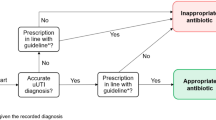
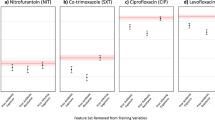
Abstract
Urinary tract infections (UTIs) often prompt empiric outpatient antibiotic prescriptions, risking mismatches. This study evaluates the impact of “UTI Smart-Set” (UTIS), an AI-driven decision-support tool, on prescribing patterns and mismatches in a large outpatient organization. UTIS integrates machine learning forecasts of antibiotic resistance, patient data, and guidelines into a user-friendly order set for UTI management. From 6/1/2021–8/31/2022, 171,010 UTI diagnoses were recorded, with UTIS used in 75,630 cases involving antibiotic prescriptions. Overall acceptance rate of UTIS recommendations was 66.0%. Among 19,287 cases with urine cultures, antibiotic mismatch rate was significantly lower when UTIS recommendations were followed (8.9% vs. 14.2%, p < 0.0001). Among women over 18, mismatch rate was 47.5% lower, and among women over 50, 55.6% lower (p < 0.001). Additionally, an overall reduction of 80.5% in ciprofloxacin usage (6.4% vs 32.9%, p < 0.0001) was observed. UTIS improved prescribing accuracy, reduced mismatches, and minimized quinolone use, highlighting AI’s potential for personalized infection management.
Similar content being viewed by others
Introduction
Urinary tract infections (UTIs) are among the most common reasons for outpatient visits and antibiotic prescriptions in adults1,2. However, the increasing incidence of antibiotic resistance among uropathogens, particularly against quinolones, challenges disease management3,4,5. The choice of antibiotics for UTI is usually made empirically since urine culture results are not immediately available at the point of care6. The selection of empiric antibiotic therapy requires careful consideration of several factors, including patient background characteristics such as age, gender, pregnancy status, past infectious histories such as prior UTIs, previous antibiotic use and culture susceptibilities, local susceptibility patterns, and patient preferences7,8,9. However, even when considering these parameters, the potential risk of antibiotic mismatch in the management of UTIs remains10,11,12. In some cases, the initial antibiotic regimen may need modification based on later culture data.
Artificial intelligence (AI) holds promise in predicting UTI resistance and suggesting suitable antibiotic regimens13,14,15,16. These tools analyze diverse data sources, including patient characteristics and microbiological data, to predict antibiotic resistance likelihood and guide treatment decisions. Consequently, AI tools may assist clinicians in selecting the most appropriate antibiotics and minimizing the use of broad-spectrum antibiotics17,18. This can reduce the risk of developing antibiotic resistance, lower the risk of adverse effects, and improve patient outcomes.
UTI management and antibiotic therapy guidelines have been published globally19. Yet studies have shown inappropriate antibiotic use for UTI in the outpatient setting and suboptimal adherence to local guidelines20,21,22,23,24. This inappropriate therapy includes the misuse of broad-spectrum antibiotics and inappropriate dose or duration of therapy, which are major drivers for the development of drug-resistant bacteria25,26.
With rising antibiotic resistance among uropathogens and concerns about antibiotic availability for UTIs, antimicrobial stewardship programs are increasingly crucial. These programs ensure appropriate antibiotic use by favoring narrow-spectrum and shorter-duration regimens, balancing risk and benefit for optimal effectiveness27,28,29. Implementing an electronic health record (EHR)-based antimicrobial stewardship program can provide a timely clinical decision support system (CDSS)30,31,32,33. However, there is limited data on the utility of EHR-based CDSS in improving antimicrobial prescribing behaviors, especially in the community setting and for UTI34,35.
In this study, we describe and evaluate the nationwide deployment of “UTI Smart order- set” (UTIS), a machine-learning model incorporating interpretable CDSS for empiric treatment of UTI. UTIS generates antibiotic recommendations for UTI based on personalized AI-driven predictions of antibiotic resistance13, adapted to the healthcare organization’s practice guidelines for appropriate antibiotic usage. This study aimed to evaluate the tool’s real-world performance, accuracy, and impact on prescription patterns of antibiotics for the treatment of UTI.
Results
Between June 1st, 2021, and August 31st, 2022, 171,010 UTI diagnoses were recorded in MHS (Fig. 1). In 58,517 cases, UTIS was not applicable because the preliminary criteria required for initiating the tool were not followed. Of the 112,493 cases where UTIS popped up, physicians prescribed antibiotics in 75,630 cases (67.2%). The overall acceptance rate of UTIS recommendation for antibiotic selection was 66.0% and was stable during the study period (the lowest monthly value was 61%, the highest was 72%, and the overall variance was <0.1%). The demographics and clinical characteristics of patients whose physicians accepted and did not accept the UTIS recommendation are shown in Table 1.
