Abstract
We introduce a proof-of-concept extended reality (XR) environment for discussing cancer, presenting genomic information from multiple tumour sites in the context of 3D tumour models generated from CT scans. This tool enhances multidisciplinary discussions. Clinicians and cancer researchers explored its use in oncology, sharing perspectives on XR’s potential for use in molecular tumour boards, clinician-patient communication, and education. XR serves as a universal language, fostering collaborative decision-making in oncology.
Similar content being viewed by others
Introduction
Genomic data holds extraordinary promise for transforming our understanding of cancer, in the laboratory and the clinic. As the scientific community endeavours to resolve the multitude of remaining questions in cancer, we will rely on layers of information coming from diverse sources and must capitalise on the collective expertise of multidisciplinary teams working at the intersections of disciplines1. In the clinic, interpreting genomic data in its spatial and temporal context is essential for making treatment decisions for cancer patients2,3. Interpreting genomic data with respect to time and space is critical for a deeper understanding of tumour evolution during metastatic dissemination, with the majority of cancer deaths caused by metastases4. Yet, current tools are not equipped to integrate genomic, radiological and therapeutic information5, nor is the human brain effective at holding this information for discussing such complex cases6. Generating data on multiple tumours from an individual is becoming increasingly common in research and the clinic, to face the clinical realities of tumour heterogeneity7,8. Therefore, interpreting genomic data in light of other important system components9, and by multidisciplinary teams, becomes increasingly critical.
A person who had a primary lung neuroendocrine tumour and ninety metastases disseminated through their body came to their oncologist requesting to donate their tumour tissues to research after death. Accepting this invaluable donation set us on a journey, a high-resolution n = 1 study10, where we sought to distil new knowledge about tumour evolution that may be missed in studies of more patients with fewer samples. Research autopsies are emerging internationally as an unmatched resource for studying tumour evolution, the emergence of treatment resistance, immune evasion and optimal drug targeting strategies, driven by selfless patients as partners in research11. We generated a large body of multi-omic data from 42 spatially distinct tumour sites, which presented a significant comprehension and analysis challenge. We aimed to integrate genomic information from multiple sites within metastatic tumours across their body collected at autopsy, complemented by circulating tumour DNA assays, clinical biopsies, radiological information showing tumour growth and shrinkage across a ten-year timeline, and therapeutic information, to produce a detailed understanding of the drivers and processes of tumour evolution in this person’s disease8.
Turning to extended reality (XR) technology for visualising this complex data provided opportunities to rethink and reimagine how cancer data can be presented and interrogated by teams of people. XR is an umbrella term that describes technologies that allow the merging of digital and physical information and encompasses virtual reality (VR), augmented reality (AR) and mixed reality (MR). The latter two are immersive technology that projects 3D digital elements into the physical world, usually through headsets12 or smartphones, that enable both digital and face-to-face interactions primed to enhance collaboration13. Although potential clinical avenues for XR have been explored, including telemedicine14, surgical planning15, psychological treatments16, and medical education17, alongside research-grade bioinformatic tools18,19, integrating multimodal oncology datasets for multidisciplinary investigation hasn’t yet been explored in XR.
Results
Design overview
Our multidisciplinary team of scientists, clinicians, architects, digital spatial design specialists and software developers worked through an iterative design process over two years to produce an advanced extended reality (XR) environment bringing together layers of data. We aimed to produce a tool to integrate genomic, radiology and therapeutic data from across time and space for this single patient with an unusually large research dataset. In doing so, we hoped to better understand the evolution of this patient’s tumours8, and consider the future of novel collaborative and immersive tools beyond this one patient.
