Abstract
We explore biases present in publicly available fetal ultrasound (US) imaging datasets, currently at the disposal of researchers to train deep learning (DL) algorithms for prenatal diagnostics. As DL increasingly permeates the field of medical imaging, the urgency to critically evaluate the fairness of benchmark public datasets used to train them grows. Our thorough investigation reveals a multifaceted bias problem, encompassing issues such as lack of demographic representativeness, limited diversity in clinical conditions depicted, and variability in US technology used across datasets. We argue that these biases may significantly influence DL model performance, which may lead to inequities in healthcare outcomes. To address these challenges, we recommend a multilayered approach. This includes promoting practices that ensure data inclusivity, such as diversifying data sources and populations, and refining model strategies to better account for population variances. These steps will enhance the trustworthiness of DL algorithms in fetal US analysis.
Similar content being viewed by others
Introduction
Fetal ultrasound (US) imaging serves as a critical screening tool in prenatal care, providing visualization and monitoring of fetal growth within the womb. By using high-frequency sound waves, this imaging technique generates real-time images of the fetus, placenta, and surrounding maternal structures, playing a crucial role in assessing fetal health, detecting abnormalities, and guiding medical interventions during pregnancy1.
In recent years, there has been a transformative shift in fetal US image analysis due to the introduction of artificial intelligence (AI) for medical image analysis. AI can expand the capabilities of diagnostic support tools, offering clinicians valuable assistance in the decision-making processes involved in prenatal care2. In particular, machine learning (ML) and deep learning (DL) have made remarkable strides in this area, increasing the efficacy and efficiency of fetal US assessments. This progress has been well documented by the growing body of research, including research studies, international challenges, and surveys, which demonstrate the transformative impact of these technologies in the field3,4,5,6,7.
The rapid growth and success of DL algorithms are largely due to the availability and accessibility of extensive clinician-annotated data8,9. Nevertheless, the collection, annotation, and distribution of fetal US data present significant challenges, due to the inherently sensitive nature of the images and the need for ethical and legal considerations, such as patient or caregivers’ consent10. The sheer complexity of obtaining a heterogeneous dataset can lead to data gaps in the information available and variances in data quality and format, limiting the breadth and depth of data required for training effective DL models11.
To cope with data scarcity, the medical image analysis community has taken proactive steps by organizing international challenges and releasing large international benchmark datasets (https://grand-challenge.org/challenges/). These initiatives aim to stimulate research interest and engagement in addressing critical clinical tasks. At the same time, the released datasets allow researchers to compare algorithms' performance and contribute to the continuous improvement of DL techniques in the realm of medical imaging12,13,14,15. Fetal US image analysis is not an exception, and researchers have been actively utilizing such benchmark datasets to evaluate the effectiveness and generalizability of their algorithms.
While the relevance of publicly available fetal US datasets for evaluating and improving DL algorithms is unquestionable16, it is equally essential to assess the biases that these datasets may harbor, which in turn affect the algorithms trained on them. In fact, the scientific literature has often focused on biases that may arise during routine fetal US scans, such as biases in measurements17,18, or biases in the identification of fetal malformations19. However, biases embedded in benchmark public datasets used for training DL models have not yet been investigated, nor have the potential ethical implications, such as the perpetuation of social inequalities, the discrimination against minority groups, and any other related issues have been thoroughly examined in the domain of fetal US20. This lack of investigation can be attributed to the relatively recent birth of this field of research, with the first suitable public fetal US image dataset released in 2018. Nevertheless, in the clinical domain, where AI systems are increasingly employed to support decision-making processes that directly impact patient outcomes, trustworthiness is crucial. Trustworthy AI in healthcare means systems that are not only technically robust and reliable but also ethically sound and socially responsible21. To ensure trustworthiness in clinical AI applications, it is essential to evaluate and mitigate algorithmic bias, ensuring fairness. Developing and deploying AI models that operate equitably is the foundational step toward achieving this goal.
Recent studies across various domains22,23 have highlighted the issue of algorithmic biases in healthcare and medicine, especially within medical imaging24. Such biases may lead to unfair health treatment and exacerbate health disparities when the data training DL algorithms lack a comprehensive representation of diverse population groups. These biases could also steer clinicians towards inappropriate and flawed medical interventions, negatively impacting patient outcomes and amplifying the risk of harm.