Antibiotic mismatch rate
Only 34% of UTI diagnoses were followed by Urine Culture lab results. Of 75,630 cases when UTIS was opened and data was available, urine samples with relevant antibiotic sensitivity results were available in 19,287 (25.5%) cases for further analysis. Among the general adult population, physicians who accepted the UTIS recommendation for antibiotic choice had a mismatch rate of 1124 out of 12,626 (9%). In contrast, when the physicians did not accept the UTIS recommendation, the mismatch rate was 991 out of 6972 (16%). This represents a 37.4% lower mismatch rate when the UTIS recommendations were followed (Fig. 2). Among women aged 18 years and above, the mismatch rate was 47.5% lower (n = 824 out of 10,975 vs. 927 out of 6479, respectively, p < 0.001) and 55.6% lower (n = 397 out of 5389 vs. 630 out of 3798, respectively, p < 0.001) among women over 50 years of age. For pregnant women, rates were 36.6% lower, although not significant due to the small sample size (n = 34 out of 653 vs 17 out of 207, respectively, NS).
The percentage of resistant bacteria to the antibiotic prescribed in cases where antibiotic prescribed was one of the recommended by UTIS or not evaluated for: all cases, women over 18 years of age, women over 50 years of age, and pregnant women. Differences were calculated as changes in percentage between proportions. Asterisk indicates a significant difference at p < 0.001.
Antibiotic mismatch rate per regimen
Mismatch rates were specifically analyzed across antibiotic regimens in adult women (over 18 years of age), women over 50, and pregnant women (Fig. 3). Among women, adherence to UTIS significantly reduced mismatch rates for all regimens except Fosfomycin (p < 0.001), with a non-significant decrease noted for Cephalexin (Fig. 3a). Significantly lower mismatch rates (p < 0.001) were observed among women over 50 for Nitrofurantoin, Cefuroxime, and Ciprofloxacin (Fig. 3b). Among pregnant women, significantly lower mismatch rates (p < 0.001) were found for Nitrofurantoin and Cefuroxime, with a non-significant lower rate for Cephalexin (Fig. 3c).
Differences were calculated as changes in percentage between proportions. Asterisk indicates a significant difference at p < 0.001. a Women over 18 years of age. The number of cases where the antibiotic prescribed was recommended/ not by UTIS, respectively: Nitrofurantoin 3644, 642; Fosfomycin 2525, 1444; Trimethoprim/ Sulfamethoxazole 1210, 310; Cefuroxime 1923, 1013; Cephalexin 737, 578; Ciprofloxacin 936, 2462. b Women over 50 years of age. Number of cases where the antibiotic prescribed was recommended/ not by UTIS, respectively: Nitrofurantoin 1082, 379; Fosfomycin 1648, 525; Trimethoprim/ Sulfamethoxazole 775, 225; Cefuroxime 557, 735; Cephalexin 111, 307; Ciprofloxacin 496, 1627. c Pregnant women. Number of cases where the antibiotic prescribed was recommended/ not by UTIS, respectively: Nitrofurantoin 138,15; Fosfomycin 1601, 100; Cefuroxime 325, 41; Cephalexin 66, 40.
Rates of target-concordant antibiotic prescriptions among women
Figure 4 shows the difference in the total number of prescriptions between the target value (i.e., the designated target values for six antibiotics; see methods section, intervention, part 2) and prescriptions given. The difference was calculated per antibiotic regimen, with vs. without the use of UTIS. For all regimes except Cefuroxime, a significantly smaller distance from the target value was observed when UTIS was used (p < 0.0001).
Asterisk indicates a significant difference at p < 0.001. a Overall rate of prescriptions given per antibiotic and desired predetermined target percentages (i.e., the designated target values for six antibiotics, see methods section, intervention, part 2). b Comparison between number of prescriptions per antibiotic regimen and predetermined target percentages. A positive number indicates over-prescription; Negative indicates sub-usage of antibiotic positive number indicates over-prescription; a Negative indicates sub-usage of antibiotics. Zero indicates complete concordance. The total number of prescriptions was 69,856, of which 25,484 were without UTIS recommendation and 44,372 following recommendation. Target Number of cases for each antibiotic prescribed, number when recommended and not by UTIS where, respectively: Nitrofurantoin 27,942, 13,862, 2677; Fosfomycin 20,957, 13,301, 6687; Trimethoprim/ Sulfamethoxazole 8383, 3819, 1278; Cefuroxime 6287, 6892, 3977; Cephalexin 3493, 3322, 2405; Ciprofloxacin 2794, 3177, 8460.