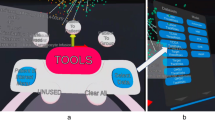
Our model takes the form of an immersive extended reality arena, merging physical and digital elements (Fig. 1). It enables the presentation of genomic data, in its anatomical context, alongside tumour scans and other relevant clinical information. Using Microsoft HoloLens II MR headsets, teams of up to ten users may collaboratively interact with the data projected into the physical room through the headsets, including interacting with different parts of the model simultaneously. At its heart, stands an accurate patient body model (skeleton, organs) derived from CT scans, forming the base for other information to be displayed. Overlaid are 3D representations of the tumours derived from the CT scans, enabling users to scroll through the timeline and observe tumours growing, shrinking in response to treatment, and ultimately spreading around the body. The timeline also indicates therapies received (chemotherapy and radiotherapy, including the field of radiotherapy).
a Overview of the information displayed in the model, including interactive phylogenetic tree (left), central body model showing tumours (red) overlaid on organs (blue), with genomic data sites labelled and coloured according to shared genomic information in the phylogenetic tree, and interactive annotated timeline (right). b See tumours changing through time, with respect to other clinical information. Users may slide through a timeline, annotated with therapeutic information, to access radiology-based tumour information displayed on the main body model (in red, shown behind participants). c Recognise genomic tumour heterogeneity. The lung has been isolated from the main body model for detailed inspection. Sampled tumour sites for which genomic data was generated are annotated. Labels relate to sample site codes, with colouring representing the genomic clades of tumour-relatedness. d Workspace for multidisciplinary collaboration. The tool was designed to be used by up to ten participants simultaneously, allowing multidisciplinary teams to discuss different facets of the data and integrate their personal understanding of the layers of data together. For example, major tumours can be ‘pulled’ out of the main skeletal model, as can be seen here, where the participants on the left are examining the pancreatic tumour while the participants on the right are examining the primary lung tumour. A further participant is discerning extra information about tumour genomic data on a large monitor at the back of the room. A phylogenetic tree based on DNA variants shown to the left of the main patient model allows users to view the spatial distribution of tumours that share common genomic changes and indicate relatedness. Users may highlight a clade on the phylogenetic tree and observe these samples highlighted on the main model. Many tasks can be completed simultaneously by small groups of people, or alternatively, one user may guide all other participants through the dataset, with all users seeing the same information through their headsets. b–d shows the view through one participant’s headset, while they are interacting with other participants.
Simplistic representations of the genomic data are included at each of the 42 sampled sites8. Genomic relatedness is indicated by colour scheme, where groups of tumours carrying common variants are highlighted in the same colour, allowing the genomic data to be interpreted in the context of tumour changes detected on CT scans and treatment information. An interactive 3D evolutionary tree presents information on the relatedness of tumours based on shared DNA variants, and users may highlight clades (groups of related tumours) on the main body model via the evolutionary tree. Users may reach into the model and ‘pull’ a tumour towards them, to have the tumour enlarged and overlaid with representative genomic information. We also produced a tablet application that provided detailed genomic variant annotations, with the ability to highlight the anatomical position of all tumours carrying that variant in the XR model (viewed on a large monitor in Fig. 1d). The multiplayer XR visualisation allows teams of users to interact with the data together, each member coming from a different speciality and perspective, facilitating the work of a translational multi-disciplinary team. This encourages detailed interdisciplinary discussions and enables a deeper understanding and integration of the datasets than is possible with traditional tools. In addition to designing the XR model, the physical space was carefully designed to facilitate bringing together multiple users to discuss the data, through furniture supporting the use of laptops and other traditional tools, and additional monitors for data display (Fig. 2). A video demonstration of the model can be viewed in Supplementary Video 1.
a Digital information accessible via Microsoft HoloLens MR headset, including patient model (within yellow circle at centre), model controls including time-slider and evolutionary tree (pink circle), and static overview graphics on the wall (green circle). b Physical elements enable best use of the model, including space to use traditional tools such as laptops and whiteboards, and ample space for members of the team to explore the data individually or together.