In light of this, the importance of ensuring the development of fairness in algorithms cannot be overstated. The implementation of fairness in DL for the fetal US is a broad, multi-step process that must begin with a focus on the biases present in the collected or annotated datasets. Our research aims to fill this critical gap by focusing on this initial stage. Guided by this objective, our study is centered on three main methodological questions:
-
Are there biases present in fetal US datasets that are publicly available today? If so, what kind of biases (Q1)?
-
What are the ethical issues and risks involved (Q2)?
-
Which types of mitigation (actions) can be foreseen to address these biases (Q3)?
To address these questions comprehensively, we conducted an in-depth review of all publicly available datasets within the fetal US domain. Building upon established research on biases in healthcare and medical imaging, we adapted methodologies to investigate and analyze potential biases in these datasets.
By shedding light on these biases, our work highlights the importance of fostering fairness and reliability in AI systems in the domain of fetal well-being. At the same time, the implications of this study may go beyond fetal US imaging. It provides the first systematic framework and practical approach for identifying and mitigating biases that can also be applied to a broader context of medical imaging, paving the way for more equitable and trustworthy AI solutions in healthcare.
Results
Datasets
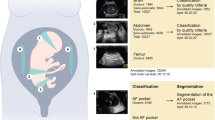
We identified five publicly available datasets, whose relevant characteristics are summarized in Fig. 1 and Table 1 (details of the A-AFMA dataset are only available to participants of the associated challenge and are therefore excluded from Table 1). The datasets are grouped based on the main tasks in fetal US imaging analysis using DL, aligning with common steps in benchmark fetal well-being assessment practice (see section Methods):
-
1.
Two datasets related to fetal standard-plane detection (SPD):
-
FETAL_PLANES_DB(https://zenodo.org/record/3904280)
-
Maternal-fetal US planes in African countries (MFUSPAC)(https://zenodo.org/records/7540448)7
-
2.
Two datasets for analysis of anatomical structures (AAS):
-
Fetal Abdominal Structures Segmentation Dataset (FASSD)(https://data.mendeley.com/datasets/4gcpm9dsc3/1)
-
3.
One dataset for fetal biometry parameter estimation (BPE):
Percentages derived from studies utilizing the FETAL_PLANES_DB, MFUSPAC, and HC18 datasets. The FASSD dataset has not yet been utilized. a Boxplot showing the interquartile ranges of dataset citation impact scores (CiteScore 2022). b Donut-style pie chart depicting the overall distribution of publication years (2018–2024). c Donut-style pie chart representing the type of contributions (journal/conference) involving the datasets.
All analyses conducted on such datasets were performed in accordance with the Declaration of Helsinki.
FETAL_PLANES_DB includes 12,400 US images from 1792 pregnant women attending routine screenings during their second and third trimesters at Hospital Clinic and Hospital Sant Joan de Deu, both located in Barcelona, Spain. An experienced sonographer organized the dataset by collecting US images of fetal standard planes categorized into “abdomen”, “brain”, “maternal cervix”, “femur”, “thorax”, and a miscellaneous class labeled “Other”. Notably, the fetal “brain” category is further segmented into transventricular (TV), transcerebellar (TC), and transthalamic (TT) planes. The US images were captured using six different US machines—three Voluson E6, one Voluson S8, one Voluson S10 (GE Medical Systems, Zipf, Austria), and one from Aloka (Aloka Co., Ltd) and a 3–7.5 MHz curved transducer for abdominal US and a 2–10 MHz vaginal probe for cervical US screening.
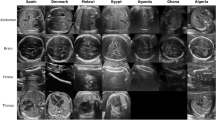
MFUSPAC consists of US images corresponding to the four most common fetal planes—abdomen, brain, femur, and thorax. It is composed of five datasets collected using various US machines and transducers across different African countries, each encompassing data from 25 patients for a total of 450 US images. First dataset was collected using a Mindary DC-N2 US machine (Shenzhen Mindray Bio-Medical Electronics Co., Ltd, China/Germany) with a 3.5 MHz curved transducer at Malawi’s Queen Elizabeth Central Hospital, focusing on second and third trimesters US images; the second dataset was acquired at the Sayedaty center in Egypt capturing second trimester scans using a Voluson P8 (GE Medical Systems, Zipf, Austria) with a 7 MHz curved transducer; the third dataset comprises third trimester images acquired using an ACUSON X600 (Siemens) and a 3–7.5 MHz curved transducer at Mulago National Referral Hospital in Uganda; the fourth dataset, obtained at Accra’s KBTH Polyclinic Center in Ghana with an EDAN DUS 60 (Edan Instruments, Inc., Shenzhen, China), includes second and third trimester images using a curved transducer with a frequency range of 3.5–5 MHz; and finally, the last dataset was collected at the EPH Kouba and Clinique Des Lilas centers in Algeria using a Voluson S8 machine (GE Medical Systems, Zipf, Austria) and a 3–7.5 MHz curvilinear transducer.
