Abstract
Intratumoral heterogeneity poses a significant challenge to the diagnosis and treatment of recurrent glioblastoma. This study addresses the need for non-invasive approaches to map heterogeneous landscape of histopathological alterations throughout the entire lesion for each patient. We developed BioNet, a biologically-informed neural network, to predict regional distributions of two primary tissue-specific gene modules: proliferating tumor (Pro) and reactive/inflammatory cells (Inf). BioNet significantly outperforms existing methods (p < 2e-26). In cross-validation, BioNet achieved AUCs of 0.80 (Pro) and 0.81 (Inf), with accuracies of 80% and 75%, respectively. In blind tests, BioNet achieved AUCs of 0.80 (Pro) and 0.76 (Inf), with accuracies of 81% and 74%. Competing methods had AUCs lower or around 0.6 and accuracies lower or around 70%. BioNet’s voxel-level prediction maps reveal intratumoral heterogeneity, potentially improving biopsy targeting and treatment evaluation. This non-invasive approach facilitates regular monitoring and timely therapeutic adjustments, highlighting the role of ML in precision medicine.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) exhibits pronounced intratumoral heterogeneity, which can confound diagnosis and clinical management, and is a leading driver of tumor recurrence1,2. Treatment-induced reactive changes further exacerbate intratumoral heterogeneity3,4. Because histopathological and molecular analyses are limited by sparse biopsy sampling, there is a significant need to develop non-invasive approaches to map the heterogeneous landscape of histopathological alterations throughout the entire lesion. Such advancements would improve surgical targeting of confirmatory biopsies and non-invasive assessment of neuro-oncological treatment response, thereby informing subsequent therapeutic strategies. Radiogenomics is a growing research field, which seeks to develop machine learning (ML) models to predict cellular, molecular and genetic characteristics of tumors based on Magnetic Resonance Imaging (MRI) and other imaging types2,5,6. Radio(gen)omics methods have been shown to accurately predict not only diversity in tumor cell density associated with diffuse invasion into the brain parenchyma peripheral to the frank lesion seen on MRI7,8,9, but also abnormalities in hallmark genes such as EGFR, PDGFRA, and PTEN10,11,12,13, IDH mutation status14,15,16,17,18, and MGMT methylation status based on radiographic features13,14,15. Histopathology-validated machine learning models have been developed to discriminate between true progression and pseudo-progression in GBM by using MRI19,20,21,22,23,24. These studies represent examples in which the prediction provides a single categorical label per tumor using imaging features that span the entire lesion.
However, precise representations of intratumoral heterogeneity require voxel-wise labels (e.g., image-localized biopsies) that reflect local or regional characteristics of the lesion. A major challenge for such prediction is the lack of large image-localized biopsy datasets25,26 to train deep learning (DL) models that are well-known to be heavily-parameterized and data-hungry. Creation of large training datasets is limited by various factors such as the invasiveness and high expense of sample acquisition, need of highly-specialized experts to create accurate labels, and difficulty in patient recruitment25. Moreover, the lack of large datasets has severely limited the number of studies focusing on predicting regional characteristics within each lesion, which are crucial for revealing intratumoral heterogeneity. A few studies have developed MRI-biology fusion models to predict regional cell density27,28,29,30,31,32 or regional copy number variation of individual driver genes such as EGFR, PDGFRA, and PTEN, in untreated, primary GBM33,34. In recurrent GBM (recGBM), however, treatment-induced reactive changes lead to additional intratumoral heterogeneity and the related additional complexity in tissue composition makes prediction of gene modules more difficult35.
In this study, we compiled a unique dataset that included multi-region biopsy samples and MRI from recGBM patients. The dataset consisted of derived measurements for three gene modules, from each biopsy, by combining data of individual gene expressions from RNA sequencing and cellular composition patterns from immunohistochemistry (IHC). The three gene modules identified through gene ontology analysis include: proliferative (Pro), associated with proliferation and cell cycle ontologies indicative of recurrent tumor; inflammatory (Inf), linked to cytokine production and immune response, representative of treatment-induced reactive cells; and neuronal (Neu), related to neuronal signaling, reflecting infiltrated brain tissue. Assessing the gene modules of GBM has significant clinical value and has drawn much attention recently36. For recGBM, the ability to differentiate proliferative/recurrent tumor and treatment-induced reactive/inflammatory cells (two primary gene modules in our dataset) is crucial for evaluating treatment effectiveness. However, such differentiation is notoriously difficult in clinical practice due to their indistinguishable appearances on MRI. Even among seasoned practitioners, accurately distinguishing between proliferative/recurrent tumors and treatment-induced reactive/inflammatory cells remains an elusive task. Currently, the sole method for distinguishing between these two gene modules is obtaining biopsies and conducting comprehensive transcriptomic and immunohistochemical profiling. However, biopsies, the gold-standard approach, can only cover a few sparse regions, leaving substantial regions within the lesion unexamined and the differentiation in these regions is nearly equivalent to a random guess. Therefore, our unique dataset, comprising a development cohort and a test cohort, facilitated the first-ever development of a non-invasive approach based on MRI and DL to predict voxel-level gene modules throughout the entire lesion for each patient.
To tackle the inherent challenge of limited training data from biopsy samples, we proposed BioNet, a novel unified framework whose learning capacity is significantly augmented by integrating multiple implicit and qualitative biological domain knowledge. The integration of biological/biomedical domain knowledge, such as biological principles, empirical models, simulations, and knowledge graphs, can provide a rich source of information (pseudo data) to help alleviate the data shortage in training DL models. Various approaches have been proposed for integrating domain knowledge, depending on its form. For example, some researchers proposed to use the knowledge of biological pathways to guide the design of DL architecture37,38. In certain biomedical domains, knowledge exists in the form of algebraic equations that capture biological principles, which were integrated with DL architecture or loss functions29,39. Some researchers proposed to integrate knowledge about feature behavior as attribution priors into DL training40. However, existing methods lack the ability to simultaneously incorporate multiple implicit, qualitative domain knowledge that is difficult to describe in mathematical formulations, making them unsuitable for our problem41. To fully harness the potential of this type of domain knowledge, BioNet integrated several strategies. Firstly, it creates large virtual biopsy datasets based on domain knowledge to pre-train the DL model, enabling it to learn generalizable feature representations that can be transferred to the downstream task based on real biopsy samples. Secondly, BioNet adopts a hierarchical design inspired by domain knowledge, considering the interaction between gene modules and their conditional relationships. Lastly, BioNet employs a knowledge attention loss function that combines data-driven and knowledge-driven components, penalizing violations of domain knowledge on unlabeled samples. These strategies collectively empower BioNet to effectively integrate domain knowledge into the learning process.
In summary, by leveraging two real clinical datasets, this study is the first of its kind that developed a non-invasive approach for quantifying regional distributions of gene modules representing proliferative/recurrent tumor and treatment-induced reactive/inflammatory cells using MRI in the recGBM setting. The mapping of these gene module distributions throughout the entire lesion for each patient offers valuable clinical benefits. It can assist in the identification of locations within a lesion for confirmatory biopsy sampling. With BioNet’s guidance, the likelihood of sampling locations with the desired gene modules is significantly improved, offering a more reliable alternative to the current sampling approach. It can also assist clinicians in evaluation of treatment effectiveness. The non-invasive nature of the approach can potentially facilitate regular monitoring of the gene modules over time, enabling identification of treatment response or resistance and making timely therapeutic adjustment. By gaining granularity from regional assessment, clinicians may gain better understanding of patient-specific nuances and tailor treatment more individually.
Results
Figure 1 presents an overview of the application of BioNet in assisting the assessment of treatment responses and informing subsequent therapy decisions.
Our datasets comprise two types of data: sparsely labeled data from biopsy locations, and abundant unlabeled data from all locations throughout the entire brain. The labels for biopsy samples (\({y}_{i}\)) are obtained through comprehensive transcriptomic and immunohistochemical profiling. The input features (\({x}_{i}\)) for all samples, both labeled and unlabeled, which are utilized in the training and testing of BioNet, are extracted from multiparametric MRI. Within the tumoral Area of Interest (AOI) of each patient (blue outline), local regions (small squares) were created by sliding windows according to the physical size of surgical biopsies. Statistical and texture features \(\left({x}_{i}\,\right)\) were computed based on multiparametric MRI within the sliding windows at biopsy locations (red, a few) and remaining unlabeled locations (yellow, abundant). Labeled samples, along with selectively chosen unlabeled samples, are employed in the training of BioNet. Once adequately trained, BioNet is capable of generating voxel-level prediction maps for proliferative/recurrent tumors (Pro) and treatment-induced reactive/inflammatory cells (Inf), respectively, within AOI. These prediction maps yield crucial insights into the gene status at the voxel level throughout the entire tumor.
