = 0.52, p < 0.001).
A Related characteristics of metabolic subtypes IMCS1, IMCS2, and IMCS3. B Redundancy analysis (RAD) among IMCS1, IMCS2, and IMCS3. C–G Differences in clinical metabolic characteristics among IMCS1, IMCS2, and IMCS3. H Related immune infiltration characteristics of IMCS1, IMCS2, and IMCS3. I, J Quantification of differential expression of p53 and Ki-67 in IMCS1, IMCS2, and IMCS3. K Immunohistochemical staining of differential expression of p53 and Ki-67 in IMCS1, IMCS2, and IMCS3. L Survival curve analysis between IMCS1, IMCS2, and IMCS3. (*p < 0.05, **p ≤ 0.01, ***p < 0.001, ****p ≤ 0.0001, indicating statistical significance).
We also performed RAD analysis based on a linear model to investigate the associations of IMCSs with environmental and metabolic factors and further examine the relationships among environmental/metabolic factors, samples, and intratumoral microbiota, as well as their pairwise interactions. This analysis demonstrated that IMCS1, IMCS2, and IMCS3 were related to sugar, protein, and lipid metabolism, respectively, and correlated with corresponding environmental factors. These three subtypes were effectively distinguishable in the dimensionality reduction analysis (Fig. 6B). When we next compared the metabolic profiles of patients with different IMCSs, we observed a progressive decrease in fasting blood glucose levels from IMCS1 to IMCS2 to IMCS3. Moreover, IMCS2 showed significantly elevated albumin and globulin levels, while IMCS3 exhibited markedly increased triglyceride and cholesterol concentrations (Fig. 6C–G). These findings indicate that the three IMCSs in patients with colorectal cancer liver metastasis are associated with distinct host metabolic states, and the patients with IMCS1 and IMCS2 display distinct metabolic profiles.
Regarding the host immune responses (Fig. 6H), IMCS1 was positively correlated with T cell activation (r = 0.19, p < 0.05) and was thus designated the T cell-activated immunity subtype. IMCS2 was positively correlated with NK cell activation (r = 0.33, p < 0.01) and was thus designated the NK cell activated-immunity subtype. In contrast, IMCS3 was neither positively nor negatively correlated with T cells, B cells, T helper cells, T suppressor cells, or NK cells and was therefore classified as the pauci-immune subtype. To investigate the proliferation and invasion of colorectal cancer, we compared the immunohistochemical markers p53 and Ki-67 across the three IMCSs (Fig. 6I–K). We found that their expression progressively increased in IMCS1, IMCS2, and IMCS3. This finding indicates that IMCS3 has increased invasion and proliferation rates compared with IMCS1 and IMCS2; IMCS3 may be associated with high malignancy and poor prognosis. In contrast, IMCS1 appears to be associated with a more favorable prognosis.
Moreover, we tracked the occurrence of liver metastasis in colorectal cancer patients, including those without initial liver metastasis who achieved clinical remission after initial treatment. We continued to follow up with the patients without liver metastasis to record any development of liver metastasis using pathological analysis, computed tomography, or positron emission tomography-computed tomography, the standard methods of metastasis monitoring. When we collected follow-up data and performed a stratified log-rank test to evaluate disease-free survival (DFS), we found significant differences in DFS among IMCS1, IMCS2, and IMCS3 (log-rank P = 0.0176). The median DFS (mDFS) was 22 months for IMCS1, 12 months for IMCS2, and 10 months for IMCS3. These findings suggest a progressive decline in prognosis from IMCS1 to IMCS3, as indicated by decreasing survival durations. IMCS3 exhibited a higher degree of malignancy in colorectal cancer liver metastasis compared with IMCS1. The mDFS between IMCS2 and IMCS3 differed by only two months, indicating a similarly poor prognosis for both subtypes. In contrast, IMCS1 demonstrated the most favorable prognosis in terms of malignancy and survival, indicating greater therapeutic benefits. Building on these findings, we propose a clinicomolecular and prognostic subtyping system based on intratumoral microbiota for patients with colorectal cancer liver metastasis.