A-AFMA was part of the ISBI 2021 challenge, which aimed to automatically detect two anatomies within each frame of a US video—amniotic fluid and the maternal bladder. Detailed information about this dataset is not publicly available, and access is limited to request only. Therefore, further analysis of this dataset is not feasible for this study.
FASSD comprises nearly 1500 fetal US images from 169 subjects of the abdomen circumference, acquired and annotated by two sonographers between September 2021 and September 2023. The dataset was collected at the Polydoro Ernani de São Thiago University Hospital in Florianópolis, Brazil, including full-term pregnant women, comprising those awaiting labor initiation, cesarean delivery, and those with certain pregnancy-related problems such as gestational diabetes and pre-eclampsia. Preterm pregnancies, multiple gestations, and pregnancies with fetal anomalies, instead, were not included. Images were acquired with Siemens Acuson, Voluson 730 (GE Healthcare Ultrasound), and Philips-EPIQ Elite (Philips Healthcare) equipment, all using 2–9 MHz curved linear transducers.
HC18 consists of 1334 fetal US images from 551 women, including skull US images during the first, second, and third trimesters of pregnancy for head circumference (HC) identification. For every image, HC is marked by one sonographer by drawing an ellipse that best represents the fetus’s skull section. Images were captured at the Obstetrics Department of Radboud University Medical Center in Nijmegen, Netherlands, using Voluson E8 and Voluson 730 US machines (GE Medical Systems, Zipf, Austria).
Dataset biases
We rely on benchmark studies on the ethics of AI in healthcare and medicine, drawing specific insights from refs. 25,26,27, whose merit is to map prominent controversial biases in alike domains to ours. Given the specificity of the field investigated, the inquiry carried out allowed us to not only align with established frameworks but also to critically evaluate additional estimations of data-related biases inherent to US technology.
The following biases, summarized in Table 2, have been identified: label bias, cohort bias, missing data bias, minority bias, informativeness bias, device variability bias, temporal bias, probe maneuver bias, and algorithm-clinician interaction bias. Each of these is commented on both in terms of its significance and its specific implications for the problem at hand.
Label bias is the consequence of the way data is labeled which can introduce skewed perspectives. As pointed out in ref. 9, in medical image analysis, the challenges of annotation are especially pronounced. This field often grapples with limited data, significant differences in ratings among experts, ambiguous labeling practices, and the unique annotation styles of individual medical professionals. As regards the analyzed datasets, this bias arises from the fact that the annotations necessary for algorithm development and evaluation are provided by a relatively small group of experienced sonographers, as evident from Table 1. As shown in Table 2, the label bias is present in HC18 and FETAL_PLANES_DB datasets, for which annotations were provided by a single experienced sonographer, resulting possibly conditioned by subjective expertise. The issue is more evident with MFUSPAC datasets, where the lack of reported information on the precise number of annotators adds another layer of uncertainty.
Cohort bias arises when research or analysis disproportionately focuses on traditional or easily measured groups, neglecting finer levels of granularity. This bias can lead to an incomplete understanding of diverse populations and potentially overlook critical variations within subgroups. Here, it arises from the disproportionate representation of certain classes over others, especially minorities and vulnerable groups, posing significant challenges to the development of fair AI models. This imbalance can hinder the model’s ability to accurately detect and classify instances of the less-represented classes and distort the assessment of model performance, necessitating mitigation strategies for a balanced and accurate evaluation. FETAL_PLANES_DB dataset exemplifies notable class imbalance, with the “fetal brain” category being significantly over-represented compared to others, such as “fetal femur” or “fetal abdomen”. In contrast, the MFUSPAC datasets, while showing more balance among classes compared to FETAL_PLANES_DB, only include four fetal standard planes—thorax, femur, brain, and abdomen—highlighting another aspect of cohort bias—the omission of certain classes, of certain standard planes.