This XR tool allowed us to better visualise the complex genomic information generated for this patient’s 42 metastatic tumours and the hypothetical evolutionary paths these tumours could have followed. While this information was described in the format of a traditional peer-reviewed journal paper recently8, the static two-dimensional figures that constrain traditional publications provide only limited opportunities to visualise and understand the anatomical and temporal relationships between tumours. However, by using XR, we can better understand the anatomical relationships between genomic clades with respect to tumour growth and spread (Supplementary Fig. 1), and visualise extensive tumour heterogeneity within large tumours where multiple samples were analysed (Supplementary Fig. 2).
Enhanced ‘asymmetric communication’ emerged during development
While our team built this model to meet an immediate need within our research, we realised there might be further applications for this work. We completed a formal qualitative evaluation of an early version of the model, where we invited eighteen cancer clinicians and scientists to interact with the model in pairs while being observed by the research team, and complete an interview on the experience (described in the supplement). We thematically analysed observations and interview responses, coding according to dominant themes (Supplementary Fig. 3). During this testing, clinicians spontaneously described how they would use it in an oncology clinic20. Through presentations at national and international medical conferences, and exhibiting at the international design and technology festival Ars Electronica, Linz, we could informally test hypotheses around how multi-modal cancer data can be integrated and interrogated by teams of people, and share our research with a wide audience, educating them about cancer, tumour evolution and metastasis in the process. The Ars Electronica audience included diverse technical backgrounds spanning engineering, science, medicine, and art, alongside non-specialists, yet their questions were similar, centred around applying this model to other patients and using a tool like this in clinical care. These interactions inspired our team to consider our project as a proof-of-concept for future medical XR tools and investigate potential areas where this collaborative model may add value.
To explore additional views, leaders in oncology, translational research and cancer biology joined the research group and spent two days immersed in the model; their impressions were gathered informally. They were tasked with exploring the model through different lenses, including research, clinical and educational applications. The remainder of this article describes the shared conclusions of this meeting.
The fundamental strength of the XR model was in its role as a communication tool, priming discussion between disciplines. When participants with different areas of expertise can work together on a problem, gains in understanding are made. The XR model provides a universal language and facilitates ‘asymmetric communication’, creating a level playing field when each contributor has a different specialist background.
Cancer data, or indeed any data, holds the most meaning when interpreted in ‘context’21,22,23; in this case, with respect to treatment, disease stage, genomic data from different tumour sites, and more. There are many emerging tools for visualising cancer genomic data24, including other endeavours to visualise cancer data in XR18. However, our approach is distinct as it is designed for use in multidisciplinary environments to achieve a greater understanding at the resolution of a single patient. The XR model provides a fast, powerful and efficient way to transmit complex information and facilitate communication around the results.
Our approach provided users with a holistic global perspective of the patient’s cancer, rather than a reductionist view. This, in turn, offers a visual representation of tumour stage, an important parameter of prognosis25. Users quickly grasp the natural history of these tumours, including sample relatedness and tumour evolution under treatment pressure. By placing genomic findings from single samples in their anatomical context, layered among other clinical data, interpreting tumour heterogeneity comes naturally.
Summarising datasets visually is a balancing act between retaining enough granularity or depth of information (particularly in the genomic data), and interpretability by a range of specialists. Some of our research team reasoned that the simplification of the data limited its use. However, the purpose is not to replace the subspecialist tools but to present the information in a way that can be interpreted by the non-specialist, to facilitate interdisciplinary communication. Therefore access to summarised data and specialised technical data is required in the model. Due to the augmented nature of the technology, XR is easily used in combination with existing subspecialist tools such as genome browsers and bioinformatic tools, as we do in our research group, or radiology browsers as we do in the clinic.
Advancing our understanding of tumour evolution, or improving cancer patient care, requires the coordinated knowledge of multidisciplinary teams, across the intersection of disciplines, each possessing a small piece of the puzzle—i.e., the concept of asymmetric communication. Beyond research, asymmetric communication scenarios that our approach is directly relevant to include multidisciplinary clinical decision-making, clinician-patient communication, and medical education, each explored below.
