Abstract
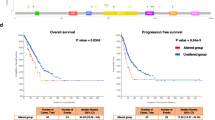
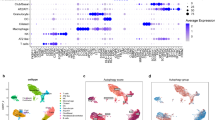
Genetic interactions (GIs) drive carcinogenesis and treatment resistance via non-additive phenotypic effects between genes. Traditional bulk-based methods fail to capture cell-type-specific interactions in heterogeneous tumors like lung adenocarcinoma (LUAD), limiting precision oncology. Resolving cell-type-specific GIs at single-cell resolution persists as a major hurdle, hindered by limited analytical methodologies. Here, we develop scPGI-finder, a computational framework that identifies gene pairs whose coordinated high expression is associated with higher proliferation-related fitness at single-cell resolution, which we refer to operationally as single-cell positive genetic interactions (scPGIs). Using scPGI-finder, we identify 49,808 and 15,896 scPGIs spanning epithelial cells and T cells in LUAD, respectively. The predicted scPGIs display tighter junctions in the protein interaction network compared to non-scPGIs. Furthermore, we demonstrate the predictive power of scPGIs for malignancy and immunotherapy response through multi-omics validation across diverse cohorts. Notably, with a mean area under the ROC curve (AUROC) of 0.974 in bulk tissue validation, the epithelial-derived scPGI classifier enables concordant malignancy identification across scales ranging from epithelial single cells and lung cancer cell lines, through spatial transcriptomic maps, to bulk LUAD tissue profiles. Additionally, a six-scPGI T cell signature reliably forecasts immunotherapy efficacy, with AUROC values exceeding 0.80 across multiple datasets. Together, our research advances the understanding of underlying cancer-positive GIs at the single-cell level. scPGIs of epithelial and T cells serve as robust biomarkers for malignancy evaluation and treatment response, offering a translational framework for precision oncology.
Similar content being viewed by others
Data availability
All data analyzed during this study can be downloaded from public databases or retrieved from associated files of papers, such as The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/), Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), CCLE, DepMap (https://depmap.org/portal/) and the molecular signatures database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb), CTRP (https://portals.broadinstitute.org/ctrp/), GDSC (https://www.cancerrxgene.org/). Detailed information was provided in the Supplementary Tables.
Code availability
The scPGI-finder pipeline is available at GitHub (https://github.com/chencccchen/scPGI-finder-pipeline) with an archived version on Zenodo (https://zenodo.org/records/17817076). Because scPGI-finder is implemented as an analysis workflow rather than a standalone software package, the scripts can be executed directly in a standard R environment without installation. The repository provides step-by-step instructions to reproduce the analyses reported in this study. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
Wang, J. et al. Computational methods, databases and tools for synthetic lethality prediction. Brief Bioinform. https://doi.org/10.1093/bib/bbac106 (2022).
Setton, J. et al. Synthetic lethality in cancer therapeutics: the next generation. Cancer Discov. 11, 1626–1635 (2021).
Fong, S. H. et al. A multilineage screen identifies actionable synthetic lethal interactions in human cancers. Nat. Genet. 57, 154–164 (2025).
van de Haar, J. et al. Identifying epistasis in cancer genomes: a delicate affair. Cell 177, 1375–1383 (2019).
O’Meara, C. P. et al. Genetic landscape of T cells identifies synthetic lethality for T-ALL. Commun. Biol. 4, 1201 (2021).
Liu, M. et al. Synthetic viability induces resistance to immune checkpoint inhibitors in cancer cells. Br. J. Cancer 129, 1339–1349 (2023).
Sahu, A. D. et al. Genome-wide prediction of synthetic rescue mediators of resistance to targeted and immunotherapy. Mol. Syst. Biol. 15, e8323 (2019).
Gallagher, M. D. & Chen-Plotkin, A. S. The post-GWAS era: from association to function. Am. J. Hum. Genet. 102, 717–730 (2018).
Zhou, P. et al. Single-cell CRISPR screens in vivo map T cell fate regulomes in cancer. Nature 624, 154–163 (2023).
Kuzmin, E. et al. Systematic analysis of complex genetic interactions. Science https://doi.org/10.1126/science.aao1729 (2018).