Patient characteristics
This study involved a developmental cohort (A) and a test cohort (B). Cohort A was acquired as part of a retrospective, observational study designed to study patients who had undergone repeat surgical resection for recurrence of high-grade gliomas following chemotherapy and radiation therapy36. This cohort included 84 biopsies harvested from 37 patients (mean age = 56, 63% male, 1–3 biopsies per patient). Cohort B was acquired as part of a prospective, clinical trial for convection enhanced delivery of topotecan chemotherapy42. It comprised 31 biopsies from five patients (median age = 56, 60% male, 1–10 biopsies per patient). Biopsy samples from each patient were obtained according to IRB-approved protocol from the operating room (see details in Methods). Patient information was de-identified and maintained by a tissue broker who has designated clinical information. Both cohorts received standard of care neuroimaging performed at Columbia University Irving Medical Center within one month prior to surgery (Cohort A) or one day prior to surgery (Cohort B). The neuroimaging exam of each patient produced multiparametric MRI data including T1-weighted+Gd (T1Gd), T2-weighted (T2), FLAIR, apparent diffusion coefficient (ADC), and susceptibility weighted imaging (SWI). Detailed MRI parameter setting can be found in Methods and Supplementary Data 2.
Biologically-informed design principles for BioNet
Our goal is to develop a model, which can accurately predict labels of Pro (\({y}_{i,{pro}}\)) and Inf \(({y}_{i,\inf })\) for each region \(i\) within a tumoral Area of Interest (AOI) based on regional MRI features (\({x}_{i}\)), for individual patients. The labels of three gene modules Pro (\({y}_{i,{pro}}\)), Inf \(({y}_{i,\inf })\), and Neu (\({y}_{i,{neu}}\)) are determined by comparing the raw scores to zero. Specifically, scores above zero are labeled as “high”, while those below zero are labeled as “low”. Given the limitations on the small sample size of labeled biopsies, BioNet is proposed to leverage the substantial volume of unlabeled AOI samples by integrating domain knowledge through its unique framework design.
Specifically, the domain knowledge reveals two key relationships between Pro and Inf conditional on the status of Neu (Fig. 2a): (1) Genes in the Neu module are enriched in normal brain tissue and depleted in lesional brain tissue. Thus, samples with high Neu tend to have low Pro and low Inf; (2) In samples with low Neu, the lesional component of the tissue comprises a mixture of Pro and Inf. Thus, if a sample has more proliferative tumor, i.e., Pro high, it is likely to have less inflammatory response, i.e., Inf Low, and vice versa. This implies that samples with low Neu are inclined to exhibit a negative correlation between Pro and Inf. High Pro indicate regions of active tumor cell proliferation, which may suppress the local inflammatory response, resulting in low Inf. Conversely, areas with high Inf may represent regions where the inflammatory response is more pronounced, potentially inhibiting tumor cell proliferation, resulting in low Pro. The two relationships are evident when dividing biopsy samples into two groups based on the enrichment scores of Neu: above-zero (high) and below-zero (low). Figure 2b shows that samples with high Neu have significantly lower average scores of Pro and Inf, compared to samples with low Neu. This empirical evidence confirms the relationship (1). Figure 2c shows that samples with low Neu have a significant negative correlation between Pro and Inf, which confirms the relationship (2). These findings are consistent with the complex interactions between tumor growth and the immune environment within the GBM microenvironment. While these relationships have been demonstrated in biopsy samples, they are also presumed to exist in unlabeled samples as well. This presumption is based on fundamental understanding of the spatial landscape of neuropathological alterations in GBM36. As described in36, the three gene modules, determined by distinct patterns of cellular composition, reveal these relationships in the analysis. Incorporation of these knowledge-based relationships leads to a hierarchical design of BioNet (Fig. 2d).
a Revealing biological relationships from domain knowledge to inspire BioNet design. b Bar chart for the group mean over the average scores of Pro and Inf in the group with high Neu in comparison to that in the group with low Neu. c Scatter plot of Pro and Inf for the group with low Neu. The Pearson correlation coefficient is represented by \(r\). d Hierarchical design of BioNet inspired by two biological relationships (1) and (2) between Pro and Inf given the high/low status of Neu.
Construction of BioNet_Neu to predict Neu using transfer learning and uncertainty quantification
The overall architecture of BioNet_Neu is presented in Fig. 3. We adopted several strategies to tackle the challenge of a small biopsy sample size: (1) Employed transfer learning by pre-training the network using a large number of unlabeled samples who have noisy Neu labels informed by biological knowledge, then fine-tuning it using real biopsy samples. (2) Incorporated Monte Carlo dropout43 to enable uncertainty quantification (UQ) of the predictions. (3) Applied data augmentation by including neighboring samples of each biopsy sample in training. BioNet_Neu achieved an Area Under the Curve (AUC) of 0.77 based on Cohort A using 5-fold cross validation (CV). Without Monte Carlo dropout, transfer learning and data augmentation, the AUC was reduced to 0.70, 0.64 and 0.56 (Supplementary Fig. 3a). In the data augmentation approach, including samples within a 5-voxel radius of the biopsy sample yielded the most optimal performance.
BioNet consists of two networks: BioNet_Neu to predict Neu using MRI; BioNet_ProInf to simultaneously predict Pro and Inf using MRI. a BioNet_Neu is a feedforward neural network pre-trained using a large number of unlabeled samples with noisy Neu labels informed by biological knowledge, and fine-tuned using biopsy samples with data augmentation. It also corporates Monte Carlo dropout to enable uncertainty quantification for the predictions. The role of BioNet_Neu is to stratify unlabeled samples with high predictive certainty, which were then incorporated into the training of BioNet_ProInf. b BioNet_ProInf is a multitask semi-supervised learning model with a custom loss function. The architecture consists of a shared block and task-specific blocks. The loss function combines a prediction loss and a knowledge attention loss that penalizes violation of the knowledge-based relationships on unlabeled samples.
In the hierarchical design of BioNet, BioNet_Neu played an important role in stratifying unlabeled samples into high and low predicted Neu groups, denoted as \(\left\{i\in {{Neu}}^{+}\right\}\) and \(\left\{i\in {{Neu}}^{-}\right\}\), respectively. As the subsequent model was dependent on this sample stratification, we aimed to select unlabeled samples which had high predictive certainty. This highlighted the importance of the UQ capability of BioNet_Neu. To evaluate the UQ capability of a DL model, a common strategy is to examine if the model satisfies the “more certain more accurate (MCMA)” criterion44, indicating that predictions with higher certainty are more accurate. To evaluate this criterion for BioNet_Neu, we first computed the predictive entropy (PE)45 as an uncertainty score for each biopsy sample. We then computed the accuracy on subsets of samples above increasingly stringent PE thresholds. Supplementary Fig. 3b shows that BioNet_Neu satisfied the MCMA criterion. The accuracy increased from 71% to 90% when computed on the top certain samples. To ensure the selected samples have relatively high accuracy, we set a threshold, PE*, corresponding to a 90% accuracy level, and retained only those samples for which PE is less than PE*.
Construction of BioNet_ProInf to predict Pro and Inf by combining multitask learning, semi-supervised learning, and domain knowledge
BioNet_ProInf is a multitask, semi-supervised learning model with a custom loss function. The loss function imposes penalties on prediction errors \({{\mathcal{L}}}_{{prediction}}\) as well as violation of the knowledge-based relationships \({{\mathcal{L}}}_{{knowledge}}\). There are three components in \({{\mathcal{L}}}_{{knowledge}}\). The first two are tailored to correspond with two knowledge-based relationships, for Neu high and Neu low respectively. The third component is a barrier loss46 defined on all unlabeled samples, aiming to discourage the predicted \({\hat{y}}_{i,{pro}}\) and \({\hat{y}}_{i,\inf }\) from both being high. The input for BioNet_ProInf consists of biopsy/labeled samples, along with unlabeled samples that are selected and stratified by BioNet_Neu. The overall architecture of BioNet_ProInf is presented in Fig. 3.