Multidisciplinary clinical decision-making
Multidisciplinary decision-making in a clinical environment shares commonalities with the research scenario we faced: teams of specialists must gather to integrate knowledge. For example, in a tumour board, such as the molecular tumour board (MTB) at our centre, this information covers treatment history, histopathology, radiology, and genomics (which may include samples from different tumours, different time points, and/or liquid biopsy samples)26. Integrating this data informs genomics-led care for improved patient outcomes27,28. The MTB is an advanced communication exercise at the intersection of many disciplines, serving to integrate domain knowledge held by different specialists. This task, by nature, must occur in teams; therefore, it needs tools optimised for use by teams. Overlaying genomic information at the sites of tumour samples within 3D tumour models encourages users to make connections between previous treatments, tumour growth patterns and genomic profiles. This approach helps to highlight potential genomic tumour heterogeneity and evolution, uncover truncal driver variants suitable for longitudinal monitoring and/or targeted therapy, and shape future treatment decisions29.
An XR tool integrating these layers of information enhances communication between the disciplines participating in an MTB and may act as a 'language bridge', ensuring content is accessible and easily digestible, and promoting a shared conversation where each participant plays an equal role. For example, non-clinician scientists often have little prior experience in interpreting CT scans, and many clinicians are not genomic specialists, so a visual reference to these datasets benefits both parties20. Even for those familiar with CT scans, XR has been shown to improve spatial understanding and time to synthesise information30, and we found that specialists would naturally guide others on information outside their speciality, aided by the model. Better communication produces informed treatment decisions integrating all information27, particularly important in complex cases brought to an MTB. Improving the efficiency of an MTB by bringing complex information together quickly helps to improve accessibility and scalability31, in turn helping to bring the MTB into routine care32. An immersive educational environment33,34, without the distractions present in other learning environments, helps to fulfil the educational mandate of the MTB and provides genomic information at a superficial level suitable for non-specialists. It was the shared inference of our research team that the XR model facilitated the full participation of all members of a multidisciplinary clinical team, helping to ensure that the most appropriate treatment decision would be reached. Participants remarked on the ease of inferring connections between treatment information (e.g., position of radiotherapy beam), tumour spread and genomic heterogeneity, for example (another example demonstrated in Supplementary Fig. 1). However, further studies would be needed to formally measure this. Finally, with XR it is possible to facilitate remote participation in an MTB34,35, particularly in centres lacking international expertise36 (however, we have not included this feature in our prototype). It may enable experts in any location to dial into a treatment meeting via HoloLens and participate in an immersive discussion with all patient data presented. Using XR to augment multidisciplinary clinical communication has the potential to benefit patients from many angles; through better clinical decision-making, upskilled clinicians and the ease of facilitating international expert collaboration.
Elevating clinician-patient communication
When sharing the XR model with our patient’s family, they noted a deep connection to the representation of their deceased family member and requested to spend time alone in the XR model as a family. They noted a greater understanding of their mother’s disease and wished that they could have used the model during her clinical care. This sentiment was shared by members of the public at the Ars Electronica festival. The clinicians in our team echoed this response, recognising that many patients and their families are eager to see, understand and follow their disease data, but noting that current tools (such as 2D computer software for viewing radiological data) are not optimised for easy interpretation by non-specialists. One used a poignant analogy; “the ocean swimmer is more scared of sharks than the scuba diver.” We should give patients the chance to go ‘underwater’ and see with clarity what is going on in their disease, via tools optimised for interpretation by non-specialists. Representing tumours identified in medical imaging in 3D, as opposed to their native grey and black 2D format, may provide information to a patient and their family in a more accessible form. Past studies have highlighted that while patients generally gain a stronger understanding of their disease from seeing their own CT scans37, the use of 3D images/models may aid patients in forming a detailed and accurate understanding of their disease, information recall and trust in their diagnosis38. This may have important flow-on benefits, such as revolutionising consent to surgical procedures through an increased understanding of tumour positioning and surgical consequences39,40,41, and may work towards the democratisation of data, where patients can access their own medical data in a non-specialist format.