Discussion
Liver metastasis represents a significant challenge in the management of colorectal cancer, highlighting the importance of early detection of colorectal cancer liver metastases in clinical practice. By detecting the abundance of intratumoral microbiota in colorectal cancer patients with and without liver metastasis, we identified differential intratumoral microorganisms. Based on a retrospective analysis of patient systemic metabolism and immune regulation, we established clinically stratified subtypes of patients with colorectal cancer liver metastasis based on intratumoral microbiota, a stratification that holds significant prognostic value.
We identified four intratumoral bacteria at the genus level, Fusobacterium, Faecalibacterium, Methylobacterium, and Agrobacterium, which are associated with early detection of liver metastasis. Previous studies have reported that multi-species microbiota from the genera Fusobacterium, Bacteroides, Proteus, and Faecalibacterium serve as potential biomarkers for colorectal cancer diagnosis. Furthermore, both fecal and tissue microbiota have shown significant correlations with tumor immune cell infiltration, metastasis, and survival prognosis in colorectal cancer6,14.
Fusobacterium, as an oral anaerobic opportunistic pathogen, is significantly enriched in both fecal and tumor tissues of colorectal cancer patients15,16. Fusobacterium has a sustained positive correlation with regional lymph node and distant metastasis. The presence of invasive Fusobacterium has been detected in both liver metastasis and lymphatic metastasis of colorectal cancer, demonstrating the stability of the microbiome between paired primary and metastatic tumors17. Its product, Fap2, can resist the acidic environment of the gastrointestinal tract. Fap2 binds to the Gal-GalNAc expressed on tumors, which further facilitates the gastrointestinal translocation and colonization of Fusobacterium during metastasis18,19. Fusobacterium targets the long non-coding RNA (lncRNA) ENO1-IT1, promoting glycolysis and tumorigenesis in colorectal cancer20. Additionally, Fusobacterium recruits tumor-infiltrating immune cells, activates the NF-κB pathway, upregulates the expression of pro-inflammatory cytokines, and selectively recruits immune cells such as myeloid-derived suppressor cells (MDSCs) and dendritic cells. This contributes to the establishment of an immune microenvironment that promotes tumor progression. Fusobacterium creates a pro-inflammatory microenvironment that favors colorectal tumor formation and progression, while also remodeling the tumor microenvironment through immune evasion, leading to tumor immune suppression21. Faecalibacterium is one of the most important bacteria in the human gut microbiota and a major producer of butyrate, which has anti-inflammatory effects, maintains bacterial enzyme activity, and protects the digestive system from intestinal pathogens22. The production of its metabolic product, butyrate, can shape the gut microbiota and maintain intestinal homeostasis by releasing anti-inflammatory molecules such as IgA, B vitamins, and microbial anti-inflammatory molecules23,24. Faecalibacterium inhibits the proliferation of HCT116 colorectal cancer cells and significantly reduces the frequency and formation of abnormal crypt foci in AOM-induced colorectal cancer in rats25. A decrease in Faecalibacterium abundance is significantly associated with a reduction in the circulating DP8α Treg cell population, suggesting that reduced DP8α T cell activity may contribute to colorectal cancer development by affecting the immune microenvironment. Methylobacterium is a Gram-negative rod bacterium, belonging to the family Methylobacteriaceae (Alpha-Proteobacteria). Previous studies have suggested that it can cause varying degrees of colonization and infection in immunocompromised patients, including those with solid or hematologic malignancies, organ transplants, renal failure, HIV infection, and tuberculosis26,27. Analysis of the microbiota from tumor sites, adjacent tumor regions, and non-tumor tissue from colorectal cancer patients, as well as from healthy controls, revealed that Methylobacterium is one of the bacteria driving cancer development and is associated with a higher risk of colorectal cancer28. High enrichment of Methylobacterium was also observed in the gastric mucosa microbiota of gastric cancer patients, where its presence was significantly correlated with poor prognosis. Furthermore, Methylobacterium was shown to decrease the expression of TGFβ and CD8 + TRM cells in the gastric cancer immune microenvironment29. Agrobacterium, a Gram-negative bacterium, is commonly associated with infections in tumors and elderly patients30. One of its products, succinoglycan, is an extracellular polysaccharide produced by most Agrobacterium strains. It promotes lymphocyte proliferation in peripheral blood, spleen, and lymph nodes in sarcoma mouse models, and reduces the expression of Blc-2 family proteins and Caspase-3 proteins, thereby promoting tumor apoptosis31. These four intratumoral bacteria, by regulating host metabolism and immune modulation, influence the occurrence and progression of cancer, providing important evidence for further exploration in this study.