We compared BioNet_ProInf with a range of existing models: (1) Supervised learning models such as feed-forward neural network (NN), support vector regression (SVR), and random forest (RF), which used only biopsy/labeled samples. (2) Semi-supervised learning (SSL) that utilized both labeled and unlabeled samples. We included a recent method called AdaMatch47, a refined model for the highly-cited FixMatch48. (3) Multitask learning (MTL) that exploited the relationship between multiple outputs such as Pro and Inf in our case. We included a supervised MTL-NN and a recent semi-supervised MTL model called multitask adversarial autoencoder (MTL-AAE)49. The hyperparameters of BioNet and all competing methods were systematically tuned based on the same data-driven criterion through random search and grid search techniques.
Excluding 15 samples due to corrupted files or missing scans, there are 69 biopsy/labeled samples from 31 patients in cohort A (1–3 per patient). As shown in Fig. 4a, BioNet_ProInf achieved AUCs of 0.80 and 0.81 for predicting Pro and Inf, respectively, on cohort A using leave-one-patient-out CV (LOPO CV). The classification accuracies (ACCs) by dichotomizing the scores into high and low were 80% and 75% with standard deviation 0.014 and 0.013. In contrast, despite extensive hyperparameter tuning, competing methods demonstrated limitations, achieving only modest performance metrics: AUCs lower or around 0.6 and ACCs lower or around 70%. This outcome not only underscores the intrinsic challenges posed by the task but also reflects the complexity of the underlying data. Presently, even experienced experts face difficulties in differentiating between Pro and Inf. Similarly, current data-driven approaches fall short in accurately classifying these two gene states, indicating a significant gap in the predictive capability of existing models.
Results are shown for developmental cohort A under leave-one-patient-out cross-validation (LOPO CV) in a, and for test cohort B in b. c A table summarizes the key performance metrics of BioNet and competing methods. The standard deviation of accuracy for cohort A, calculated using leave-one-patient-out cross-validation (LOPO CV), is shown in brackets.
Testing of BioNet in an independent cohort B
The primary objective of our study was to evaluate the effectiveness of BioNet in its capability to precisely predict the Pro and Inf gene states in unseen datasets. This assessment aimed to verify the model’s generalizability across different datasets, underscoring its potential applicability in broader gene state classification tasks. We employed cohort B as a blind test dataset, comprising 31 biopsy/labeled samples from 5 patients (1–10 per patient). As shown in Fig. 4b, BioNet achieved AUCs of 0.80 and 0.76 for predicting Pro and Inf, respectively, on cohort B. The ACCs by dichotomizing the scores into high and low were 81% and 74%. In comparison, the competing methods achieved AUCs ranging in 0.51–0.69 and 0.53–0.60, ACCs ranging in 51%–65% and 58%–65%, for predicting Pro and Inf. BioNet outperformed all the competing methods.
Evaluating the generalizability of BioNet on both cohorts
The most robust assessment of model performance is derived from the prediction accuracy of biopsy samples, as previously indicated. However, due to their sparse nature, biopsy samples are not suitable for evaluating the generalizability of each method. To overcome this, BioNet_ProInf and its competing methods were utilized to predict the Pro and Inf scores for unlabeled samples within each AOI. To assess the models’ generalizability, we proposed to leverage two known relationships between gene modules. This analysis served as an effective method to validate the predictive accuracy of each model beyond the biopsy locations. Specifically, we computed two knowledge concordance (KC) metrics, denoted as \({{KC}}_{{{neu}}^{+}}\) and \({{KC}}_{{{neu}}^{-}}\), to quantify the concordance between the model’s predictions and the relationships. The results were presented in Fig. 5b.
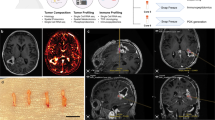
a The patient’s MRI images. b Knowledge concordance (KC) metrics, denoted as \({{KC}}_{{{neu}}^{+}}\) and \({{KC}}_{{{neu}}^{-}}\), for the predictions on unlabeled samples with Neu high and Neu low, respectively from the tumoral area of interest (AOI) of each patient on developmental cohort A and test cohort B. Prediction maps of c recGBM_Pro and d recGBM_Inf within the tumoral AOI by BioNet and the best-performing ML and DL methods (color maps overlaid on (a)). Three purple boxes denote the locations of three biopsies on this MRI slice. The maps indicate that BioNet attains the lowest absolute errors in predicting both Pro and Inf.
To provide a visualization for the regional distributions of gene modules across the AOI, prediction maps of two gene modules were generated for each patient. Figure 5 displays the prediction maps on a selected MRI slice, specifically chosen for having the most biopsies in a single slice. The prediction maps in Figure. 5c, d were from BioNet and the best-performing ML and DL competing methods (color maps overlaid on the patient’s T1Gd MRI image Fig. 5a). The average prediction entropy for BioNet is 0.588, markedly lower than that of RF (0.683) and AdaMatch (0.667).
Discussion
In this study, we utilized a unique dataset and developed BioNet to facilitate non-invasive quantification of regional distributions of gene modules in recGBM patients. The differentiation of proliferative/recurrent tumor (Pro) and treatment-induced reactive/inflammatory cells (Inf) is crucial for evaluating treatment effectiveness and guiding subsequent therapeutic strategies, but is very challenging in clinical practice due to their indistinguishable appearances on MRI. Biopsy, as the gold-standard approach, is invasive and only samples a few sparse regions. Our approach enabled voxel-level mapping of the regional distribution of these gene modules across the entire lesion for each patient.
We systematically evaluated the performance of our proposed method through a developmental cohort A and a blind test cohort B. On cohort A, biopsy/labeled samples are sparse, with each patient having an average of approximately 2 biopsies. BioNet achieved AUCs of 0.80 and 0.81 for predicting Pro and Inf, respectively, using leave-one-patient-out CV. The ACCs were 80% and 75%. It is worth mentioning that there are 31 biopsy/labeled samples from 5 patients in cohort B, with 1–10 samples per patient. A notable advancement in cohort B is the average of approximately 6 biopsies per patient, significantly higher than that in cohort A. This increase in the number of samples per patient enhances the ability to assess whether the model can generalize across different regions of the brain without overfitting to specific areas. On cohort B, BioNet achieved AUCs of 0.80 and 0.76 for predicting Pro and Inf, respectively. The ACCs were 81% and 74%. For comparison, we applied a range of supervised learning, semi-supervised learning, and multitask learning algorithms to the same dataset, and their AUCs and ACCs were much lower on both cohorts. All these existing methods have partial considerations of utilizing biopsy samples, unlabeled samples, and knowledge, while BioNet considered all these aspects in an integrated framework. Also, even though multitask learning algorithms were intended to account for the relationships between multiple outputs, they account for them in a general sense, explaining their worse performance than BioNet which considered the specific relationships between Pro and Inf using a custom loss function.
The strong generalizability of BioNet is further validated on unlabeled samples through knowledge concordance. BioNet_ProInf demonstrated moderately high KC metrics (Fig. 5b), exceeding 60% in both cohorts. In contrast, the \({{KC}}_{{{neu}}^{+}}\) values for all competing methods did not surpass 50% in either cohort. These values suggest that the incorporation of knowledge regularization into these models was effective. However, it is important to recognize the inherent uncertainty in domain knowledge. Consequently, overly strong regularization, potentially leading to very high concordance, was not desirable. Instead, we aimed for a balanced approach where models harmonize knowledge regularization with prediction losses. From this perspective, achieving moderately high concordance aligns well with our objectives. In addition, the observation that all methods demonstrated moderately high \({{KC}}_{{{neu}}^{-}}\) suggests that this relationship might be inherently more learnable for models, even without explicit regularization.
Furthermore, the prediction maps (Figure. 5c, d) illustrate that BioNet exhibits higher confidence in its predictions compared to competing methods. The majority of locations in the prediction maps generated by competing methods, especially by RF, are with light color, which suggests a tendency of these models to predict most samples with gene module scores around zero. Such a pattern indicates a degree of uncertainty in the classification, reflecting a potential limitation in the models’ discriminative capabilities. Supporting this, the average prediction entropy for BioNet is 0.588, markedly lower than that of RF (0.683) and AdaMatch (0.667).