While increasing patient understanding may have many benefits, we note that not all patients would want to confront their disease in immersive XR, a point raised during the evaluation of an early prototype with clinicians20. Not all people with cancer have the same psychological coping style, with a spectrum from ‘monitoring’ to ‘blunting’42. Those identifying with ‘monitoring’ typically want all the information about their disease that they can get, whereas those identifying with ‘blunting’ are more likely to avoid this information. It is recognised that patients fare better (both psychologically and physiologically) when the information they receive about their disease is tailored to their psychological coping style42—something clinicians would assess before showing an XR model to their patients.
Medical and scientific education
Whereas the complexities of using XR technologies for patient communication may be solved long-term (via new software development pipelines), we see education as an immediate opportunity for XR technologies. In biomedical science education, grasping the clinical context is difficult for students without direct clinical exposure, and in the clinic, further genomics education is needed. Without any modification, our XR model may facilitate transdisciplinary education to better equip scientists to solve clinical challenges and lift genomics literacy in the clinic, bridging these educational silos. When we conducted a formal qualitative evaluation of an early prototype of our XR tool, many participants (irrespective of career seniority or field of expertise) came away with a greater appreciation for tumour heterogeneity and evolution, the form that metastatic cancer can take, the limitations of assaying single biopsies, and the value of circulating tumour DNA for capturing tumour heterogeneity20. This is critical understanding for interpreting single tumour biopsies in the absence of additional information. Participants also noted a greater appreciation for the patient face of biomedical research, for the selfless person behind the data. The deep individual focus on a single patient in this XR model mirrors the personalised focus during ideal clinical care. Further investigation is needed on the effectiveness of immersive virtual environments in educational scenarios, as some but not all previous studies have found a benefit; however, noting that past studies finding no significant educational benefit to the immersive technology have focussed on virtual rather than augmented technologies43,44.
Pitfalls, perils and possibilities
There are very few examples of new health technology reducing inequity45,46. An XR model, therefore, requires careful implementation in the right clinical scenario to protect against this substantial risk, and development with diverse input, including expertise from Indigenous people. In addition to asserting rights-based (UNDRIP Article 31) Indigenous oversight and stewardship of data belonging to Indigenous patients and whānau incorporated into XR environments, Indigenous knowledge is best placed to shape processes involving such data (including images) in culturally appropriate ways47. We suggest that adopters of this XR concept globally should engage early with Indigenous experts local to the implementation site. Furthermore, implementation must prioritise a clinical scenario where this technology has the greatest potential to benefit patients; where care is multidisciplinary, provided by a network of clinicians, where disease is complex and metastatic, and where genomics might add value. In most clinical cases currently, at our MTB for example, genomic sequencing is available on only one or a small number of tumours and/or blood plasma samples27, however, an XR model still provides an opportunity to integrate this data with serial medical imaging and other clinical information.
Any new clinical tools must be optimised for a time-poor clinical system, noting the learning curve and time to use an XR model in existing clinical workstreams. A major barrier for adoption, particularly in resource-stretched health systems like New Zealand’s, is the cost of the XR technology and its implementation. Currently available XR headsets have several technical limitations, including those related to ergonomics, display resolution and field of view, and processing power. HoloLenses or other XR headsets are not currently used in most hospitals, although this will change as the technology improves and becomes mainstream. The COVID-19 pandemic facilitated the widespread adoption of digital technology in healthcare48—“a crisis provides an opportunity, and this crisis … provides a great opportunity for digital technology”49. Policy supporting the development and adoption of digital healthcare technologies has facilitated the uptake of emerging digital technologies internationally48,50,51. This can already be seen with the emergence of virtual or extended reality tools for surgical planning12,15,52,53 and medical education17,54,55,56,57, but XR tools have not yet been created for multidisciplinary decision-making or cancer patient consultation. Future tools incorporating the learnings of this proof-of-principle project, alongside technical and operational refinements, will allow practical implementation in appropriate clinical and educational environments. Critical will be usability refinements fit for clinical utility. These may include new ways to record discussions and outcomes occurring in XR (including video recording and integrated note-taking), speciality-dependent data views, incorporating domain analytical and decision support tools including AI tools, and integrating other data types such as histopathology58. These will ensure that the benefits of the XR approach are realised in the real world.