In addition to uneven representation, the granularity of categories should be taken into account. FETAL_PLANES_DB groups images into six coarse-grained categories—abdomen, brain, femur, thorax, maternal cervix, and a miscellaneous class called “other”—and includes also a further subdivision of the “brain” category into three classes—TV, TT, and TC planes—enabling exploration of fetal brain conditions across different levels of granularity. As discussed in ref. 3, DL models developed using datasets with this bias may be overly proficient in identifying conditions within certain US planes, on which they were trained, while failing to recognize conditions or anomalies that manifest in planes not included in the training set. This discrepancy in performance could result in unequal quality of care, where some conditions are detected more reliably than others. For example, a model trained primarily on sagittal plane images might excel at identifying spinal anomalies but struggle with detecting cardiac anomalies that are better visualized in the transverse plane. Regarding the HC18 and FASSD datasets, they only provide data focused on specific anatomy (skull and abdomen, respectively). Thus, cohort bias and the misrepresentation of classes can also be noticed in these datasets.
It is also necessary to consider cohort bias resulting from the disproportionate representation of pregnancy trimesters. Given the dynamic nature of human fetal development, it is crucial to include data from the entire gestation period. As the fetus grows, significant changes occur in organ development and overall physiological status. This continuous growth means that the medical and developmental context of a fetus can vary greatly from one trimester to another. The omission of trimester-specific information, as seen in FASSD, could lead to incomplete or inaccurate interpretations of fetal health and development. For instance, a model trained predominantly on second-trimester data might miss early signs of hypoplastic left heart syndrome that are more apparent in the first trimester. Additionally, it might fail to detect late-developing issues such as arrhythmias that become noticeable in the third trimester. This gap in detection can result in critical missed diagnoses and potentially inadequate medical care. FETAL_PLANES_DB and MFUSPAC datasets focus exclusively on fetal US images obtained during the second and third trimesters of gestation. As detailed in section “Methods”, the process of identifying standard planes, crucial for precise fetal evaluation, varies significantly across trimesters. Consequently, when datasets are restricted to specific trimesters, they can lead to notable cohort bias if the derived DL algorithms are used throughout the entire pregnancy. It is essential that the DL systems are clearly employed only within the trimesters for which they are validated and applicable. Expanding the dataset to encompass all trimesters would significantly enhance our understanding of fetal development, leading to more accurate assessments and better clinical outcomes. This comprehensive approach ensures that the model can detect a broader range of conditions throughout the entire gestation period, ultimately enabling more timely and effective medical interventions. While HC18 stands out as the sole dataset offering image acquisition across all stages of gestation, it is important to note that in clinical practice, HC measurements are predominantly conducted during the second and third trimesters. HC measurements are predominantly conducted during the second and third trimesters. Integrating first-trimester biometric data would fill a crucial gap, providing insights into early fetal growth patterns and developmental indicators. This integration could lead to the early detection of potential developmental issues, such as neural tube defects or chromosomal abnormalities, which may be indicated by atypical growth patterns in the first trimester. Additionally, it would allow for more accurate baseline measurements, improving the monitoring of growth trajectories and potentially leading to earlier and more effective interventions if deviations from expected growth patterns are detected.
Missing data bias emerges in this context from limitations in dataset acquisition concerning patient nationality, the involvement of hospital centers, and their geographical location (whether they are in the same or different city, nation, or continent). Datasets, such as FETAL_PLANES_DB, HC18, and FASSD, are exclusively obtained from national centers, displaying a significant vulnerability to missing data bias. Models trained on a dataset predominantly representative of one population, when used to evaluate or predict certain relevant fetal biometries for a diverse population, risk yielding inaccurate growth assessment or misdiagnosis (an effect known as population target bias or training serving skew). Additionally, the absence of information on subjects’ nationality further compromises the broader applicability and generalizability of models trained with these data. Previous research has consistently highlighted the variation in fetal US measurements across different nationalities28,29,30,31,32. Such differences are particularly evident for parameters such as HC, AC, femur length, and estimated fetal weight. For HC18, which is primarily used to assess methodologies for HC estimation, the bias can be particularly concerning. Incorrect HC measurements could lead to misjudgments regarding fetal health, gestational age, or potential abnormalities, with tangible implications for clinical decisions regarding prenatal care and interventions33. For example, if