Compared with competing methods, BioNet not only delivers more accurate predictions on biopsies, but also exhibits significantly stronger generalizability and higher confidence in its predictions on unlabeled samples. It helps enhance the accuracy of surgical targeting for confirmatory biopsies, assists in evaluating treatment effectiveness, enables regular monitoring of the gene modules for timely identification of treatment response or resistance, and provides a deeper understanding of patient-specific nuances to tailor treatment more effectively.
Due to the limited availability of labeled/biopsy samples, leveraging abundant unlabeled samples and the biological relationships among three gene modules were crucial to achieving a clinically usable model. We would like to discuss key highlights in the architecture and loss function design of BioNet.
In the architecture design, domain knowledge indicates a pronounced relationship between Pro and Inf, conditioned on their corresponding Neu. Consequently, the discriminative features identified for Neu are posited to be beneficial in the classification tasks for Pro and Inf. Inspired by this insight, we integrated the auxiliary task Neu into the BioNet_ProInf architecture as a regularization strategy. Instead of explicitly providing the model with labels for Neu, we designed it to independently discern the relationships between MRI features and Neu. The linear classification layer of Neu (indicated in deep blue in Fig. 3) compels the model to embed discriminative features of Neu into the final shared layers’ outputs, which enriches the feature representation for two primary classification tasks. This approach enabled the model to align closely with expert understanding, thereby enhancing its predictive accuracy and generalizability.
In the loss function design, prediction errors for Pro and Inf are computed using biopsy/labeled samples. Conversely, prediction errors for Neu are derived from both labeled samples and unlabeled samples with high certainty predicted labels \({\widetilde{y}}_{i,{neu}}\) from BioNet_Neu. This approach strengthens the generalization of discriminative features for Neu. Violations of relationships are characterized by a knowledge attention loss, and defined on unlabeled samples. To subtly incorporate Neu labels into the model, we propose using predicted labels \({\hat{y}}_{i,{neu}}\) from BioNet_ProInf for calculating the knowledge loss, rather than the true labels. This strategy stems from the observation that learning the biological relationships is relatively straightforward. In contrast, mapping MRI features to these gene modules is significantly more complex and challenging. Employing true labels in the knowledge loss will lead the model to prioritize learning the simpler task and neglect the more critical yet difficult ones. Consequently, after perfectly learning the easier tasks, the model starts to trivially predict Pro and Inf based solely on Neu, disregarding the MRI features. This is a scenario we aim to avoid. On the other hand, employing predicted Neu labels can encourage the model to explore which relationships should be followed, and to better understand the intricate relationships between MRI features and the gene modules.
This study has several limitations. One limitation of this study is the constrained sample size of biopsy samples in both cohorts. Although BioNet is innovatively designed to mitigate this limitation by leveraging unlabeled samples and domain knowledge, the pivotal role of biopsy/labeled samples in training an accurate and robust predictive model cannot be overstated. Increasing the number of biopsy samples has the potential to significantly improve the prediction accuracy of BioNet. Nevertheless, the invasive nature of biopsies restricts the feasibility of acquiring large sample sizes from individual patients. The second bottleneck of this work is the model’s explainability. Although BioNet has demonstrated effectiveness in predicting regional distributions of gene modules using MRI, the complexity of the model poses challenges in understanding the underlying mechanisms driving the predictions. The complex architecture and numerous parameters of DL models frequently result in a lack of interpretability.
Future research should aim to expand patient enrollment across multiple centers, facilitating a more substantial collective sample base. Different treatments can significantly impact the tumor microenvironment and imaging characteristics. However, due to the limited sample size and the variability in treatments received, our current dataset is not large enough to support a study dividing patients into different treatment groups. In the future, as we collect more data and the sample size increases, we plan to study how these treatments impact tumor progression and imaging characteristics. To more robustly evaluate the proposed method, we are actively working on collecting a new dataset from Mayo Clinic. By leveraging this dataset, we plan to not only evaluate BioNet’s performance on the three gene modules but also to test its adaptability to related tasks by predicting different cell types. Additionally, considering recent findings on sex differences in GBM50, the development of demographic-specific models, such as sex-specific variants, may further refine prediction accuracy. Additionally, future efforts should therefore prioritize enhancing the interpretability and explainability of BioNet. Investigating methodologies such as advanced visualization tools, and model-agnostic interpretability approaches can provide valuable insights into the specific image features and patterns that BioNet leverages for its predictions. Such advancements will not only guarantee that decisions are made based on accurate and comprehensible information, but will also offer critical insights into the development of different gene modules, thereby paving the way for future research in GBM.
In summary, we introduced BioNet, a biologically-informed model designed to predict the regional distributions of three tissue-specific gene modules: proliferating tumor, reactive/inflammatory cells, and infiltrated brain tissue. BioNet offers valuable insights into the integration of multiple implicit and qualitative biological domain knowledge, which are challenging to describe in mathematical formulations. It significantly outperforms a range of existing methods in both cross-validation and blind test datasets. The voxel-level prediction maps generated by BioNet help reveal intratumoral heterogeneity, potentially enhancing the precision of surgical targeting for confirmatory biopsies and the assessment of neuro-oncological treatment effectiveness. Additionally, the non-invasive nature of this approach could enable regular monitoring of gene module development over time, facilitating timely therapeutic adjustments.
Methods
Biopsy acquisition
Samples for the developmental cohort (A) and test cohort (B) were acquired from the brain tumor bank at Columbia University Irving Medical Center. Study protocols were approved by the Columbia University Institutional Review Board (IRB). All samples were de-identified prior to analysis. Analyses were carried out in alignment with the principles outlined in the World Medical Association (WMA) Declaration of Helsinki and the Department of Health and Human services Belmont Report. Informed written consent was provided by all patients. None of the participants were compensated for participating in this study. Tissue acquisition followed the same procedure that has been used in our prior publications32,42,51,52. A flowchart of the acquisition procedure is shown in Fig. 6. The tissue sampling was performed during the normal surgical plan, and posed no additional risk to the patient. Samples were taken from contrast-enhancing and contrast-negative, FLAIR-positive regions of tumor within the planned surgical trajectory. The locations of these biopsies were chosen at the neurosurgeon’s discretion and the locations of the biopsies were documented intraoperatively. Frameless stereotactic guidance was provided by a volumetric T1+Gd scan uploaded to a neuronavigation interface (Brainlab, Feldkirchen, Germany). Biopsy location was recorded by screen captures of the neuronavigation interface, allowing the downstream determination of the Cartesian coordinates of each biopsy. A custom registration software written in MATLAB was used to acquire the coordinates of each biopsy on the T1-post-contrast 1 mm slice MRI image. FSL software was used to confirm biopsy position, co-register all MRI-sequences to the base T1-post-contrast thin slice MRI, and for any segmentation of FLAIR and contrast-enhancing regions. Fresh tissue samples were divided into two pieces by trained neuropathologists and technicians. One piece was embedded in paraffin and underwent traditional H&E staining, while the second piece was stored for later use in a −80°C freezer.
Spatial alignment between biopsy samples and imaging data
All biopsies were collected at the time of recurrence. Supplementary Fig. 1 shows the distribution of times between initial resection and recurrence. Biopsies were collected before surgical debulking, use of mannitol, cerebrospinal fluid diversion, or hyperventilation to minimize the effects of brain shift and deformation. We have tested the accuracy of the spatial correlation in Fig. 4 of ref. 32. Specifically, we showed that the radiomic-histomic correlation changes as a function of distance from the true biopsy location. The correlation between the MR signal and cell counts drops to zero beyond 3 mm of the true biopsy location, indicating that, on average, we have a spatial resolution of approximately 3 mm or better. The window size used to extract the textural features for BioNet is 5 mm \(\times\) 5 mm, thus spatial alignment between biopsy samples and imaging data should have minimal impact on the model’s predictions.
Identification of three tissue-specific gene modules: Neu, Pro, Inf
To reduce complexity and improve prediction accuracy, we amplified the signal-to-noise ratio of genetic and cellular heterogeneity signal in the tissue by combining individual gene expressions and cellular composition patterns into three gene modules. While the radiogenomic signal associated with individual genes can be noisy, there is a great potential to improve the accuracy of ML/DL models by targeting on predicting clusters of correlated genes. The detailed analysis procedure is as follows:
-
(1)
SOX2, CD68, Ki67, and NeuN were used as (non-comprehensive) proxies for different cell populations in the glioma microenvironment. SOX2 is described as a robust marker for glioma cells53; CD68 is a known marker of macrophages; Ki67 is a known marker of proliferation; NeuN, also known as RBFOX3, is a canonical marker of neurons. The majority of biopsies (48/84; 57%) in cohort A had both RNA sequencing and IHC staining for SOX2, CD68, Ki67, and NeuN.