In this study on intratumoral microbiota associated with colorectal cancer liver metastasis, and through the analysis of differential intratumoral microbiota correlations and diagnostic capabilities, we explored the combined analysis of the four aforementioned intratumoral bacteria with patients’ clinical characteristics to further predict microbiota that may influence colorectal cancer liver metastasis. These four bacteria overall drive liver metastasis through complex mechanisms. By performing cross-linkage analysis with metabolism-related and immune-related indicators, we identified three distinct intratumoral microbiota subtypes: IMCS1, IMCS2, and IMCS3. Among these, IMCS1 subtype accounted for the highest proportion (57%) of liver metastasis patients. In this group, the tumor tissue expressed little to no Fusobacterium, Faecalibacterium, Methylobacterium, and Agrobacterium, or only Methylobacterium was detected. IMCS1 patients showed low expression of Ki67 and p53, with relatively low levels of proliferation and malignancy. This suggests that tumors in the IMCS1 liver metastasis subtype grow slowly, and the lack of high abundance expression of these bacteria may indicate that their high expression could, to some extent, promote the onset of colorectal cancer. IMCS1 liver metastasis is also associated with glucose metabolism, with increased glucose content detected in the body, and correlated with T-cell immune activation. IMCS2, which accounts for one-third of liver metastasis patients, is characterized by high expression of two out of the four bacteria. Ki67 and p53 are moderately expressed in this group, and the subtype is related to protein metabolism, with elevated NK cell expression observed in these patients. IMCS3, which represents 11% of liver metastasis patients, often shows an increased abundance of three or all four of the intratumoral bacteria. These patients exhibit high expression of Ki67 and p53, which are associated with lipid metabolism, and present an immune-suppressed profile. Each intratumoral microbiota subtype, IMCS1, IMCS2, and IMCS3, has its own characteristics, including distinct host metabolic profiles and varying degrees of immune cell activation or suppression. Therefore, the interplay between the microbiota, microbial metabolites, and the immune system constitutes a triad of complex regulatory systems in the colorectal cancer microenvironment, participating in the process of colorectal cancer development and progression32.
When we examined the correlations between intratumoral microbiota and the expression of p53 and Ki67, we observed a progressive increase in malignancy across patients with IMCS1, IMCS2, and IMCS3, with IMCS3 showing high expression levels and the greatest degree of malignancy. A significant number of observations also indicated that immunohistochemical positivity for p53 and Ki-67 was associated with early relapse and distant metastasis in colorectal cancer patients. p53 and Ki-67 have been suggested as independent predictors of survival and may serve as reliable indicators for colorectal cancer liver metastasis, independent of factors such as gender, age, or prior treatments33,34. In this study, the combination of host intratumoral microbiota and immunohistochemical protein markers suggests that the more intratumoral bacteria associated with liver metastasis are detected as positive, the higher the degree of malignancy. This indicates that malignancy and the presence of liver metastasis-related intratumoral microbiota may be linked and serve as potential markers for prognosis.