an HC measurement inaccurately suggests that the fetus is smaller than expected for the gestational age, this could lead to unnecessary interventions such as early delivery or increased surveillance for intrauterine growth restriction. Conversely, overestimating HC could result in missed diagnoses of microcephaly, delaying crucial interventions or preparations for potential neurological complications after birth. The FASSD dataset, aimed at abdominal structure segmentation rather than direct biometric extrapolation, also contends with biases related to demographic or clinical characteristics. Such biases undermine the precision and utility of segmentation models based on this dataset, potentially compromising the evaluation of fetal health and prenatal care planning. On the other hand, FETAL_PLANES_DB is focused on identifying the optimal US plane, a fundamental step preceding any measurement or segmentation. In the presence of missing data bias in this dataset, the potential implications are also significant. Models trained on a non-representative dataset may consistently misidentify the correct plane when applied to different populations, leading to inherently flawed subsequent measurements (e.g., HC, AC). Similar concerns extend to MFUSPAC as well, however, this dataset distinguishes itself by aggregating data from multiple national centers, which inherently enhances its potential to mitigate missing data bias. Collecting data from diverse sources increases the dataset’s representativeness, reducing the likelihood of models trained on it suffering from biases not reflective of a wider demographic. Nevertheless, it is important to acknowledge that the MFUSPAC dataset focuses on the African continent introduces a geographical limitation. This approach helps address certain missing data biases, particularly in comparison to more narrowly focused datasets such as FETAL_PLANES_DB. However, it is crucial to note that applying models trained on the MFUSPAC dataset to populations outside of Africa may still introduce some degree of bias and produce a population target bias effect.
Minority bias occurs when a model’s performance is skewed due to the under-representation or misrepresentation of minority groups within the training data, such as members of social, ethnic, and health minorities, including those affected by rare pathologies or infrequent conditions. This bias prevents us from providing equally accurate health treatment and empowers those mostly marginalized or oppressed from a social, economic, cultural, or health standpoint. In this regard, fetal US datasets raise concern due to the evident under-representation of fetal twins, which inherently present unique physiological and anatomical variations distinguishing them from singleton pregnancies, carrying substantial clinical implications. Notably, none of the datasets include twin pregnancies. Consequently, any data-driven model not accounting for these differences may fall short in its predictions and interpretations. One pressing concern is the potential for misdiagnosis or oversight of complications that are unique to twin pregnancies, such as twin-twin transfusion syndrome (TTTS)34. TTTS, a serious condition wherein blood circulates unevenly between twins who share a placenta, underscores the critical need for early US examination in identifying and managing this syndrome35,36. Timely detection can significantly impact outcomes. In the clinical context, where every decision relies on accurate and comprehensive data, such biases could jeopardize the care provided to twin pregnancies, underscoring the urgent need to address and rectify these dataset limitations.
Informativeness bias emerges when a dataset disproportionately emphasizes typical fetal anatomy and physiology, potentially overlooking the diversity of abnormalities and variations encountered in clinical practice. For the HC18 dataset, composed solely of normal fetal brain US images, this bias leads to a significant oversight. By not including pathological conditions, the dataset fails to provide a comprehensive view of the range of fetal brain conditions that sonographers or clinicians may encounter in real-world settings. Significant pathological conditions that may be overlooked include ventriculomegaly (an abnormal enlargement of the brain’s ventricular system), holoprosencephaly (a disorder where the brain does not properly divide into the right and left hemispheres), and cerebral cysts, among others37,38. Similarly, the FASSD dataset, which is focused on segmenting anatomical regions of the fetal abdomen, faces similar concerns. Notable examples of significant fetal abdominal pathologies that might be overlooked include abdominal wall defects such as gastroschisis and omphalocele, where intestines and sometimes other organs are outside the abdomen, intra-abdominal cysts, and structural anomalies such as enlarged liver (hepatomegaly) or kidney irregularities39,40. Training a model exclusively on non-pathological abdominal images from the FASSD could result in inadequate detection or complete oversight of such critical conditions, potentially impacting clinical diagnosis and decision-making processes6. Additionally, the FETAL_PLANES_DB and MFUSPAC datasets, concentrating solely on fetal standard planes, may perpetuate this bias. In real-world clinical settings, sonographers maneuver the US probe over the mother’s belly, encountering not only standard planes but a continuous video sequence subject to their movement. Moreover, various pathological conditions can alter or affect these standard planes. For instance, skeletal dysplasia may impact the appearance and dimensions of long bones41.