-
(2)
Hemotoxylin counterstain was used to label all nuclei, providing a measure of total cell density.
-
(3)
Pearson correlation was calculated between the normalized expression values for each gene and the IHC labeling index for each marker, including SOX2, CD68, Ki67, NeuN, and total cell density. A correlation matrix of all genes by 5 IHC markers was built.
-
(4)
A p value cutoff of 0.05 (un-adjusted) was set for determining the significance of the IHC-gene expression correlation for each IHC stain, and all genes with significant correlations with ≥1 marker were selected for downstream analysis. 7779 genes were identified as statistically significant at this step.
-
(5)
Based on the resulting correlation matrix, hierarchical clustering determined three distinct clusters/modules with mutually exclusive genes (Fig. 7a). Module 1–3 consisted of genes which have significant positive correlations with the labeling indices of SOX2, Ki67, and total cell density; the labeling index of NeuN; and the labeling index of CD68, respectively. For a complete list of the genes included in each module, please refer to Supplementary Data 1. Gene ontology analysis demonstrated that module 1–3 were associated with proliferative (Pro)–proliferation/cell cycle ontologies; neuronal (Neu)–neuronal signaling; inflammatory (Inf)–cytokine production/immune response, respectively (Fig. 7b).
-
(6)
These three gene clusters were used as gene sets for downstream Gene Set Variation Analysis (GSVA) on a sample-by-sample basis, which produced gene set enrichment scores for all MRI-localized biopsies (Fig. 7c).
a Heatmap depicting correlation between normalized gene expression and immunohistochemical labeling indices, with subsequent hierarchical clustering revealed three orthogonal tissue-specific gene modules. Module 1 consists of 3688 genes significantly positively correlated with SOX2/Ki67/H&E; module 2 consists of 1673 genes correlated with NeuN; module 3 consists of 2418 genes correlated with CD68. b Bar plot depicting top significant gene ontologies enriched in each of the three tissue-specific gene modules derived from the IHC-RNAseq correlation analysis. X axis is –log10 (p value) of each ontology. Module 1 is enriched in genes involved in proliferation (Pro), module 2 in neuronal-specific genes (Neu), and module 3 in genes in immune infiltration (Inf). c Heatmap depicting single-sample Gene Set Variation Analysis (GSVA) for each of 84 MRI-localized biopsies for each of the three tissue-specific gene modules. Color gradient represents magnitude/direction of tissue-specific enrichment for each biopsy.
The results shown in Fig. 7 were obtained based on analysis performed on cohort A. Upon completing the sample collection of cohort B, the gene clusters identified in cohort A were used to perform GSVA on cohort B to obtain the enrichment scores for the same gene modules. In this paper, the gene modules were named Pro, Inf, and Neu. In this study, to adhere to standard practices in machine learning, align with algorithm requirements and simplify implementation, the raw enrichment scores of three gene modules, originally ranging from −1 to 1, are transformed to a [0, 1] scale using the mapping function \(f\left(x\right)=\frac{x+1}{2}\). Consequently, the threshold distinguishing high and low gene module expression is correspondingly transformed from 0 to 0.5.
RNAseq-IHC correlation analysis, clustering, and GSVA
For samples in Cohort A that had both RNAseq and IHC quantification, Pearson correlation was calculated between the normalized expression values for each gene and the IHC labeling index for each marker, building a correlation matrix of 15,001 genes by 5 IHC markers (4 IHC markers and total normalized cellularity from H&E images). The 15,001 genes were filtered from 23,802 genes to include only protein coding genes that had more than 10 reads across all samples. A p value cutoff of 0.05 (un-adjusted) was set for determining the significance of the IHC-gene expression correlation for each stain, and all genes with significant correlations with ≥1 marker were selected for downstream analysis. Furthermore, based on the resulting correlation matrix, we performed clustering analysis. A variety of different clustering algorithms were used, which repeatedly found the optimal cluster number to be two or three. Considering the findings in a recent publication by our group that described a three-gene-module model of glioma36, we ultimately decided to proceed with the three-cluster result from hierarchical clustering with Ward’s linkage. Three major clusters with mutually exclusive genes were identified, and these gene clusters were used as gene sets for downstream Gene Set Variation Analysis (GSVA) on a sample-by-sample basis, which produced three gene set enrichment scores for each MRI-localized biopsy. We call the three gene sets “gene modules” and use the enrichment scores as estimates of gene module expression. Given that there exists a degree of noise in bulk RNAseq analysis, this technique allows us to leverage the multi-modal data (RNAseq and IHC) to link expression with biologically meaningful tissue features (IHC markers) to further strengthen the gene module approach.
RNA extraction and pooled library amplification for transcriptome expression RNA-sequencing (PLATE-Seq)
Total RNA was extracted from these tissue samples using the institutional genomic core. Samples with an RNA Integrity Number (RIN) greater than six were selected and subsequently sequenced using the PLATE-Seq protocol. PLATE-Seq is a novel sequencing platform that can produce high-throughput RNA-sequencing data through barcoding and pooling of cDNA libraries10. RNA samples from 84 MRI-localized samples were normalized to between 60 and 100 nanograms in 16.5 microliters of nuclease-free water and placed in a 96-well PCR plate. Purified mRNA was reverse transcribed and barcode segments were added to create easily identified cDNA. Multiple batches were pooled, cDNA was purified, and then underwent PCR amplification. Samples were ultimately sequenced on an Illumina NextSeq 500 sequencer10. Raw reads were mapped to the human transcriptome by the institutional sequencing center using STAR alignment to generate gene counts. All RNAseq processing and analysis was performed using the statistical computing software R. RNA sequencing counts were first trimmed to only include genes with ≥10 counts across all samples. Counts were then normalized using the DESeq2 package to generate normalized expression values for each gene.
Immunohistochemical staining (IHC) and quantification
A subset of the biopsies in Cohort A (n = 48) and all biopsies in Cohort B underwent histological staining for H&E and IHC for SOX2, Ki67, CD68, and NeuN. Five-micrometer sections from a single localized biopsy were obtained for staining with hematoxylin-eosin and immunostaining with SOX2, Ki67, CD68, and NeuN. Slides were subsequently scanned and digitized at ×40 magnification using a Leica SCN400 system (Leica Biosystems, Buffalo Grove, Illinois). Total cell density was calculated using a validated semi-automated whole slide cell-counting algorithm17.
In brief, the algorithm was trained to select all hematoxylin-stained nuclei using 9 randomly generated high-power fields (HPF) from each sample, defined as 0.225 mm × 0.225 mm (900 × 900 pixels). Subsequently, the algorithm could be used to iteratively process all HPFs until the entire H&E stained slide was counted. After collecting the total cell counts for an H&E-stained slide, the algorithm was trained to select SOX2-stained and Ki67-stained nuclei on 9 randomly generated HPFs from SOX2-stained or Ki67-stained slides, respectively. For CD68-stained and NeuN-stained samples, the algorithm was trained to select CD68-stained and NeuN-stained cells in 9 randomly generated HPFs from CD68-stained or NeuN-stained slides, respectively. Areas of the slide without tissue were excluded from calculations in all cases. Algorithm-derived cell counts were manually verified and total cell density, SOX2 cell density, Ku67 cell density, CD68 cell density, and NeuN cell density were calculated across all HPFs in a slide. The mean cell count per HPF was used as a representative measure in all cases given that intra-specimen heterogeneity was limited by the standardized size of each biopsy. A labeling index (LI) for each immunostain was computed by dividing the number of immunostain-positive cells by the total cell count in a HPF (from the same slide). For H&E-stained slides, cell counts were converted to a percentage by dividing each by the largest cell count across all samples. Algorithm-derived cell counts were manually verified by a human reviewer. For validation, 100 HPFs were chosen at random and each field as manually inspected to determine the number of nuclei present. The same field was then evaluated by the automated cell-counting algorithm. A high correlation was observed between the automated algorithm and the manual cell counts for SOX2 and Ki67 cell density as determined by a Pearson coefficient, as shown in Supplementary Fig. 2.