The overall rate of Ciprofloxacin and Nitrofurantoin prescriptions among women was examined, and cases were compared when UTIS recommendations were accepted and rejected. For Ciprofloxacin, an 80.5% overall reduction in prescriptions was observed with UTIS usage compared to when it was not used: 3176 of 49,495 prescriptions (6.4%) were prescribed following acceptance of UTIS recommendations versus 8459 out of 25,685 (32.9%) who following rejection, (p < 0.001). Conversely, for Nitrofurantoin, a 169.2% increase in prescriptions was observed with acceptance of UTIS recommendation compared to rejection: 13,861 out of 49,495 prescriptions (28.0%) were prescribed following acceptance of UTIS recommendations versus 2677 out of 25,685 (10.4%) when the recommendations were not accepted (p < 0.001).
Discussion
This large-scale study, conducted in a real-world setting, provides robust data on the performance of UTIS, a decision support tool that generates personalized recommendations tailored to individual patients and is implemented into routine clinical practice as a user-friendly set of recommendations. This innovative AI implementation into a large outpatient healthcare system’s nationwide interface successfully addresses a common challenge with the selection of empiric antibiotic therapy for UTI. Accepting the UTIS recommendation resulted in a 37.4% reduction in antibiotic mismatches and an 80.5% reduction in ciprofloxacin usage.
UTIS recommendations were developed based on the pioneering work of Yelin et al.13. They retrospectively achieved a 30% reduction in antibiotic mismatch cases by selecting the antibiotic with the highest likelihood of susceptibility. However, when choosing appropriate antibiotics, clinical guidelines, local resistance patterns, and patients’ personal factors should also be considered36. These considerations can impact the prescription pattern, leading to variations in the rate of antibiotic prescriptions8,37. Based on local antibiotic guidelines and resistance data, we incorporated predetermined thresholds of targeted antibiotics to better the selection of appropriate antibiotics. In the real-world setting, this integration of AI-driven algorithms with human expertise resulted in a similar overall reduction in antibiotic mismatch rates (37.4%, p < 0.001, Fig. 4). Nevertheless, a 9% mismatch rate was still observed when accepting UTIS recommendations. This can be attributed to algorithm limitations in complex cases, missing or incomplete patient data, or inadequate antibiotic target values. For example, Cephalexin showed higher mismatch rates, but the number of prescriptions was relatively low. Therefore, future changes in the target values of the frequency table should be reconsidered.
While another study, conducted by Kanjilal et al.38, focused explicitly on non-complicated UTIs, UTIS was aimed to apply to most patients, considering the diverse range of UTI presentations encountered in clinical practice. Among women aged 50 years and older, a 55.6% reduction in antibiotic mismatches was observed (p < 0.001). A decrease of 36.6% in antibiotic mismatch for pregnant women was also observed but was not statistically significant due to the small number of cases. Assuming that a culture is primarily recommended in complicated UTI cases6, where resistance is higher, it is plausible that if urinary cultures were obtained in all UTI cases, the performance of UTIS would improve due to the overall lower resistance rate and availability of more accurate data.
One of the barriers to achieving appropriate antibiotic prescriptions, including proper regimen, dose, and duration, is the absence of real-time point-of-care order sets that provide recommendations39. Using machine learning-based CDSS as a dynamic order-set presents a scalable solution to expand antibiotic stewardship interventions while delivering highly personalized recommendations13. We employed predetermined thresholds for targeted antibiotics to balance the use of narrow-spectrum antibiotics and maintain clinical efficacy. For example, Ciprofloxacin, a broad-spectrum antibiotic associated with potential side effects40, had a target threshold of 4%, while Nitrofurantoin had a target threshold of 40%. When accepting UTIS recommendations, an overall tendency toward the targeted thresholds was observed. Complete concordance wasn’t achieved due to multiple factors, mainly the physician’s clinical judgment, which affected the selection of the antibiotics and the availability of different pharmacological products. Another concern pertains to the integration phase of the process, which is continuously calculated for the entire population of adult MHS patients, while the results reflect only 75,630 cases where antibiotics were prescribed for UTI. This partial representation does not necessarily reflect the population’s distribution for UTIS recommendation. Nevertheless, accepting UTIS recommendations resulted in significantly lower usage rate of Ciprofloxacin and a higher usage rate of Nitrofurantoin, which may reflect the effectiveness of this CDSS tool in antibiotic stewardship while maintaining clinical efficacy.