Conclusion
Genomics is transforming our understanding of cancer. To ensure these benefits are realised for people with cancer, we require new tools that meet the needs of multidisciplinary clinical teams, people with cancer, and the future genomic workforce. Here, we have shown the opportunities for a novel XR tool bringing together tumour-specific data in the context of medical imaging, creating a level playing field for discussing complex cancer data. We offer a window into future possibilities for meeting the interpretation and communication needs of collaborative cancer teams.
We are indebted to the patient at the heart of this project and her family. Her selfless donation instigated and encouraged this research.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Sharp, P. A. & Langer, R. Promoting convergence in biomedical science. Science 333, 527–527 (2011).
Rolfo, C. et al. Multidisciplinary molecular tumour board: a tool to improve clinical practice and selection accrual for clinical trials in patients with cancer. ESMO Open 3, e000398 (2018).
Moore, D. A. et al. Prospective analysis of 895 patients on a UK Genomics Review Board. ESMO Open 4, e000469 (2019).
Dillekås, H., Rogers, M. S. & Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med 8, 5574–5576 (2019).
Perakis, S. O. et al. Comparison of three commercial decision support platforms for matching of next-generation sequencing results with therapies in patients with cancer. ESMO Open 5, e000872 (2020).
Lazebnik, Y. Can a biologist fix a radio? Or, what I learned while studying apoptosis. Cancer Cell 2, 179–182 (2002).
Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
Robb, T. J. et al. Complex patterns of genomic heterogeneity identified in 42 tumor samples and ctDNA of a pulmonary atypical carcinoid patient. Cancer Res. Commun. 3, 31–42 (2023).
Hood, L. Systems biology and p4 medicine: past, present, and future. Rambam Maimonides Med. J. 4, e0012 (2013).
Blenkiron, C. et al. Tailoring a rapid autopsy protocol to explore cancer evolution: a patient collaboration. N. Z. Med. J. 132, 83–92 (2019).
Robb, T. J., Tse, R. & Blenkiron, C. Reviving the autopsy for modern cancer evolution research. Cancers 13, 409 (2021).
Pires, F., Costa, C. & Dias, P. On the use of virtual reality for medical imaging visualization. J. Digit Imaging 34, 1034–1048 (2021).
Yuan, J. et al. Extended reality for biomedicine. Nat. Rev. Methods Prim. 3, 14 (2023).
Wang, S. et al. Augmented reality as a telemedicine platform for remote procedural training. Sensors 17, 2294 (2017).
Pratt, P. et al. Through the HoloLens™ looking glass: augmented reality for extremity reconstruction surgery using 3D vascular models with perforating vessels. Eur. Radiol. Exp. 2, 2 (2018).
Ortiz-Catalan, M., Sander, N., Kristoffersen, M., Håkansson, B. & Brånemark, R. Treatment of phantom limb pain (PLP) based on augmented reality and gaming controlled by myoelectric pattern recognition: a case study of a chronic PLP patient. Front. Neurosci. 8, 71345 (2014).
Sivananthan, A. et al. A feasibility trial of HoloLens 2™; using mixed reality headsets to deliver remote bedside teaching during COVID-19. Res. Sq. 6, e35674 (2021).
Lau, C. W. et al. Virtual reality for the observation of oncology models (VROOM): immersive analytics for oncology patient cohorts. Sci. Rep. 12, 11337 (2022).
Pirch, S. et al. The VRNetzer platform enables interactive network analysis in Virtual Reality. Nat. Commun. 12, 2432 (2021).
Robb, T. J. Tumour evolution in a single patient PhD thesis, University of Auckland, (2021).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Goldman, M. J. et al. A user guide for the online exploration and visualization of PCAWG data. Nat. Commun. 11, 3400 (2020).