Parameters for each MRI sequence
Each patient received an MRI exam, which included T1Gd, T2, FLAIR, ADC, and SWI sequences. For the T1 sequence, we used a 3D acquisition with fields of view (FOV) ranging from 240 to 260 mm. FLAIR sequences mostly used 2D acquisitions, featuring FOV typically around 220 to 240 mm. T2 sequences were executed in 2D, with FOV also in the 220 to 240 mm range. ADC mapping, crucial for diffusion measurement, used 2D acquisitions with FOV mostly around 220 to 240 mm. SWI sequences, included both 2D and 3D formats with FOV from 220 to 260 mm. All sequences conducted on MRI systems from manufacturers such as GE Medical Systems, Philips Medical Systems, and Siemens, with magnetic field strengths of either 1.5 Tesla or 3 Tesla. Detailed MRI parameters used in both cohort A and B can be found in Supplementary Data 2.
Multiparametric MRI pre-processing, segmentation, and feature extraction
All MRI images were preprocessed using a pipeline built in Python. For each patient, the multiparametric image set was affine registered (12 degrees of freedom) to the DTI B0 image using SimpleElastix54. Registration transforms were then applied to the associated tumor segmentation masks for each image. Next, inhomogeneity correction was performed using the N4 algorithm55 implemented in SimpleITK56. In addition, a brain mask was extracted for each patient using MONSTR, a multi-contrast brain-stripping tool57. The brain mask was used to normalize the image intensities to a mean of zero and standard deviation of one for all images except the computed ADC. Finally, all images and segmentations were resampled to a common voxel size of 1.05 mm × 1.05 mm for data consistency across patients.
For each patient, the enhancing tumor and infiltrating tumor were manually segmented based on T1Gd and T2/FLAIR images. The infiltrated tumor defined on T2/FLAIR MRI scans refers to hyperintense regions of the brain that show evidence of tumor cell infiltration beyond the clearly defined, solid tumor boundaries typically visible on T1Gd scans. Images were segmented by trained technicians using a rule set for selecting hyperintense signal caused by tumor for each image sequence. All segmentations are then reviewed for quality assurance and consistency by a segmentation supervisor (Lisa Paulson). A board-certified neuroradiologist (Leland Hu) was available for consultation as needed. A grouped segmentation was then determined by combining the T1Gd and T2/FLAIR segmentations. This group segmentation was then dilated by a margin of 7 mm to create our AOI.
A sliding window of 5 × 5 pixel2 was placed at each pixel within the AOI. From each sliding window, we extracted textural features from the ADC, FLAIR, SWI, T1Gd, and T2. Specifically, two commonly used texture analysis algorithms, Gray-Level Co-occurrence Matrix (GLCM) and Gabor Filters, were used to generate a total of 38 textural features for each of the five MRI images, for a total of 190 features. In addition, 18 commonly used 1st-order statistical features such as mean, standard deviation, and energy were extracted, for a total of 90 1st-order statistical features from the five MRI images. Collectively, our texture analysis pipeline generated 280 features for each sliding window. These features were also used in our prior radiomic study of GBM and shown to be effective for capturing imaging phenotypic information correlative with genetic and histopathological characteristics of the tumor27,33,34.
Extraction of regional features from multiparametric MRI
The regions were defined as 5 × 5 pixel2 windows on the axial view of MRI. This size approximated the physical size of biopsy samples. For each biopsy sample, image features were extracted from a window at the biopsy location. This resulted in a labeled dataset \({\left({x}_{i},{y}_{i}\,\right)}_{i=1}^{l}\), \({y}_{i}=({y}_{i,{neu}},{y}_{i,{pro}},{y}_{i,\inf })\). Additionally, image features were extracted from regions beyond the biopsy locations within a tumoral AOI, to generate unlabeled samples. To do this, we first defined the AOI of each patient by combining the segmented enhancing tumor portion on T1Gd and infiltrating tumor portion on T2/FLAIR plus a 7 mm margin. Then, we placed sliding windows with a size of 5 × 5 pixel2 and a stride size of 1 throughout the AOI, and extract image features from each window (Fig. 1 Texture feature extraction). This resulted in an unlabeled dataset \({\left({x}_{i}\,\right)}_{i=l+1}^{l+u}\) with about 1.82e6 samples (5e3 to 9e4 per patient). The regional image feature set consisted of 280 statistical and texture features computed from T1Gd, T2, FLAIR, ADC and SWI.
Training, validation, and testing datasets
We utilized cohort A for training and validation, and cohort B for testing. Cohort A comprises 69 biopsy/labeled samples. By employing leave-one-patient-out cross-validation (LOPO CV), the validation set for each fold includes 1 to 3 labeled samples from the validation patient. In contrast, cohort B contains 31 biopsy/labeled samples. Consequently, in our study, each training set consisted of 66 to 68 labeled samples (cohort A), the validation sets included 1 to 3 labeled samples (cohort A), and the testing set comprised 31 labeled samples (cohort B). The distribution of biopsy/labeled samples across the three gene modules is shown in Table 1.
Construction of BioNet using cohort A
BioNet includes two networks: (1) BioNet_Neu is to predict Neu; (2) BioNet_ProInf is to simultaneously predict Pro and Inf.
1) BioNet_Neu
Architecture
This network included two 2048-dimension hidden layers with ReLU as the activation function. To incorporate UQ, Monte Carlo dropout was adopted with a dropout rate of 1e-3.
Knowledge informed pre-training
We created a large, noisy labeled dataset, denoted as \(\{{Noisy}\}\), to pre-train the network. The noisy labeled dataset consisted of unlabeled samples that were likely to have high or low Neu based on domain knowledge, denoted as class \(\widetilde{1}\) or class \(\widetilde{0}\), respectively. The overhead ‘\(\sim\)’ indicates uncertainty and potential labeling errors, which were acceptable for pre-training. In detail, we have the knowledge that Neu tends to be high on the boundary of AOI and outside the AOI (i.e., in the normal brain areas), as the genes included Neu are predominantly involved in neuronal signaling. To avoid unwanted brain structures, we chose to include samples located on the AOI boundary as class \(\widetilde{1}\) samples. Furthermore, we have the knowledge that Neu tends to be low within the enhancing tumoral area on T1Gd, as this region is known to involve tumor proliferation or immune response58 rather than neuronal signaling. Thus, we included samples from the enhancing tumoral area as class \(\widetilde{0}\) samples. As a result, the noisy labeled dataset included about 7500 samples in class \(\widetilde{1}\) and \(\widetilde{0}\), respectively, from 31 patients. We used this dataset to pre-train the network with cross-entropy loss.
Labeled data guided fine-tuning
The pre-trained network was then fined-tuned using biopsy samples under the soft cross-entropy loss. Data augmentation was used by incorporating neighbor samples of each biopsy sample.
Prediction and stratification of unlabeled samples
The network was used to predict the Neu scores for unlabeled samples within the ROI of each patient. Recall that the unlabeled samples corresponded to 5 × 5 pixel2 windows with a stride of 1, sliding over the AOI. Using zero as a cutoff, the predicted scores were dichotomized into two classes, \({\{{Neu}}^{+}\}\) or \(\{{{Neu}}^{-}\}\). Furthermore, the network’s UQ capability made it possible to generate an uncertainty score for each prediction, measured by Predictive Entropy (PE)45. To filter out unlabeled samples whose predictions have high certainty, we applied a threshold PE**, corresponding to a 90% accuracy level, and only retained samples with PE < PE*. The retained samples were then divided into two subsets: \(\left\{i\in {{Neu}}^{+}\right\}\) and \(\left\{i\in {{Neu}}^{-}\right\}\), which included unlabeled samples predicted to be \({{Neu}}^{+}\) or \({{Neu}}^{-}\) with high certainty, respectively. These subsets would be used in training BioNet_ProInf as discussed in the following section.