Metabolic regulation plays a critical role in the development and progression of colorectal cancer. Cellular metabolism of glucose, amino acids, and lipids is reprogrammed, altering the tumor microenvironment and modulating the composition and activity of the microbiome35. Additionally, studies have shown that metabolites are strongly associated with microbial pairs, suggesting a strong coupling between the microbiome and metabolomics, and indicating that part of the metabolic characteristics in CRC can be attributed to microbial interactions, ultimately promoting the development of CRC36. In this study, we innovatively combine host metabolism and intratumoral microbiota in patients with colorectal liver metastasis, and observe that the three microbiome subtypes (IMCS1, IMCS2, and IMCS3) exhibit strong positive correlations with glucose metabolism, protein metabolism, and lipid metabolism, respectively. After excluding diabetes and detection errors from the blood glucose measurements, we found that IMCS1 liver metastasis patients have higher baseline blood glucose levels compared to IMCS2 and IMCS3 subtypes, which are positively correlated with serum glucose metabolism. IMCS2-type liver metastasis patients showed a strong positive correlation with albumin and globulin levels, indicating a link to protein metabolism. Furthermore, we observed a negative correlation between albumin and IMCS1, and a negative correlation between IMCS2 and glucose metabolism, suggesting an antagonistic relationship between these two subtypes. IMCS3, on the other hand, was found to be associated with lipid metabolism. This indicates that the positive or negative expression of different intratumoral bacteria in these microbiome subtypes may lead to distinct metabolic profiles, influencing the degree of invasiveness, malignancy, and survival prognosis. Jie Hong et al. demonstrated that Fusobacterium nucleatum activates lncRNA ENO1-IT1 transcription through upregulation of the transcription factor SP1, promoting glycolysis and tumorigenesis in CRC20. The increased risk of CRC has been shown to be associated with alterations in the gut microbiome and reduced production of short-chain fatty acids (SCFAs). CRC cells primarily rely on aerobic glycolysis for energy, and targeting SCFA transporters to modulate lactate production and butyrate synthesis has been implicated in CRC development37. Butyrate, through the GPR109a-AKT signaling pathway, significantly inhibits glucose transport and glycolysis in CRC cells by reducing the abundance of membrane GLUT1 and cytoplasmic G6PD38. We also found that IMCS3 was associated with lipid metabolism markers such as cholesterol, triglycerides, and low-density lipoprotein (LDL). CRC risk is mainly driven by environmental factors, particularly diet. A high dietary fat intake is considered a risk factor for the formation of premalignant lesions or exacerbating colon tumorigenesis. The pro-tumor activity of high-fat diets is attributed to their impact on gut microbiota composition and bile acid metabolism39,40. Additionally, Aguirre-Portolés C et al. have demonstrated that lipid regulation in CRC participates in ATP synthesis and the activation of essential cellular signaling pathways, influencing processes such as membrane tissue and plasticity. In both primary tumors and distant metastases, lipid metabolism affects a wide range of tumorigenic processes41. Targeting lipid metabolism is of significant importance in the diagnosis, treatment, and prognostic evaluation of CRC.
The development of colorectal cancer is accompanied by immune cells that collaboratively create an immunosuppressive tumor microenvironment, which plays a crucial role in CRC progression, immune evasion, and responses to immunotherapy. The microbiome shapes the immune microenvironment in CRC through diverse mechanisms. Research has shown that microbiome-derived short-chain fatty acids (SCFAs), such as butyrate, promote cellular metabolism and enhance the memory potential of activated T cells42. Fusobacterium nucleatum-derived succinate inhibits the cGAS-interferon-β pathway, thus suppressing the anti-tumor response by limiting CD8 T cell trafficking into the tumor microenvironment43. The resident microbiota in CRC tissues can promote liver metastasis through lactylation and immune modulation. Retinoic acid-inducible gene 1 (RIG-I) lactylation inhibits NF-κB recruitment to the Nlrp3 promoter in macrophages, reducing its transcription. This deficiency in Nlrp3 impacts the immunosuppressive activity of regulatory T cells (Tregs) and the anti-tumor activity of T cells44. Microsatellite instability (MSI) is associated with higher tumor immunity and mutational burden, and there is evidence suggesting that the intratumoral microbiota may vary depending on MSI status. The intratumoral microbiota in CRC is strongly correlated with MSI and genes related to MSI-associated tumor molecular characteristics, including Dialister and Casatella45,46. The tumor-associated microbiome plays an important role in cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) blockade, programmed cell death protein 1 (PD-L1) mediation, and T cell stimulation47. Our research revealed that IMCS1 and IMCS2 were associated with the activation of T cells and NK cells, respectively, whereas IMCS3 did not result in immune cell activation. These findings suggest that patients with IMCS1, characterized by significant T cell activation, may exhibit improved response to systemic immunotherapy, potentially resulting in enhanced therapeutic benefits. Patients with IMCS2, characterized by NK cell activation, may benefit from the natural anti-tumor properties of NK cells, which are capable of recognizing and killing tumor and infected cells, as well as modulating the functions of T cells and B cells. In contrast, patients with IMCS3 showed a suppressed immune response and potentially limited benefits from immunotherapy. These findings indicate that specific activation of immune cells by intratumoral microbiota can shape the immune microenvironment, with therapeutic choices potentially leading to different benefits and survival outcomes.