Restrictions in infrastructure and systems may pose barriers that impede the application of innovative operations41. Indeed, implementing UTIS presented challenges that were addressed with specific solutions. Firstly, a batch-oriented system architecture was adopted to maintain real-time access to current medical information. This architecture scheduled daily processing to generate recommendations for the relevant population, resulting in more than 2.6 million daily recommendations. Secondly, integrating the system with operational legacy systems used by physicians required strict adherence to security, availability, and performance requirements. Thirdly, providing real-time recommendations during medical encounters necessitated a separation between model training and prediction processes, with flexible, dynamic tables-driven post-processing. Finally, a monitoring infrastructure was established to track physicians’ responses to meet evolving user needs, enabling ongoing analysis and system enhancements.
In 26,123 cases where a diagnosis of UTI was made by a physician and UTIS was opened, no antibiotic treatment was prescribed (Fig. 1). The decision not to prescribe antibiotics could have been based on the spontaneous resolution of symptoms, where the infection was likely self-limiting or treated only with symptomatic treatment and did not require antibiotic intervention, in line with Hoffmann et al.42. The decision to prescribe antimicrobial treatment, if needed, requires further research to support the development of a future AI-based model that could assist physicians in making antibiotic -prescribing decisions.
Notably, in about one-third of cases, physicians did not accept UTIS recommendations, and in cases they did there is uncertainty as to whether clinicians’ choices were directly influenced by the tool or if they coincided with the CDSS recommendations regardless. Further analysis is planned to profile physicians who either accepted or declined UTIS recommendations, and understand and solve possible barriers. One possible explanation for rejecting the recommendation is that some doctors may have concerns about the algorithm’s accuracy or reliability or may not fully understand how it works43. Another reason could be that some physicians may favor treatment based on their clinical experience and hesitate to alter their practices. The goal of UTIS deployment was to enhance decision-making by supporting clinicians without undermining their autonomy or compromising patient safety. If clinicians were to follow AI recommendations strictly without applying their own clinical judgment, there could be a risk of over-reliance on technology, potentially leading to suboptimal patient outcomes, as physicians are best suited to evaluate each patient’s unique context. Conversely, disregarding UTIS’ recommendations could result in missed opportunities for improved patient care, given that the system is designed to provide valuable insights and streamline aspects of UTI management, thereby reducing clinician workload. To address these risks, UTIS was designed as a supportive tool to assist, not replace, clinical judgment. Additionally, UTIS is continuously monitored and refined through regular feedback from physicians. Regular audits and updates based on real-world performance are also conducted to enhance accuracy and minimize potential risks, ensuring the system remains a safe and practical tool for patient care. Indeed, this large-scale implementation of UTIS in MHS systems could have applicability and relevance to other health services. A CDSS-based machine learning algorithm is also planned to address other infections, such as skin infections.
There are some limitations: First, this study was conducted in an Israeli HMO and thus may have limited generalizability to other countries or settings, such as inpatient or long-term care facilities. Second, as this study demonstrates the feasibility and benefits of such an AI support system focused on microbiological data and antibiotic mismatch, it did not address clinical cure, the need for recurrent culture, or antibiotic switch. This further information would probably improve the performance of the model and the results. Moreover, as UTIS was triggered based on UTI diagnosis, some cases did not invoke it. Cases of asymptomatic bacteriuria were waived as UTIS was not presented if culture was in the system. Finally, the susceptibility data is subject to changes in clinical breakpoints and shifts in the underlying distribution over time, potentially reducing the accuracy of models trained on historical data.
In conclusion, UTIS, a decision support tool for UTI management based on incorporating an AI model with clinical guidelines and personal patient characteristics optimized antibiotic prescribing practices, reduced antibiotic mismatches, and lowered usage of quinolones. This study represents an important step towards leveraging technology to enhance clinical decision-making processes and deliver personalized care in infection management.
Methods
Setting
The study was a prospective longitudinal observational study conducted in Maccabi Healthcare Services (MHS), Israel’s second-largest Health Maintenance Organization (HMO). MHS offers outpatient care for more than 2.6 million members nationwide, with over 6000 physicians and approximately 22 million medical encounters annually. MHS’ central data repository retains patient demographic data, physician data, and laboratory results using each patient’s unique national identification number. All healthcare providers within MHS use a unified electronic medical record (EMR) system. Before implementing UTIS, the organizational guidelines recommended Nitrofurantoin as the first-line antibiotic and Fosfomycin as the second-line antibiotic.