2) BioNet_ProInf
Architecture
The network used to predict Pro and Inf is a multitask semi-supervised learning model with a custom loss function. It comprises a shared block and task-specific blocks. The shared block consists of three layers with dimensions of 256, 128, and 128, respectively. The task-specific blocks for Pro and Inf each consist of three layers with dimensions of 128, 128, and 64, respectively. As the relationships between Pro and Inf are conditional on the status of Neu, discriminant features of Neu provided significant guidance for the main tasks. To enforce the shared latent representations to encode the discriminant features, the auxiliary task block corresponding to Neu was kept simple, comprising one layer of 128 units. Input to the network included both biopsy/labeled samples and unlabeled samples in \(\left\{i\in {{Neu}}^{+}\right\}\bigcup \left\{i\in {{Neu}}^{-}\right\}\) which were selected by BioNet_Neu as previously described. The output was used to define a custom loss function, introduced as follows:
Loss function design
There are two parts in the loss function to penalize (1) prediction errors on biopsy/labeled samples, and (2) violation of domain knowledge, i.e.,
To compute \({{\mathcal{L}}}_{{prediction}}\), the network generated three predicted scores of Pro, Inf, Neu for each biopsy sample, which were compared with the true scores using the L2 norm for two main tasks and Kullback–Leibler divergence for the auxiliary task. In addition, prediction errors for Neu are also derived from unlabeled samples with high certainty predicted labels \({\widetilde{y}}_{i,{neu}}\) from BioNet_Neu, i.e.,
The knowledge attention loss \({{\mathcal{L}}}_{{knowledge}}\) is defined on unlabeled samples. To implicitly incorporate the labels for Neu into the model, we utilize the predicted labels \({\hat{y}}_{i,{neu}}\) in \({{\mathcal{L}}}_{{knowledge}}\), which is designed to include three components:
Here,
Minimizing this loss encourages the predicted \({\hat{y}}_{i,{pro}}\) and \({\hat{y}}_{i,\inf }\) to be low for unlabeled samples in \(\left\{i\in {{Neu}}^{+}\right\}\), where \({\hat{y}}_{i,{neu}}\) acts as a weight, promoting a higher occurrence of such predictions for samples with higher \({\hat{y}}_{i,{neu}}\).
Minimizing this loss encourages the predicted \({\hat{y}}_{i,{pro}}\) and \({\hat{y}}_{i,\inf }\) to be negatively correlated for unlabeled samples in \(\left\{i\in {{Neu}}^{-}\right\}\), where \((1-{\hat{y}}_{i,{neu}})\) acts as a weight, promoting a higher occurrence of such predictions for samples with lower \({\hat{y}}_{i,{neu}}\).
\({{\mathcal{L}}}_{{neu}}\) is a barrier loss46 defined on all unlabeled samples \(\left\{i\in {{Neu}}^{+}\right\}\bigcup \left\{i\in {{Neu}}^{-}\right\}\), aiming to discourage the predicted \({\hat{y}}_{i,{pro}}\) and \({\hat{y}}_{i,\inf }\) from both being high. This loss can help strengthen the effect of the other two losses, with its utility controlled by \(\beta\). The form of \({{\mathcal{L}}}_{{neu}}\) follows the standard log barrier function commonly found in optimization literature, i.e.,
\({\rm{c}}\) is an upper bound that can be treated as a tuning parameter. However, for simplicity of the design, we set c = 1.2, which is the maximum value of the summation of scaled Pro and Inf scores in biopsy samples.
Statistical analysis of model performance on cohort A
To assess the statistical significance of the performance gain for BioNet, we performed a one-sided paired t-test to compare BioNet against the competing methods with the average best accuracy using leave-one-patient-out cross-validation (LOPO CV). According to the p values from one-sided t-test as shown in Table 2, BioNet significantly outperformed all competing methods on both Pro and Inf.
Testing of BioNet using cohort B
The MRI scans in cohort B were acquired at a different resolution compared to cohort A. Using this test cohort could provide valuable insights into the generalizability of BioNet on a less ideal yet more realistic dataset, reflecting the common practice that MRI scans can be obtained under varying conditions for different patients. However, the resolution discrepancy created challenges in directly applying the trained BioNet from cohort A to cohort B. In DL, approaches that address input discrepancy when deploying a model from one domain to another domain have been explored in the subfield of domain adaptation59.
To address this discrepancy, we implemented an approach inspired by domain adaptation, which replaced the unlabeled samples from cohort A with those from cohort B to re-train BioNet_ProInf. The unlabeled samples were abundant and contained only image features. Using the unlabeled samples from cohort B had the effect of biasing BioNet_ProInf toward the image representation in cohort B. Notably, this re-training process did not include any biopsy sample from cohort B. Thus, the re-trained BioNet_ProInf was still “blind” to the ground-truth scores of the biopsy samples in cohort B.
Computation of the knowledge concordance (KC) metrics overall unlabeled samples
Recall that the domain knowledge indicates two key relationships: (1) Pro and Inf are likely to be negatively correlated for samples with low Neu; (2) Pro and Inf are likely to be low for samples with high Neu. To compute the KC metrics, we first used the trained BioNet_Neu model to stratify unlabeled samples into two groups with low (<0) and high (>0) predicted scores of Neu. Denote these groups by \(\left\{i\in {{Neu}}^{-}\right\}\) and \(\left\{i\in {{Neu}}^{+}\right\}\). Note that these groups included all unlabeled samples within each AOI, not just the unlabeled samples included to train BioNet_ProInf which are samples with high certainty.Furthermore, we computed the KC metric with respect to relationship (1), \({{KC}}_{{{neu}}^{-}}\), as the percentage of unlabeled samples in \(\left\{i\in {{Neu}}^{-}\right\}\) whose predicted Pro and Inf satisfy one being below 0 and the other above 0. We computed the KC metric with respect to relationship (2), \({{KC}}_{{{neu}}^{+}}\), as the percentage of unlabeled samples in whose predicted Pro and Inf are both below 0.
Data availability
The datasets for Cohort A and Cohort B are accessible on Figshare through the project page: https://figshare.com/projects/Texture_features_of_Multiparametric_MRI_-_Recurrent_Glioblastoma/193223, with the dataset available at the following https://doi.org/10.6084/m9.figshare.23950584.v1. RNAseq and enrichment analysis of these datasets have been published in36.
Code availability
The code used to extract texture features, perform experiments, and analyse data is available at: https://github.com/hairongw/BioNet.git.
References
Ene, C.I. & Fine, H. A. Many tumors in one: a daunting therapeutic prospect. Cancer Cell 20, 695–697 (2011).
Hu, L. S., Hawkins-Daarud, A., Wang, L., Li, J. & Swanson, K. R. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 477, 97–106 (2020).
Qazi, M. A. et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann. Oncol. 28, 1448–1456 (2017).
Parker, N. Renee et al. Molecular heterogeneity in glioblastoma: potential clinical implications. Front. Oncol. 5, 55 (2015).
Kazerooni, A. F., Bakas, S., Rad, H. S. & Davatzikos, C. Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review. J. Magn. Reson. Imaging 52, 54–69 (2020).
Singh, G. et al. Radiomics and radiogenomics in gliomas: a contemporary update. Br. J. Cancer 125, 641–657 (2021).
AL, B. et al. Patient-specific metrics of invasiveness reveal significant prognostic benefit of resection in a predictable subset of gliomas. PLoS One 9, E17–E18 (2014).
Neal, M.L. et al. Discriminating survival outcomes in patients with glioblastoma using a simulation-based, patient-specific response metric. PLoS One 8, e51951 (2013).
Rockne, R.C. et al. A patient-specific computational model of hypoxia-modulated radiation resistance in glioblastoma using 18F-FMISO-PET. J. R. Soc. Interface 12, 20141174 (2015).
Akbari, H. et al. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro Oncol. 20, 1068–1079 (2018).
Chen, H. et al. Deep learning radiomics to predict PTEN mutation status from magnetic resonance imaging in patients with glioma. Front. Oncol. 11, 734433 (2021).
Kickingereder, P. et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology 281, 907–918 (2016).
Tykocinski, E. S. et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro Oncol. 14, 613–623 (2012).
Zhang, X. et al. IDH mutation assessment of glioma using texture features of multimodal MR images. Med. Imaging Comput.-Aid. Diagn. 10134, 462–469 (2017).
Zhang, B. et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol. 19, 109–117 (2017).
Combs, S. E. et al. Prognostic significance of IDH-1 and MGMT in patients with glioblastoma: one step forward, and one step back? Radiat. Oncol. 6, 1–5 (2011).
Chang, X. P. et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am. J. Neuroradiol. 39, 1201–1207 (2018).