Device variability bias arises in fetal US imaging when machines from different vendors introduce significant inconsistencies, creating challenges for DL algorithms. Models trained on datasets from specific vendor machines may struggle to adapt to data from others (i.e., data interoperability issues), which can present variations in image quality and appearance due to variability in hardware, software, and imaging protocols followed. Furthermore, the variability extends to the US probes or transducers used in imaging. Different types of US probes (e.g., curved array, 3D/4D curved array, endocavitary, and 3D/4D endocavitary) can be employed to capture fetal images, also, US probes can work at different frequencies. Generally, the higher the frequency range, the more shallow the penetration. For fetal examinations, low-frequency convex probes operating in the 1–5 MHz range are deemed most suitable due to their deeper penetration capabilities, crucial for comprehensive fetal assessment42. This equipment and setup variability can significantly impact image quality, necessitating the development of DL algorithms that are adaptable across various resolutions and setups and robust enough to handle data from different vendor machines to ensure generalizability. To effectively mitigate these discrepancies, integrating data from multiple vendors and specifying clearly the imaging protocols adopted is crucial. Consequently, all fetal US datasets are acquired using US machines of at least two vendors. To account for this bias, the FETAL_PLANES_DB dataset was acquired using devices from four different vendor devices. However, a bias strictly inherent to probe types used can be identified—an endocavitary probe was used for cervical US screening, while a curved probe was used for all other plane acquisitions. This might have introduced a bias in distinguishing the maternal cervix plane from all other classes. To develop generalizable solutions, datasets should not only work properly with high-quality images and similar acquisition protocols but also with lower-resolution images from less advanced types of equipment. The MFUSPAC datasets approach to SPD task also takes into account diverse US devices from five different vendors and with different working frequency ranges, including lower resolution images, to better adapt to the nuanced complexities of real-world clinical environments. This inclusion demonstrates the potential of DL algorithms to enhance diagnostic capabilities under less favorable conditions. However, not all datasets provide comprehensive information on the imaging equipment used. This is the case of the HC18 dataset, which lacks details on the US probe. Such omission underscores the importance of transparency and thorough documentation in dataset creation to better understand and mitigate device variability bias.
Although currently unexplored in the context of fetal US analysis, specific biases may emerge and strictly depend on the interaction of automated algorithms with clinicians. The ability of ML systems to process and interpret large volumes of fetal US data may enable the detection of fetal anomalies with potentially greater precision than traditional methods while also expediting the process of fetal follow-ups. Despite these advancements, the increasing reliance on ML solutions necessitates a careful examination of their impact on clinical decision-making. The presence of human cognitive biases can lead clinicians to over-depend on algorithmic recommendations and automate bias and errors (i.e., automation bias), particularly in nuanced or ambiguous cases, calls for a balanced approach that values human expertise as much as technological innovation. Moreover, addressing the challenges posed by data biases and the opaque nature of some ML processes is crucial to ensure ethical and responsible use.
To navigate these challenges, a robust framework for the development and integration of ML systems in fetal diagnostics is today essential but still absent. This framework should emphasize ethical considerations, transparency, and the interpretability of ML processes, ensuring they support and augment rather than replace the critical judgment of medical professionals. Involving clinicians in the development and evaluation of these technologies is key to aligning ML’s capabilities with the real-world demands of prenatal care. By fostering a collaborative environment, we can leverage ML to enhance diagnostic processes while maintaining the high standards of care and ethical responsibility essential in healthcare.
Two forms of bias that have not been considered in the context of fetal US imaging are temporal bias and probe maneuver bias. Both biases stem from the interaction between the probe and clinicians, specifically reflecting the inability of existing datasets to capture aspects influenced by human expertise in US acquisition. Indeed, US imaging is not only about capturing a static moment; it is a dynamic process. Beyond the inherent biases related to fetal anatomy or clinical parameters, the accuracy of US is significantly influenced by the operator’s skill. As emphasized in ref. 3, obtaining an accurate assessment of biometry parameters and anatomical analysis hinges on the precise positioning and maneuvering of the US probe. This repositioning and adjustment is not instantaneous—it unfolds over time. As the operator searches for the optimal scan plane, he or she continuously adjusts the angle, depth, and orientation of the probe. An automated algorithm, to perform effectively in this context, should aim to closely replicate the decision-making process followed in clinical practice. However, the loss of information caused by frame selection may significantly impact its real-world applicability. If key intermediate frames containing relevant anatomical details or probe adjustments are omitted, the algorithm may struggle to generalize when deployed in a clinical setting, where image acquisition is inherently dynamic. Ensuring that the algorithm accounts for the sequential nature of US scanning is therefore crucial to improving its robustness and reliability in real-world applications. Using video-based datasets instead of static images could help mitigate these biases by preserving the temporal progression of probe adjustments and capturing the scanning maneuver as a continuous process rather than an isolated snapshot. This approach provides richer contextual information, enabling models to learn not only from the sampled frames but also from the sequence of movements that led to their acquisition. Consequently, when considering datasets derived from such scans, it is critical to account for this temporal progression, recognizing that it is not just about where the probe ends up, but also the journey it takes to get there. For this reason, the datasets analyzed are prone to probe maneuver bias, which may cause the missing of important information.