Qu, Z., Lau, C. W., Nguyen, Q. V., Zhou, Y. & Catchpoole, D. R. Visual analytics of genomic and cancer data: a systematic review. Cancer Inform. 18, 1176935119835546 (2019).
Saadatmand, S., Bretveld, R., Siesling, S. & Tilanus-Linthorst, M. M. A. Influence of tumour stage at breast cancer detection on survival in modern times: population based study in 173 797 patients. BMJ 351, h4901 (2015).
Erdmann, J. All aboard: will molecular tumor boards help cancer patients? Nat. Med. 21, 655–656 (2015).
Kato, S. et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat. Commun. 11, 4965 (2020).
Repetto, M. et al. Molecular tumour board at European Institute of Oncology: report of the first three year activity of an Italian precision oncology experience. Eur. J. Cancer 183, 79–89 (2023).
Fittall, M. W. & Van Loo, P. Translating insights into tumor evolution to clinical practice: promises and challenges. Genome Med. 11, 20 (2019).
Pelanis, E. et al. Use of mixed reality for improved spatial understanding of liver anatomy. Minim. Invasive Ther. Allied Technol. 29, 154–160 (2020).
Pishvaian, M. J. et al. A virtual molecular tumor board to improve efficiency and scalability of delivering precision oncology to physicians and their patients. JAMIA Open 2, 505–515 (2019).
Russo, A. et al. The challenge of the Molecular Tumor Board empowerment in clinical oncology practice: a position paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIC-SIF-SIGU-SIRM Italian Scientific Societies. Crit. Rev. Oncol. Hematol. 169, 103567 (2022).
Dodds, H. E. Immersive learning environments: designing XR into higher education.A Practitioner’s. Guide Instr. Des. High. Educ. 1, 69–79 (2021).
Zweifach, S. M. & Triola, M. M. Extended reality in medical education: driving adoption through provider-centered design. Digital Biomark. 3, 14–21 (2019).
Lee, Y. & Yoo, B. XR collaboration beyond virtual reality: work in the real world. J. Comput. Des. Eng. 8, 756–772 (2021).
Khader, J. Improving cancer outcomes through international collaboration in developing countries: King Hussein Cancer Center as a unique experience. J. Glob. Oncol. 4, 161s–161s (2018).
Carlin, L. E., Smith, H. E. & Henwood, F. To see or not to see: a qualitative interview study of patients’ views on their own diagnostic images. BMJ open 4, e004999 (2014).
Phelps, E. E., Wellings, R., Griffiths, F., Hutchinson, C. & Kunar, M. Do medical images aid understanding and recall of medical information? An experimental study comparing the experience of viewing no image, a 2D medical image and a 3D medical image alongside a diagnosis. Patient Educ. Couns. 100, 1120–1127 (2017).
Mulsow, J. J. W., Feeley, T. M. & Tierney, S. Beyond consent—improving understanding in surgical patients. Am. J. Surg. 203, 112–120 (2012).
Falagas, M. E., Korbila, I. P., Giannopoulou, K. P., Kondilis, B. K. & Peppas, G. Informed consent: how much and what do patients understand? Am. J. Surg. 198, 420–435 (2009).
Perin, A. et al. Informed consent through 3D virtual reality: a randomized clinical trial. Acta Neurochir. 163, 301–308 (2021).
Miller, S. M. Monitoring versus blunting styles of coping with cancer influence the information patients want and need about their disease. Implications for cancer screening and management. Cancer 76, 167–177 (1995).
Stepan, K. et al. Immersive virtual reality as a teaching tool for neuroanatomy. Int. Forum Allergy Rhinol. 7, 1006–1013 (2017).
Makransky, G., Terkildsen, T. S. & Mayer, R. E. Adding immersive virtual reality to a science lab simulation causes more presence but less learning. Learn. Instr. 60, 225–236 (2019).