Baldock, A. L. et al. Invasion and proliferation kinetics in enhancing gliomas predict IDH1 mutation status. Neuro Oncol. 16, 779–786 (2014).
Akbari, H. et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer 126, 2625–2636 (2020).
Lee, J. et al. Discriminating pseudoprogression and true progression in diffuse infiltrating glioma using multi-parametric MRI data through deep learning. Sci. Rep. 10, 20331 (2020).
Sun, Y.Z. et al. Differentiation of pseudoprogression from true progression in glioblastoma patients after standard treatment: a machine learning strategy combined with radiomics features from T1-weighted contrast-enhanced imaging. BMC Med. Imaging 21, 17 (2021).
Jang, B.S., Jeon, S.H., Kim, I.H. & Kim, I.A. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci. Rep. 8, 12516 (2018).
Moassefi, M. et al. A deep learning model for discriminating true progression from pseudoprogression in glioblastoma patients. J. Neurooncol. 159, 447–455 (2022).
A. S. McKenney et al. Radiomic analysis to predict histopathologically confirmed pseudoprogression in glioblastoma patients. Adv. Radiat. Oncol. 8, 100916 (2023).
Urcuyo, J. C. et al. Image-localized biopsy mapping of brain tumor heterogeneity: a single-center study protocol. medRxiv https://doi.org/10.1101/2022.11.14.22282304 (2022).
Gill, B. J. et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl Acad. Sci. USA 111, 12550–12555 (2014).
Hu, L. S. et al. Multi-parametric MRI and texture analysis to visualize spatial histologic heterogeneity and tumor extent in glioblastoma. PLoS One 10, e0141506 (2015).
Gaw, N. et al. Integration of machine learning and mechanistic models accurately predicts variation in cell density of glioblastoma using multiparametric MRI. Sci. Rep. 9, 1–9 (2019).
Wang, L., Hawkins-Daarud, A., Swanson, K. R., Hu, L. S. & L,i J. Knowledge-infused global-local data fusion for spatial predictive modeling in precision medicine. IEEE Trans. Autom. Sci. Eng. https://doi.org/10.1109/TASE.2021.3076117. (2021).
Wang, C. H. et al. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res. 69, 9133–9140 (2009).
Swanson, K. R., Bridge, C., Murray, J. D. & Alvord, E. C. Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J. Neurol. Sci. 216, 1–10 (2003).
Chang, P. D. et al. A multiparametric model for mapping cellularity in glioblastoma using radiographically localized biopsies. Am. J. Neuroradiol. 38, 890–898 (2017).
Hu, L. S. et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 19, 128–137 (2017).
Hu, L. S. et al. Uncertainty quantification in the radiogenomics modeling of EGFR amplification in glioblastoma. Sci. Rep. 11, 1–14 (2021).
Wen, P. Y. et al. Response assessment in neuro-oncology clinical trials. J. Clin. Oncol. 35, 2439–2449 (2017).
Al-Dalahmah, O. et al. Re-convolving the compositional landscape of primary and recurrent glioblastoma reveals prognostic and targetable tissue states. Nat. Commun. 14, 2586 (2023).
Elmarakeby, H. A. et al. Biologically informed deep neural network for prostate cancer discovery. Nature 598, 348–352 (2021).
Azher, Z.L., Vaickus, L.J., Salas, L.A., Christensen, B. C. & Levy, J.J. Development of biologically interpretable multimodal deep learning model for cancer prognosis prediction. In: Proceed. 37th ACM/SIGAPP Symp. Appl. Comput. 636–644. https://doi.org/10.1101/2021.10.30.466610 (2022).
Deist, T. M. et al. Simulation-assisted machine learning. Bioinformatics 35, 4072–4080 (2019).
Erion, G., Janizek, J. D., Sturmfels, P., Lundberg, S. M. & Lee, S. I. Improving performance of deep learning models with axiomatic attribution priors and expected gradients. Nat. Mach. Intell. 3, 620–631 (2021).
L. Mao et al. Knowledge-informed machine learning for cancer diagnosis and prognosis: a review. arxiv https://arxiv.org/abs/2401.06406 (2024).
Spinazzi, E. F. et al. Chronic convection-enhanced delivery of topotecan for patients with recurrent glioblastoma: a first-in-patient, single-centre, single-arm, phase 1b trial. Lancet Oncol. 23, 1409–1418 (2022).
Srivastava, N., Hinton, G., Krizhevsky, A. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Kendall, A. & Gal, Y. What uncertainties do we need in Bayesian deep learning for computer vision? Adv. Neural Inf. Process Syst. 30 (2017).
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948).
Luenberger, D.G. & Ye, Y. Linear and nonlinear programming (Springer International Publishing, 2016).
Berthelot, D., Roelofs, R., Sohn, K., Carlini, N. & Kurakin, A. AdaMatch: a unified approach to semi-supervised learning and domain adaptation. In: The Tenth International Conference on Learning Representations (2022).
Sohn, K. et al. FixMatch: simplifying semi-supervised learning with consistency and confidence. Adv. Neural Inf. Process Syst. 33, 596–608 (2020).
Latif, S. et al. Multi-task semi-supervised adversarial autoencoding for speech emotion recognition. IEEE Trans. Affect. Comput. 13, 992–1004 (2022).
Bond, K.M. et al. Glioblastoma states are defined by cohabitating cellular populations with progression-, imaging- and sex-distinct patterns. bioRxiv. https://doi.org/10.1101/2022.03.23.485500 (2022).
Bowden, S. G. et al. Local glioma cells are associated with vascular dysregulation. Am. J. Neuroradiol. 39, 507–514 (2018).
Petridis, P. D. et al. BOLD asynchrony elucidates tumor burden in IDH-mutated gliomas. Neuro Oncol. 24, 78–87 (2022).
Yuan, J. et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 10, 57 (2018).
Marstal, K., Berendsen F., Staring, M. & Klein, S. SimpleElastix: a user-friendly, multi-lingual library for medical image registration; simpleElastix: a user-friendly, multi-lingual library for medical image registration. In: Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 134–142, https://doi.org/10.1109/CVPRW.2016.78 (2016).
Tustison, N. J. et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010).
Beare, R., Lowekamp, B. & Yaniv, Z. Image segmentation, registration and characterization in R with SimpleITK. J. Stat. Softw. 86, 8 (2018).
Roy, S., Butman, J. A. & Pham, D. L. Robust skull stripping using multiple MR image contrasts insensitive to pathology. Neuroimage 146, 132–147 (2017).
Kersch, C. N., Ambady, P., Hamilton, B. E. & Barajas, R. F. MRI and PET of brain tumor neuroinflammation in the era of immunotherapy, from the AJR special series on inflammation HHS public access. AJR Am. J. Roentgenol. 218, 582–596 (2022).
Ganin, Y. & Lempitsky, V. Unsupervised domain adaptation by backpropagation. arXiv https://arxiv.org/abs/1409.7495 (2014).
Acknowledgements
This work was supported by NIH grant U01CA250481-01A1, NSF grant DMS-2053170, NIH/NINDS R01NS103473, NIH/NCI R01CA161404 and the NIH/NCI Cancer Center Support Grant P30CA013696. We would like to acknowledge the Herbert Irving Comprehensive Cancer Center Molecular and Pathology Core.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.W., H.Y., J.G., K.R.S., P.C., J.L. Data acquisition: M.G.A., D.B., A.S., P.P., W.S., O.A.D., J.N.B. Data analysis: H.W., M.G.A., H.Y., D.B., A.S., P.P., W.S., P.J., A.H.D., N.T., L.H., K.W.S., L.P., O.A.D., J.N.B. Original draft: H.W., M.G.A., J.G., K.R.S., P.C., J.L. Draft revisions: H.W., M.G.A., H.Y., P.J., A.H.D., N.T., L.H., K.W.S., J.G., K.R.S., P.C., J.L. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, H., Argenziano, M.G., Yoon, H. et al. Biologically informed deep neural networks provide quantitative assessment of intratumoral heterogeneity in post treatment glioblastoma. npj Digit. Med. 7, 292 (2024). https://doi.org/10.1038/s41746-024-01277-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-024-01277-4
This article is cited by
-
Advances in surrogate modeling for biological agent-based simulations: trends, challenges, and future prospects
Journal of Mathematical Biology (2026)