Costanzo, M. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science https://doi.org/10.1126/science.aaf1420 (2016).
Lee, J. S. et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell 184, 2487–2502 e2413 (2021).
Jerby-Arnon, L. et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 158, 1199–1209 (2014).
Avila Cobos, F., Vandesompele, J., Mestdagh, P. & De Preter, K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics 34, 1969–1979 (2018).
Bravo Gonzalez-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
Su, C. et al. Cell-type-specific co-expression inference from single cell RNA-sequencing data. Nat. Commun. 14, 4846 (2023).
Wilk, A. J., Shalek, A. K., Holmes, S. & Blish, C. A. Comparative analysis of cell-cell communication at single-cell resolution. Nat. Biotechnol. 42, 470–483 (2024).
Lafzi, A., Moutinho, C., Picelli, S. & Heyn, H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 13, 2742–2757 (2018).
Salcher, S. et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 40, 1503–1520 e1508 (2022).
Gu, Y. et al. A landscape of synthetic viable interactions in cancer. Brief. Bioinform. 19, 644–655 (2018).
Magen, A. et al. Beyond synthetic lethality: charting the landscape of pairwise gene expression states associated with survival in cancer. Cell Rep. 28, 938–948 e936 (2019).
Zhang, Y. et al. A T cell resilience model associated with response to immunotherapy in multiple tumor types. Nat. Med. 28, 1421–1431 (2022).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Lu, S. & Keles, S. Dozer: Debiased personalized gene co-expression networks for population-scale scRNA-seq data. bioRxiv https://doi.org/10.1101/2023.04.25.538290 (2023).
Yang, L. et al. Single-cell transcriptome analysis revealed a suppressive tumor immune microenvironment in EGFR mutant lung adenocarcinoma. J. Immunother. Cancer https://doi.org/10.1136/jitc-2021-003534 (2022).
Moreno Leon, L. et al. The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA contributes to an aggressive phenotype in lung adenocarcinoma through regulation of oxidative stress. Oncogene 38, 7146–7165 (2019).
Lu, T. P. et al. Identification of regulatory SNPs associated with genetic modifications in lung adenocarcinoma. BMC Res. Notes 8, 92 (2015).
Lu, T. P. et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Biomark. Prev. 19, 2590–2597 (2010).
Selamat, S. A. et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 22, 1197–1211 (2012).
Chang, I. S. et al. Genetic modifiers of progression-free survival in never-smoking lung adenocarcinoma patients treated with first-line tyrosine kinase inhibitors. Am. J. Respir. Crit. Care Med. 195, 663–673 (2017).
Chen, K. Y. et al. Estrogen receptor gene polymorphisms and lung adenocarcinoma risk in never-smoking women. J. Thorac. Oncol. 10, 1413–1420 (2015).
Okayama, H. et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111 (2012).
Yamauchi, M. et al. Epidermal growth factor receptor tyrosine kinase defines critical prognostic genes of stage I lung adenocarcinoma. PLoS ONE 7, e43923 (2012).
Robles, A. I. et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thorac. Oncol. 10, 1037–1048 (2015).
Girard, L. et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin. Cancer Res. 22, 4880–4889 (2016).
Tomida, S. et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 27, 2793–2799 (2009).
Tang, H. et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin. Cancer Res. 19, 1577–1586 (2013).
Rousseaux, S. et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 5, 186ra166 (2013).
Schabath, M. B. et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 35, 3209–3216 (2016).
Takano, Y. et al. Spatially resolved gene expression profiling of tumor microenvironment reveals key steps of lung adenocarcinoma development. Nat. Commun. 15, 10637 (2024).
Hu, J. et al. Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Med. 15, 14 (2023).
Jung, H. et al. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun. 10, 4278 (2019).
Kim, J. Y., Choi, J. K. & Jung, H. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin. Epigenetics 12, 119 (2020).
Ravi, A. et al. Genomic and transcriptomic analysis of checkpoint blockade response in advanced non-small cell lung cancer. Nat. Genet. 55, 807–819 (2023).
Fehrenbacher, L. et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387, 1837–1846 (2016).