Among them, we found significant differences in the mDFS, which were 22, 12, and 10 months for IMCS1, IMCS2, and IMCS3, respectively. The fact that the mDFS of IMCS1 was approximately twice as long as that of IMCS2 and IMCS3 may be explained by the relatively low detection rate of liver metastasis-associated intratumoral microorganisms such as Fusobacterium, Faecalibacterium, Methylobacterium, and Agrobacterium. Only one or none of these genera was detected in patients with IMCS1. Furthermore, patients with IMCS1 demonstrated low expression of p53 and Ki-67, indicating minimal proliferation and invasion. Notably, patients with IMCS1 showed T cell activation, indicating improved immune activation and inherent anti-tumor potential. Therefore, intratumoral microbiota may have contributed to the better prognosis observed in patients with IMCS1. IMCS2 and IMCS3 colorectal cancers exhibited malignant proliferation, and immune cell activation was absent in IMCS3, resulting in poor response to immunotherapy. These properties contributed to the rapid disease progression in these two subtypes. The significant differences between normal and tumor tissues are associated with prognosis, with tumor tissues from patients with shorter survival showing greater disparities compared with normal tissues48. Mouradov et al. characterized and classified colorectal cancer patients based on fecal microbiota profiles and clinical features, revealing significantly different five-year survival outcomes among patients with distinct microbial compositions49. These findings suggest the potential utility of microbiota in studying colorectal cancer prognosis. Dysbiosis, metabolomic changes, and microbial variation have potential for non-invasive early diagnosis or prognostic assessment of colorectal cancer with and without metastasis50,51.
In conclusion, the intratumoral microbiota plays a crucial role in the development, progression, and treatment of colorectal cancer. For the first time, we investigated the correlations among the intratumoral microbiota, host metabolism, and immune microenvironment in patients with colorectal cancer with liver metastasis. Our research identified IMCSs distinguished by unique microbial profiles and clinical characteristics. Given the heterogeneity of colorectal cancer, incorporating IMCSs into its subtyping offers a novel approach to the diagnosis and management of the disease. Investigating the biological properties of intratumoral microbiota and the host facilitates the early detection of liver metastasis, potentially leading to reduced mortality rates. Our findings highlight the importance of developing innovative therapeutic strategies targeting intratumoral microbiota in colorectal cancer, in particular, microbiota-based tumor therapies that offer promise in improving survival outcomes in patients with colorectal cancer with liver metastasis.
Methods
Study design and retrospective data collection
This study is a retrospective analysis conducted from January 2019 to December 2023, which included a total of 256 patients diagnosed with colorectal cancer. Strict screening was performed during their first visit. The inclusion criteria for this study were: (1) newly diagnosed colorectal cancer or colorectal cancer with liver metastasis; (2) histological diagnosis of colorectal cancer confirmed by electronic colonoscopy biopsy; (3) clear detection of liver metastatic lesions by enhanced CT or PET-CT in patients with colorectal cancer liver metastasis. The exclusion criteria were: (1) patients who had undergone colorectal resection; (2) patients who had received chemotherapy, targeted therapy, or immunotherapy; (3) colorectal cancer patients with metastasis to multiple other organs in addition to the liver; (4) patients diagnosed with other cancers invading the colon or rectum. Ultimately, 85 non-liver metastatic colorectal cancer patients and 44 colorectal cancer patients with liver metastasis were included in the study cohort.
Simultaneously, we conducted timely follow-up of the non-liver metastatic colorectal cancer patients included in the study. After patients underwent standard surgery and first-line standardized chemotherapy, clinical remission was achieved (considering that each patient may have received a different chemotherapy regimen, we aimed to select chemotherapy protocols according to the NCCN guidelines to achieve complete remission whenever possible). In addition to investigating various clinical and serological data (such as tumor markers, immunohistochemistry indices, and immune lymphocytes), we focused on indicators related to the three major metabolic pathways, including glucose metabolism (fasting blood glucose), lipid metabolism (triglycerides, cholesterol, low-density lipoprotein, high-density lipoprotein), and protein metabolism (albumin, globulin). Immunohistochemical markers, including Ki-67 and p53, were also examined, as these are associated with tumor proliferation and invasiveness. Furthermore, we explored the correlation between host immune cell types and the intratumoral microbiota, including T cells, B cells, Th cells, Ts cells, NK cells, etc., to better explain the biological behavior of colorectal cancer liver metastasis patients.