Discussion
Computer-aided diagnosis with fetal US imaging began in the mid-1990s. However, it was with the advent of DL in 2010 that various datasets emerged, aiming to provide a solid foundation for the robust evaluation of DL algorithms. These datasets have been instrumental in advancing automated solutions for tasks such as standard plane detection, fetal biometrics, and anomaly identification. This work outlined both the contributions and potential limitations of these datasets to the field of fetal US imaging analysis. We emphasized the importance of identifying and mitigating ethical biases stemming from inequitable, flawed, or inaccurate data collection practices, as such biases could negatively impact the accuracy, reliability, and fairness of clinical applications. By critically examining these datasets, we aimed to ensure that DL models remain equitable and generalizable.
Our research has focused on public datasets to circumvent the inherent biases associated with private datasets, which are accessible only to selected groups. The literature has shown that the implementation of fairness is of paramount importance to ensure that these systems truly benefit everyone, particularly minorities and those who have been historically marginalized and oppressed25. While public datasets may be more susceptible to security risks, such as data manipulation, an aspect beyond the scope of this work, they are essential for promoting equitable access, fostering inclusive technological progress, and ensuring that the benefits of AI are distributed fairly.
Moreover, our study proposes strategies to mitigate these biases going beyond the mere technical removal of biases in the datasets and consequently in AI systems, actions often criticized in the AI ethics debate as insufficient and, in some cases, impossible to achieve completely. Instead, we highlight that addressing bias requires a multilayered and critical approach, involving diverse stakeholders and combining both technical and non-technical practices of responsible AI management. This perspective aligns with the need to balance fairness, accessibility, and ethical considerations in advancing AI systems, particularly in fetal US imaging.
Throughout our study, we have posed three research questions, which, upon thorough analysis, we have now addressed. As regards Q1 + Q2, our analysis revealed that biases are indeed widespread within publicly available fetal US datasets, potentially and significantly impacting the robustness, accuracy, and fairness (i.e., trustworthiness) of DL models. The biases common to all the analyzed datasets, although different, mostly relate to a lack of diversity. This ranges from (i) geographic limitation due to data acquisition being limited to specific areas, primarily Europe, and often exacerbated by the lack of information on the nationalities of the mothers who underwent screening, (ii) the exclusion of less common anatomies and physiology, such as twin pregnancies and fetuses with congenital abnormalities, and (iii) few different US machines used, in terms of both vendors and device characteristics. This indicates a collection bias towards more readily available or conventional data during the phase of acquisition. When datasets are not sufficiently diverse, ML models trained on these data can develop what is called a privilege bias. This means that the models tend to perform better for the majority groups that are more consistently represented in the data, while they may fail or be less accurate for underrepresented minority groups. This issue not only arises in cases of less common fetal conditions but also when considering the equipment used, as more expensive machines might not be available in many parts of the world, introducing another layer of inequality and reinforcing existing social and economic asymmetries in fair access to and enjoyment of clinical care and services.
Label bias is another notable problem in the biomedical imaging domain43, stemming from reliance on a limited pool of experts for data annotation, and these datasets are no exception. The often subjective and culturally influenced experiences, perspectives, and cognitive biases of these experts can heavily influence data annotations, potentially distorting the representation of reality in the resulting models. This is particularly concerning in fields where accuracy and methodological impartiality are paramount.
In addition to these challenges, the FETAL_PLANES_DB dataset also has a distinct characteristic of significant class imbalance, with certain conditions or features being over-represented, inadvertently biasing clinicians’ focus toward the more accurately detected parts, potentially overshadowing concerns in other areas.