Intervention
Formerly, Yelin et al.13 built a personalized drug-specific predictive model for antibiotic resistance in UTI using a training dataset from MHS, leveraging a machine learning algorithm. As part of the antibiotic stewardship program, MHS policymakers decided to address the growing resistance of uropathogens. A sensitivity analysis for the model was conducted to validate the ML-based algorithm, enabling its deployment across the unified nationwide electronic health record (EHR) software between December 2020 and May 2021. The integrated model for antibiotic recommendations described here incorporates three key components: the core ML-based algorithm for antibiotic resistance, predefined antibiotics prioritization, and patients’ characteristics. The project (Fig. 5) involved collaboration between a dedicated data team, medical informatics specialists, and clinicians, including an infectious disease consultant.
A The core model was employed to predict the distinct bacterial resistance according to personal resistance profile and personal characteristics such as age, gender, antibiotic prescriptions and laboratory results. B Integration between the model’s output and a predetermined antibiotic prioritization framework. C Integration with personal patients’ clinical characteristics. D Incorporation into clinical workflows as a point of care clinical decision supporting system.
Core model for prediction of antibiotic resistance (Fig. 5A): MHS Data Lake serves as the primary storage for the essential data sources, functioning as a scalable big data repository that houses current and historical medical records. The model incorporated three primary repositories: 1. Antibiotic prescriptions issued and those purchased by MHS patients. 2. Urine microbiology culture laboratory results. 3. Patient records. The system included two key processes: Monthly model training and daily model-based prediction processes. Using the extensive dataset, the model continuously learned from diverse UTI cases and enhanced its predictive capabilities over time.
The model’s monthly retraining incorporated newly accumulated data, constructed for every training iteration by considering all historical UTI events (Supplementary Fig. 1). This panel encompassed the recorded microbial lab results from 2009 onwards, compromising over 1 million events with approximately 71,000 new added annually. Each event record held more than 600 features, including gender, pregnancy status, residence in a retirement home, past urine culture results with susceptibility, matrices capturing prior antibiotics issued across 16-time bins relative to the current event, and matrices for previous antibiotic resistance and sensitivity for twelve antibiotics recorded per time bin. To handle this complexity, there is a dedicated machine-learning model for each antibiotic, with the number of models reaching up to 12 models (Supplementary Fig. 2). The training population included 440,000 individuals, 86% females and 14% males; 20% had more than three events (Supplementary Fig. 3). In addition to the monthly training, a daily prediction process generated over 2.5 million recommendations for the entire MHS population based on predefined criteria (such as age 16 years and over) and accounting for background parameters such as possible pregnancy (for women of childbearing age) and residence at a retirement home. These recommendations cover a population of more than 1.9 million individuals. Finally, combining these records with the active models for six antibiotics (defining per antibiotic if the bacteria is resistant or sensitive by CLSI methods using MIC breakpoints or Fosfomycin zone diameter breakpoints) produced nearly 18 million predictions.
Next, a daily “post-process” was performed to generate the final recommendations presented to physicians. The daily recommendation preparation process included three steps: (1) filtering antibiotic recommendations based on clinical constraints in the patient’s medical record and determining the recommended dose and duration (Supplementary Fig. 4); (2) selecting the commercial drug code for each antibiotic’s main generic ingredient based on the current pharmaceutical HMO preference for generic agents available for prescription. (3) recommending the top two antibiotics as per descending sensitivity score.
Antibiotic prioritization framework (Fig. 5B): To balance between minimizing the use of broad-spectrum antibiotics and optimizing treatment outcomes11, a predetermined target ranking was created for antibiotics. The ranking was determined through a comprehensive evaluation of local guidelines for UTI management and local antibiotic susceptibility profiles, safety profiles, efficacy, and patient adherence to therapy. An agile frequency table, including six antibiotic regimens for treating UTIs, was built. The table provided target values for the percentage of each antibiotic prescribed for UTIs among MHS patients, assuming that all patients receive antibiotics on the same day. The designated target values for six antibiotics were as follows: Nitrofurantoin (40%), Fosfomycin (30%), Trimethoprim/Sulfamethoxazole (12%), Cefuroxime (9%), Cephalexin (5%), and Ciprofloxacin (4%). These values could be readily modified in response to changes in resistance patterns or further evaluation of the model performance to optimize treatment outcomes.