The follow-up endpoint was the occurrence of liver metastasis during the follow-up period. Patients were followed up every three months, with the most important aspect being long-term imaging assessments (enhanced CT or PET-CT). If liver metastasis signs or hepatic hypermetabolic lesions were detected, a diagnosis of liver metastasis was made by a radiology expert. All treatments, sample collection, and the research procedure for the patients included in this study were conducted in accordance with the regulations issued by the National Health Commission of China and the ethical standards established by the Declaration of Helsinki. Written informed consent was obtained from all patients. The retrospective study was approved by the Institutional Review Board of the Affiliated Cancer Hospital of Guangxi Medical University (KY2021279).
16S rRNA microbiome detection in colorectal cancer tumors
We selected tumor tissue samples from patients with colorectal cancer (CRC), both with liver metastasis and without, according to the inclusion criteria for this study. Microbial community DNA was extracted from these various sources using commercially available extraction kits suited to the sample type. DNA quantification was performed using Qubit (Invitrogen, USA).
PCR amplification was conducted in two rounds using specific primers for the five variable regions (V2, V3, V5, V6, and V8) of the 16S rRNA gene. The PCR products were purified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified again using Qubit (Invitrogen, USA). The purified PCR products were assessed using the Agilent 2100 Bioanalyzer (Agilent, USA) and the Illumina library quantification kit (Kapa Biosciences, Woburn, MA, USA). The libraries that met the quality standard, with concentrations ≥0.3 ng/μL, were prepared. The qualified sequencing libraries (with non-repeating index sequences) were diluted, and the samples were mixed in the required proportions for sequencing. Following denaturation with NaOH to single-strand DNA, the libraries were subjected to sequencing on the Illumina NovaSeq 6000 platform using the PE150 sequencing mode, with the NovaSeq 6000 SP Reagent Kit (500 cycles). The sequencing data from the five amplified regions were integrated to perform bacterial taxonomic identification. Contaminant bacteria introduced during sample collection, DNA extraction, and PCR amplification were removed using negative control data. Alpha diversity and beta diversity analyses were performed based on the obtained species-level abundance tables.
Alpha diversity analysis was evaluated using five indices: observed species, Shannon, Simpson, Chao1, and goods_coverage, to assess the diversity within each habitat. Beta diversity analysis was primarily carried out by calculating the Bray-Curtis distance matrix, followed by six major analytical methods to assess diversity between habitats (samples/groups). The species and their relative abundance in each sample were determined using the SMURF algorithm and abundance tables. Statistical analyses were performed to compare the abundance of species between different groups. Depending on the sample characteristics, different statistical methods were applied. Fisher’s exact test for comparisons without biological replicates. Mann–Whitney U test for comparisons between two groups with biological replicates. Kruskal–Wallis test for comparisons between multiple groups with biological replicates. The significance threshold was set at p < 0.05. To predict the functional information of the microbiota, we used PICRUSt2 software, and functional differences were analyzed using STAMP (STatistical Analysis of Metagenomic Profiles).
Statistical analysis
Differences between colorectal cancer patients with liver metastasis and those without were analyzed using non-parametric tests, with a significance level set at p < 0.05. Both groups consisted of continuous numerical variables, and data were expressed as mean ± standard error of the mean (SEM). The diagnostic ability of relevant biomarkers for identifying colorectal cancer patients with and without liver metastasis was assessed using receiver operating characteristic (ROC) curves. The diagnostic performance was quantified by the area under the curve (AUC). The optimal cutoff values were determined using the Youden index (sensitivity + specificity − 100%). This method allows for the calculation of sensitivity and specificity at known comparable threshold values. Follow-up data were collected from the patients included in the study, and the median disease-free survival (mDFS) was evaluated using the stratified Logrank test. All statistical analyses were conducted using SPSS software (SPSS 17.0, Chicago, Illinois, USA) and GraphPad Prism Version 7.0 software (La Jolla, CA, USA).
Data availability
Additional data are made available in supplementary materials of this article. Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information with the primary accession code SRP567364, and can be accessed through the following link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1230556?reviewer=d5uofl5ta0hakbl4qbvr8qc52.