Higgs, B. W. et al. Interferon gamma messenger RNA signature in tumor biopsies predicts outcomes in patients with non-small cell lung carcinoma or urothelial cancer treated with durvalumab. Clin. Cancer Res. 24, 3857–3866 (2018).
Hwang, S. et al. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci. Rep. 10, 643 (2020).
Leng, Y. et al. GDPLichi: a DNA damage repair-related gene classifier for predicting lung adenocarcinoma immune checkpoint inhibitors response. Front. Oncol. 11, 733533 (2021).
Jang, H. J. et al. Transcriptome-based molecular subtyping of non-small cell lung cancer may predict response to immune checkpoint inhibitors. J. Thorac. Cardiovasc. Surg. 159, 1598–1610 e1593 (2020).
Ayers, M. et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Doroshow, D. B. et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 18, 345–362 (2021).
Herbst, R. S. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014).
Agioti, S. & Zaravinos, A. Immune cytolytic activity and strategies for therapeutic treatment. Int. J. Mol. Sci. https://doi.org/10.3390/ijms25073624 (2024).
Lin, C. J. et al. Genetic interactions reveal distinct biological and therapeutic implications in breast cancer. Cancer Cell 42, 701–719 e712 (2024).
Birkealv, S. et al. Mutually exclusive genetic interactions and gene essentiality shape the genomic landscape of primary melanoma. J. Pathol. 259, 56–68 (2023).
Liu, F. et al. Cryo-shocked tumor cells deliver CRISPR-Cas9 for lung cancer regression by synthetic lethality. Sci. Adv. 10, eadk8264 (2024).
Pacini, C. et al. A comprehensive clinically informed map of dependencies in cancer cells and framework for target prioritization. Cancer Cell 42, 301–316 e309 (2024).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Yang, C. et al. Mapping the landscape of synthetic lethal interactions in liver cancer. Theranostics 11, 9038–9053 (2021).
Yu, G. et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Kim, N. et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 11, 2285 (2020).
Wu, F. et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12, 2540 (2021).
Gene Ontology, C. et al. The Gene Ontology knowledgebase in 2023. Genetics https://doi.org/10.1093/genetics/iyad031 (2023).
Tate, J. G. et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47, D941–D947 (2019).
Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Huang, J. K. et al. Systematic evaluation of molecular networks for discovery of disease genes. Cell Syst. 6, 484–495.e485 (2018).
Hu, F. F., Liu, C. J., Liu, L. L., Zhang, Q. & Guo, A. Y. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Brief Bioinform. https://doi.org/10.1093/bib/bbaa176 (2021).
Schwartz, L. H. et al. RECIST 1.1-update and clarification: from the RECIST committee. Eur. J. Cancer 62, 132–137 (2016).
Altman, D. G., Lausen, B., Sauerbrei, W. & Schumacher, M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J. Natl. Cancer Inst. 86, 829–835 (1994).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (No. 2023YFF1204600), the National Natural Science Foundation of China (No. 32470702 and 32270710), the Scientific Research Project of Provincial Scientific Research Institutes of Heilongjiang Province (No. CZKYF2024-1-A010), National Multidisciplinary Innovation Team Project in Traditional Chinese Medicine (No. ZYYCXTD-D-202407), and Chunyan Team Program of Heilongjiang Province (No. CYQN24043). In addition, the authors acknowledge the efforts of all the researchers who have contributed the data to the public databases of TCGA, GEO, DepMap, CTRP, GDSC, COSMIC, and MSigDB. The interpretation and reporting of these data are the sole responsibility of the authors.
Author information
Authors and Affiliations
Contributions
Y.Y.G., B.C., and J.J.L. designed the project. B.C., M.Y.L., and Q.D. wrote the paper. B.C., K.D.L., and C.L. collected data and performed bioinformatic analysis. H.M.H., L.Z.W., N.Z., and W.Y.Z. provided valuable suggestions for the paper. All authors read and approved of the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, B., Liu, M., Dong, Q. et al. Charting cell-type-specific positive genetic interaction at single-cell resolution for lung adenocarcinoma. npj Precis. Onc. (2026). https://doi.org/10.1038/s41698-026-01328-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-026-01328-x