Integration with personal patients’ characteristics (Fig. 5C): Personalized recommendations were designed to consider the most appropriate and effective treatment options while addressing potential limitations or specific patient considerations (Supplementary Fig. 5). The integrated model accounted for personal factors influencing available treatment options: age, sex, pregnancy status, and renal function. Specific recommendations based on patient characteristics were provided (Supplementary Table 1). Drug-drug interaction was not included in the model due to using an embedded external system.
Incorporation into clinical workflows (Fig. 5D): The front end was developed to establish communication between the integrated machine learning model and the MHS’ EHR software (Clicks®)). This interface facilitated real-time and interpretable recommendations to physicians within the clinical workflow, embedding a point of care CDSS into the medical encounters. During the medical encounter, after physician evaluation, if a UTI diagnosis (ICD-9 code 599.0) is selected, the CDSS appears as a pre-defined set of orders, UTIS (Fig. 6). It may appear in a face-to-face, telephone, or virtual encounter. The Criteria for initiating UTIS were as follows: patients aged 18 or older, without urine culture results in the past 96 h, and no previous UTIS activation in the past 72 h. The order set included options to choose referrals for a urinalysis and urine culture and a urine test panel for sexually transmitted infections in men under 50. In order to expand the scope of physician decision-making, two antibiotic options were presented for selection based on the integrated model output. Initial dosage and therapy duration were pre-filled, with adjustments for the patient’s kidney function. Based on clinical judgment, the physician was able to change the antibiotic chosen or close the UTIS order-set, and the medical encounter would continue. After a period of piloting in selected healthcare facilities to monitor performance and gather user feedback, MHS policymakers decided to deploy UTIS. Starting on June 1, 2021, UTIS was deployed in all MHS EHRs.
A feature in an electronic health record system displays a pre-defined set of orders for managing urinary tract infections on the screen when a physician selects the diagnosis during a patient encounter. This feature aims to streamline and standardize the management of urinary tract infections by providing physicians with recommended practices, including the antibiotic regimen, dose, and duration.
Study outcomes
Cases where the prescribed antibiotic matched one of the UTIS recommendations were considered ‘accepted.’ Conversely, cases where the prescribed antibiotic differed from the UTIS recommendations were considered ‘rejected.’ A comparison between accepted and rejected cases was then made. The study’s primary outcome was to assess the impact of UTIS usage on the overall prescribed antibiotic mismatch rate. This was defined as the percentage of cases where the prescribed antibiotic was ineffective against the resistant bacteria detected in the urine culture. Secondary outcomes included the rate of antibiotic mismatch prescribing for each specific antibiotic regimen, the rate of guideline-concordant antibiotic prescriptions, and the rate of ciprofloxacin prescriptions.
Data analysis
Data analysis and statistical tests were conducted using Excel (Microsoft™), R (R Core Team, 2020), and RStudio (Rstudio Team, 2020). Statistical analysis included descriptive statistics and a Z-score test for population proportions, with a significance level of p < 0.05. This study is reported following the “Reporting of Studies Conducted using Observational Routinely-collected Health Data (RECORD) Statement”44. The study was carried out with the prior approval of the Maccabi internal review board and the Maccabi ethics (Helsinki) committee (0038-19-MHS). Informed consent was waived by the IRB, as all identifying details of the participants were removed before the computational analysis.
Data availability
The data supporting this study are available from the corresponding author, but restrictions apply to the availability of this information, which was used under license for the current study and so is not publicly available. Data are, however, available from the authors upon reasonable request and with permission of the local ethics committee of MHS.
Code availability
Not applicable.
References
Petersen, I., Hayward, A. C. & SACAR Surveillance Subgroup. Antibacterial prescribing in primary care. J. Antimicrob. Chemother. 60, i43–i47 (2007).
Holm, A., Cordoba, G. & Aabenhus, R. Prescription of antibiotics for urinary tract infection in general practice in Denmark. Scand. J. Prim. Health Care 37, 83–89 (2019).
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Kaye, K. S. et al. Antimicrobial resistance trends in urine Escherichia coli isolates from adult and adolescent females in the United States from 2011–2019: rising ESBL strains and impact on patient management. Clin. Infect. Dis. 73, 1992–1999 (2021).
Dunne, M. W. et al. A multicenter analysis of trends in resistance in urinary Enterobacterales isolates from ambulatory patients in the United States: 2011–2020. BMC Infect. Dis. 22, 194 (2022).