Code availability
The underlying code for this study is available in the National Center for Biotechnology Information with the primary accession code SRP567364 after July 30, 2027, and can be accessed via this link https://dataview.ncbi.nlm.nih.gov/object/PRJNA1230556?reviewer=d5uofl5ta0hakbl4qbvr8qc52.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet 394, 1467–1480 (2019).
Cervantes, A. et al. ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 34, 10–32 (2023).
Biller, L. H. & Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 325, 669–685 (2021).
Wong, C. C. & Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20, 429–452 (2023).
Conde-Pérez, K. et al. The multispecies microbial cluster of Fusobacterium, Parvimonas, Bacteroides and Faecalibacterium as a precision biomarker for colorectal cancer diagnosis. Mol. Oncol. 18, 1093–1122 (2024).
Liu, W. et al. Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology 160, 2395–2408 (2021).
Galeano Niño, J. L. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022).
Yamamoto, S. et al. Heterogeneous distribution of Fusobacterium nucleatum in the progression of colorectal cancer. J. Gastroenterol. Hepatol. 36, 1869–1876 (2021).
Cao, Y. et al. Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduct. Target Ther. 9, 15 (2024).
Yang, L., Li, A., Wang, Y. & Zhang, Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct. Target Ther. 8, 35 (2023).
Zhang, J., Wang, P., Wang, J., Wei, X. & Wang, M. Unveiling intratumoral microbiota: an emerging force for colorectal cancer diagnosis and therapy. Pharm. Res. 203, 107185 (2024).
Fu, A., Yao, B., Dong, T. & Cai, S. Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. 33, 583–593 (2023).
Luu, K. et al. Fecal and tissue microbiota are associated with tumor t-cell infiltration and mesenteric lymph node involvement in colorectal cancer. Nutrients 15, 316 (2023).
Wang, N. & Fang, J. Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31, 159–172 (2023).
Barot, S. V. et al. Distinct intratumoral microbiome of young-onset and average-onset colorectal cancer. EBioMedicine 100, 104980 (2024).
Bullman, S. et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017).
Meng, Q. et al. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 22, e52891 (2021).
Abed, J. et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20, 215–25 (2016).
Hong, J. et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 70, 2123–2137 (2021).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–15 (2013).
Ferreira-Halder, C. V., Faria, A. V. S. & Andrade, S. S. Action and function of Faecalibacterium prausnitzii in health and disease. Best. Pr. Res Clin. Gastroenterol. 31, 643–648 (2017).
Sitkin, S. & Pokrotnieks, J. Clinical potential of anti-inflammatory effects of faecalibacterium prausnitzii and butyrate in inflammatory bowel disease. Inflamm. Bowel Dis. 25, e40–e41 (2019).
Singh, V. et al. Butyrate producers, “The Sentinel of Gut”: their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 13, 1103836 (2023).
Dikeocha, I. J., Al-Kabsi, A. M., Chiu, H. T. & Alshawsh, M. A. Faecalibacterium prausnitzii ameliorates colorectal tumorigenesis and suppresses proliferation of HCT116 colorectal cancer cells. Biomedicines 10, 1128 (2022).
Lai, C. C. et al. Infections caused by unusual Methylobacterium species. J. Clin. Microbiol 49, 3329–31 (2011).
Kovaleva, J., Degener, J. E. & van der Mei, H. C. Methylobacterium and its role in health care-associated infection. J. Clin. Microbiol. 52, 1317–21 (2014).
Wang, Y. et al. Analyses of potential driver and passenger bacteria in human colorectal cancer. Cancer Manag. Res. 12, 11553–11561 (2020).
Peng, R. et al. Gastric microbiome alterations are associated with decreased CD8+ tissue-resident memory t cells in the tumor microenvironment of gastric cancer. Cancer Immunol. Res. 10, 1224–1240 (2022).
Detrait, M. et al. Agrobacterium radiobacter bacteremia in oncologic and geriatric patients: presentation of two cases and review of the literature. Int J. Infect. Dis. 12, e7–10 (2008).
Yang, Y. et al. Anti-tumor activity and immunogenicity of a succinoglycan riclin. Carbohydr. Polym. 255, 117370 (2021).
Hanus, M. et al. Immune system, microbiota, and microbial metabolites: the unresolved triad in colorectal cancer microenvironment. Front. Immunol. 12, 612826 (2021).