Robertson, S. P. et al. Genomic medicine must reduce, not compound, health inequities: the case for hauora-enhancing genomic resources for New Zealand. N. Z. Med. J. 131, 81–89 (2018).
Madhusoodanan, J. Health-care inequality could deepen with precision oncology. Nature 585, S13–S13 (2020).
Henare, K. L. et al. Mapping a route to Indigenous engagement in cancer genomic research. Lancet Oncol. 20, e327–e335 (2019).
Dal Mas, F., Massaro, M., Rippa, P. & Secundo, G. The challenges of digital transformation in healthcare: An interdisciplinary literature review, framework, and future research agenda. Technovation 123, 102716 (2023).
Ting, D. S. W., Carin, L., Dzau, V. & Wong, T. Y. Digital technology and COVID-19. Nat. Med. 26, 459–461 (2020).
NHS England & NHS Improvement. Science in healthcare: Delivering the NHS long term plan. The Chief Scientific Officer’s strategy, https://www.england.nhs.uk/wp-content/uploads/2020/03/science-in-healthcare-delivering-the-nhs-long-term-plan.pdf (2020).
Vlčková, J. & Klimková, V. The digital transformation of Czech healthcare: trends and COVID-19 impact. Int. J. Electron. Healthc. 13, 15–32 (2023).
Nan, C., Pradosh, K. & Viktor, G. Augmented reality with Microsoft HoloLens holograms for near infrared fluorescence based image guided surgery. Proc.SPIE 10049, https://doi.org/10.1117/12.2251625, (2017).
Poothicottu Jacob, G. A HoloLens Framework for Augmented Reality Applications in Breast Cancer Surgery Master of Applied Science thesis, University of Waterloo, (2018).
Zafar, S. & Zachar, J. J. Evaluation of HoloHuman augmented reality application as a novel educational tool in dentistry. Eur. J. Dent. Educ. 24, 259–265 (2020).
Hillary, L. et al. HoloLens in suturing training. Proc.SPIE 10576, https://doi.org/10.1117/12.2293934, (2018).
Luck, J., Gosling, N. & Saour, S. Undergraduate surgical education during COVID-19: could augmented reality provide a solution? Br. J. Surg. 108, e129–e130 (2021).
Tu, P. et al. Augmented reality based navigation for distal interlocking of intramedullary nails utilizing Microsoft HoloLens 2. Comput. Biol. Med. 133, 104402 (2021).
Liimatainen, K., Latonen, L., Valkonen, M., Kartasalo, K. & Ruusuvuori, P. Virtual reality for 3D histology: multi-scale visualization of organs with interactive feature exploration. BMC Cancer 21, 1133 (2021).
Acknowledgements
TJR was supported by the Auckland Medical Research Foundation, and the project was funded by the Translational Medicine Trust and the Health Research Council of New Zealand. The authors thank the University of Auckland Media Productions team for capturing and preparing video footage. Above all, the authors thank the patient and their family for their generous donation to scientific research.
Author information
Authors and Affiliations
Contributions
T.J.R., Y.L., B.W., C.W., D.H., K.P., K.H., R.M., B.H., N.Y., C.G.P., M.D., U.R. and B.L. designed the extended reality model. J.R., B.H. and N.Y. segmented the CT scans. T.J.R., D.H., C.B. and C.G.P. curated the genomic data. Y.L., D.H., R.M., B.H. and N.Y. coded the XR model. Y.L., T.J.R. and U.R. prepared the video and figure. G.H., S.B.F., L.B., P.G., A.M., C.J., V.B., L.C. and S.D. evaluated the model and provided expert advice guiding future development. C.G.P., M.D., U.R. and B.L. led and secured funding for the project. T.J.R. thematically analysed the evaluations and drafted the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Robb, T.J., Liu, Y., Woodhouse, B. et al. Blending space and time to talk about cancer in extended reality. npj Digit. Med. 7, 261 (2024). https://doi.org/10.1038/s41746-024-01262-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-024-01262-x