Grigoryan, L., Trautner, B. W. & Gupta, K. Diagnosis and management of urinary tract infections in the outpatient setting: a review. JAMA 312, 1677–1684 (2014).
Ahmed, H., Farewell, D., Francis, N. A., Paranjothy, S. & Butler, C. C. Choice of empirical antibiotic therapy and adverse outcomes in older adults with suspected urinary tract infection: cohort study. Open Forum Infect. Dis. 6, ofz039 (2019).
Krishnakumar, J. & Tsopra, R. What rationale do GPs use to choose a particular antibiotic for a specific clinical situation? BMC Fam. Pract. 20, 178 (2019).
O’Brien, K., Hillier, S., Simpson, S., Hood, K. & Butler, C. An observational study of empirical antibiotics for adult women with uncomplicated UTI in general practice. J. Antimicrob. Chemother. 59, 1200–1203 (2007).
Rosa, R., Abbo, L. M., Raney, K., Tookes, H. E. & Supino, M. Antimicrobial resistance in urinary tract infections at a large urban ED: Factors contributing to empiric treatment failure. Am. J. Emerg. Med. 35, 397–401 (2017).
Dunne, M. W. et al. Impact of empirical antibiotic therapy on outcomes of outpatient urinary tract infection due to nonsusceptible enterobacterales. Microbiol. Spectr. e0235921, https://doi.org/10.1128/spectrum.02359-21 (2022).
Zhu, H. et al. Impact of inappropriate empirical antibiotic treatment on clinical outcomes of urinary tract infections caused by Escherichia coli: a retrospective cohort study. J. Glob. Antimicrob. Resist. 26, 148–153 (2021).
Yelin, I. et al. Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat. Med. 25, 1143–1152 (2019).
Anahtar, M. N., Yang, J. H. & Kanjilal, S. Applications of machine learning to the problem of antimicrobial resistance: an emerging model for translational research. J. Clin. Microbiol. 59, e0126020 (2021).
Moran, E., Robinson, E., Green, C., Keeling, M. & Collyer, B. Towards personalized guidelines: using machine-learning algorithms to guide antimicrobial selection. J. Antimicrob. Chemother. 75, 2677–2680 (2020).
Peiffer-Smadja, N. et al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin. Microbiol. Infect. 26, 584–595 (2020).
MacFadden, D. R. et al. Decision-support models for empiric antibiotic selection in Gram-negative bloodstream infections. Clin. Microbiol. Infect. 25, 108.e1–108.e7 (2019).
Corbin, C. K. et al. Personalized antibiograms for machine learning driven antibiotic selection. Commun. Med. 2, 38 (2022).
Gupta, K. et al. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: A 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 52, e103–e120 (2011).
Shively, N. R., Buehrle, D. J., Clancy, C. J. & Decker, B. K. Prevalence of Inappropriate Antibiotic Prescribing in Primary Care Clinics within a Veterans Affairs Health Care System. Antimicrob. Agents Chemother. 62, e00337 (2018).
Schmiemann, G., Hoffmann, F., Hamprecht, A. & Jobski, K. Patterns and trends of antibacterial treatment in patients with urinary tract infections, 2015-2019: an analysis of health insurance data. BMC Prim. Care 23, 204 (2022).
Shapiro, D. J., Hicks, L. A., Pavia, A. T. & Hersh, A. L. Antibiotic prescribing for adults in ambulatory care in the USA, 2007-09. J. Antimicrob. Chemother. 69, 234–240 (2014).
Kim, M., Lloyd, A., Condren, M. & Miller, M. J. Beyond antibiotic selection: concordance with the IDSA guidelines for uncomplicated urinary tract infections. Infection 43, 89–94 (2015).
Kabbani, S. et al. Opportunities to improve fluoroquinolone prescribing in the United States for adult ambulatory care visits. Clin. Infect. Dis. 67, 134–136 (2018).
World Health Organization. Antimicrobial resistance. Key facts. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (2023).
Llor, C. & Bjerrum, L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 5, 229–241 (2014).
Barlam, T. F. et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin. Infect. Dis. 62, e51–e77 (2016).
Marcelin, J. R., Chung, P. & Van Schooneveld, T. C. Antimicrobial stewardship in the outpatient setting: A review and proposed framework. Infect. Control Hosp. Epidemiol. 41, 833–840 (2020).
Sanchez, G. V., Fleming-Dutra, K. E., Roberts, R. M. & Hicks, L. A. Core elements of outpatient antibiotic stewardship. MMWR Recomm. Rep. 65, 1–12 (2016).