Taha, A. et al. Prognostic value of immunohistochemical markers for locally advanced rectal cancer. Molecules 27, 596 (2022).
Backus, H. H. et al. Rb, mcl-1 and p53 expression correlate with clinical outcome in patients with liver metastases from colorectal cancer. Ann. Oncol. 12, 779–85 (2001).
Zhou, C. B. & Fang, J. Y. The regulation of host cellular and gut microbial metabolism in the development and prevention of colorectal cancer. Crit. Rev. Microbiol 44, 436–454 (2018).
Feng, J. et al. Microbiome and metabolic features of tissues and feces reveal diagnostic biomarkers for colorectal cancer. Front. Microbiol. 14, 1034325 (2023).
Wang, G. et al. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J. Cell Physiol. 234, 17023–17049 (2019).
Geng, H. W., Yin, F. Y., Zhang, Z. F., Gong, X. & Yang, Y. Butyrate suppresses glucose metabolism of colorectal cancer cells via GPR109a-AKT signaling pathway and enhances chemotherapy. Front. Mol. Biosci. 8, 634874 (2021).
Ocvirk, S. & O’Keefe, S. J. D. Dietary fat, bile acid metabolism and colorectal cancer. Semin Cancer Biol. 73, 347–355 (2021).
Jia, W., Xie, G. & Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
Nicolini, A. & Ferrari, P. Involvement of tumor immune microenvironment metabolic reprogramming in colorectal cancer progression, immune escape, and response to immunotherapy. Front. Immunol. 15, 1353787 (2024).
Bachem, A. et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297.e5 (2019).
Jiang, S. S. et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe 31, 781–797.e9 (2023).
Gu, J. et al. Tumor-resident microbiota contributes to colorectal cancer liver metastasis by lactylation and immune modulation. Oncogene 43, 2389–2404 (2024).
Byrd, D. A. et al. The intratumor microbiome is associated with microsatellite instability. J. Natl. Cancer Inst. 115, 989–993 (2023).
Sillo, T. O., Beggs, A. D., Middleton, G. & Akingboye, A. The gut microbiome, microsatellite status and the response to immunotherapy in colorectal cancer. Int J. Mol. Sci. 24, 5767 (2023).
Temraz, S. et al. Gut microbiome: a promising biomarker for immunotherapy in colorectal cancer. Int J. Mol. Sci. 20, 4155 (2019).
Debelius, J. W. et al. The local tumor microbiome is associated with survival in late-stage colorectal cancer patients. Microbiol Spectr. 11, e0506622 (2023).
Mouradov, D. et al. Oncomicrobial community profiling identifies clinicomolecular and prognostic subtypes of colorectal cancer. Gastroenterology 165, 104–120 (2023).
An, H. J. et al. Tumor-associated microbiome features of metastatic colorectal cancer and clinical implications. Front. Oncol. 13, 1310054 (2024).
Villéger, R. et al. Microbial markers in colorectal cancer detection and/or prognosis. World J. Gastroenterol. 24, 2327–2347 (2018).
Acknowledgements
The authors are grateful for bioinformatics support from LC-Bio Technology Co., Ltd. Thanks the generous support of Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital and Guangxi Clinical Research Center for Colorectal Cancer. This research was partially supported by Natural Science Foundation of Guangxi (2023GXNSFDA026032 and 2024GXNSFBA010083), National Natural Science Foundation of China (82160495).
Author information
Authors and Affiliations
Contributions
Linhai Yan and Maosen Huang developed the concept and designed this research. Xiaoxia Wei, Fangchao Zhong, and Lihua Fu provided clinical specimens. Maosen Huang, Xiaoxia Wei, and Linhai Yan analyzed clinical and sequencing data. Maosen Huang, Haiming Ru, and Xianwei Mo revised the paper. Linhai Yan supervised this study. All authors read and approved this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. This manuscript has not been published before and is not being considered for publication elsewhere. All authors have contributed to the creation of this manuscript for important intellectual content and read and approved the final manuscript.
Consent to publish
All authors agreed on the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, L., Wei, X., Zhong, F. et al. Intratumoral microbial community profiling identifies clinicomolecular and prognostic subtypes of colorectal cancer liver metastasis. npj Precis. Onc. 9, 284 (2025). https://doi.org/10.1038/s41698-025-01075-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-025-01075-5