Forrest, G. N. et al. Use of electronic health records and clinical decision support systems for antimicrobial stewardship. Clin. Infect. Dis. 59, S122–S133 (2014).
Bremmer, D. N., Trienski, T. L., Walsh, T. L. & Moffa, M. A. Role of technology in antimicrobial stewardship. Med. Clin. North Am. 102, 955–963 (2018).
Delory, T. et al. A computerized decision support system (CDSS) for antibiotic prescription in primary care-Antibioclic: implementation, adoption and sustainable use in the era of extended antimicrobial resistance. J. Antimicrob. Chemother. 75, 2353–2362 (2020).
Gulliford, M. C. et al. Effectiveness and safety of electronically delivered prescribing feedback and decision support on antibiotic use for respiratory illness in primary care: REDUCE cluster randomised trial. BMJ 364, l236 (2019).
Catho, G. et al. Impact of interactive computerised decision support for hospital antibiotic use (COMPASS): an open-label, cluster-randomised trial in three Swiss hospitals. Lancet Infect. Dis. https://doi.org/10.1016/S1473-3099(22)00308-5 (2022).
Rawson, T. M. et al. A systematic review of clinical decision support systems for antimicrobial management: are we failing to investigate these interventions appropriately? Clin. Microbiol. Infect. 23, 524–532 (2017).
Leekha, S., Terrell, C. L. & Edson, R. S. General principles of antimicrobial therapy. Mayo Clin. Proc. 86, 156–167 (2011).
Teixeira Rodrigues, A., Roque, F., Falcão, A., Figueiras, A. & Herdeiro, M. T. Understanding physician antibiotic prescribing behaviour: a systematic review of qualitative studies. Int. J. Antimicrob. Agents 41, 203–212 (2013).
Kanjilal, S. et al. A decision algorithm to promote outpatient antimicrobial stewardship for uncomplicated urinary tract infection. Sci. Transl. Med. 12, eaay5067 (2020).
Lecky, D. M., Howdle, J., Butler, C. C. & McNulty, C. A. Optimising management of UTIs in primary care: a qualitative study of patient and GP perspectives to inform the development of an evidence-based, shared decision-making resource. Br. J. Gen. Pract. 70, e330–e338 (2020).
Food and Drug Administration. FDA drug safety communication: FDA advises restricting fluoroquinolone antibiotic use for certain uncomplicated infections; warns about disabling side effects that can occur together. 2016. https://www.fda.gov/Drugs/DrugSafety/ucm500143.htm. Accesed April 7, 2023.
Charani, E. et al. Optimising antimicrobial use in humans - review of current evidence and an interdisciplinary consensus on key priorities for research. Lancet Reg. Health Eur. 7, 100161 (2021).
Hoffmann, T., Peiris, R., Mar, C. D., Cleo, G. & Glasziou, P. Natural history of uncomplicated urinary tract infection without antibiotics: a systematic review. Br. J. Gen. Pract. 70, e714–e722 (2020).
AI Use in Healthcare lncreasing Slowly Worldwide. Marcia Frellick. https://www.medscape.com/viewarticle/912629?form=fpf (2019). Accesed July 27, 2023.
The Reporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement. PLOS Med. 2015. https://journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.1001885.
Central bureau of statistics. Publications and Media Releases 2017. Available at: https://www.cbs.gov.il/en/mediarelease/Pages/2017/Projections-of-Israel-Population-until-2065.aspx.
Shapiro Ben David, S., Goren, I., Mourad, V. & Cahan, A. Vaccination Coverage among Immunocompromised Patients in a Large Health Maintenance Organization: Findings from a Novel Computerized Registry. Vaccines 10, 1654 (2022).
Acknowledgements
We thank Dr. Idan Yelin, Prof. Roy Kishony, Dr Moshe Fisher, Hadar Asulin, Hila Tsriker and Ariel Karlinsky.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.S.B.D., R.R., S.G., B.G. and U.L.; Methodology, S.S.B.D., R.R. and U.L.; Formal analysis, S.S.B.D., R.R. and U.L.; writing—original draft preparation, S.S.B.D.; Writing—review and editing, R.R., D.R.C., J.A. and U.L.; Supervision: D.R.C., S.G. and B.G.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shapiro Ben David, S., Romano, R., Rahamim-Cohen, D. et al. AI driven decision support reduces antibiotic mismatches and inappropriate use in outpatient urinary tract infections. npj Digit. Med. 8, 61 (2025). https://doi.org/10.1038/s41746-024-01400-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-024-01400-5