Another significant limitation is the focus of the datasets. Each of them is built to resolve a specific aspect of fetal US imaging, neglecting comprehensive factors that are crucial for a well-rounded analysis. For example, the intricate details of probe maneuvering dynamics, the full spectrum of fetal biometrics or standard planes, and the inclusion of all the trimesters of pregnancy might be overlooked. This limitation might lead to models that lack a nuanced understanding of fetal health, potentially resulting in incomplete or biased assessments. The reliance on biased ML models could also lead to technological determinism, where an algorithm begins to shape clinical practices and decisions, possibly at the expense of patient-centered care. This scenario underscores the importance of maintaining a balance between technological advancements and the human aspects of prenatal care.
With respect to Q3, addressing the biases identified in the analyzed fetal US datasets requires a multi-faceted strategy, incorporating both short-term and long-term solutions. The goal is to enhance the diversity, accuracy, and comprehensiveness of these datasets, ultimately improving the accuracy, reliability, and fairness of the resulting models. Addressing biases resulting from a lack of diversity requires a methodical and structured approach. Expanding the data collection process to include a broader range of participants, settings, and conditions is essential for enhancing the inclusivity and generalizability of research findings. This long-term solution involves collecting data from diverse geographical locations to capture varied nationalities and ethnic backgrounds, as well as incorporating a wide array of pregnancy conditions to ensure comprehensive population coverage. Such an approach is fundamental to reflecting the multifaceted nature of human development.
Equally critical is addressing the variability introduced during the annotation process. Incorporating diverse annotations from a wider variety of medical experts can improve dataset completeness by integrating multiple perspectives and expertise44. Inter-observer variability, a well-documented phenomenon in medical imaging, highlights the fact that even experienced sonographers may employ slightly different diagnostic approaches and techniques. This variability can lead to inconsistencies in annotations and impact model performance, particularly in cases that fall outside the experience of the selected annotators. Addressing this challenge is crucial for building more robust and generalizable models. Moreover, in the process of data collection, careful attention must be paid to potential biases arising from manual frame selection. Variability in expertise, subjective judgment, and redundancy in adjacent frames can all contribute to inconsistencies that impact the quality of the training data and, consequently, the performance of ML models. Techniques like image or video processing could standardize this process by assessing frame consistency through structural and textural similarities, employing metrics such as peak signal-to-noise ratio or mutual information to ensure representative and diverse frame selection, thereby minimizing redundancy and bias. The importance of constructing datasets with a pronounced focus on diversity is underscored in ref. 45, which highlights the creation and application of diverse datasets in medical imaging. These principles align with the guidelines of open science and emphasize reducing biases to ensure equitable and inclusive research46. In addition to long-term strategies, a critical short-term solution involves using semi-supervised learning techniques and leveraging both labeled and unlabeled data to overcome data scarcity47. Moreover, initiatives that engage the research community to collaboratively refine existing datasets, such as those described in ref. 48, illustrate how public datasets, such as HC18 and Fetal_Planes_DB, can be enhanced by adding detailed annotations and standardizing protocols. Inspired by this approach, researchers can work together to improve the analyzed datasets by enriching metadata, addressing annotation gaps, and aligning them with current clinical guidelines. Such collaborative efforts can ensure that these datasets remain valuable resources for advancing medical imaging research while mitigating potential biases.
Another short-term strategy to address biases could be the implementation of federated learning (FL) approaches. In the context of fetal US imaging, FL provides a highly effective framework for privacy-preserving, decentralized model training, positioning it as one of the most reliable short-term solutions. In FL, raw data remains securely stored at individual sites, ensuring privacy and compliance with data protection regulations. Rather than sharing data, only model updates (e.g., weights) are transmitted to a central server during training, where they are aggregated to build a global model49. This approach not only safeguards patient privacy but also facilitates the inclusion of diverse datasets from multiple centers, encompassing a wide range of demographics and conditions that might otherwise be underrepresented. Additionally, FL contributes to standardizing protocols, promoting consistency and efficiency in data handling and analysis across institutions49. However, FL alone does not inherently eliminate cohort bias arising from geographic limitations, as model updates are still influenced by the data distributions at participating institutions. If certain centers contribute disproportionately, the global model may still exhibit biases reflecting the dominant cohorts. To mitigate this, methods such as weighted aggregation strategies or bias-aware optimization techniques can be incorporated to ensure that underrepresented populations receive appropriate model weighting50. Furthermore, few-shot learning (FSL) can help address biases related to data scarcity in underrepresented populations by enabling models to generalize from limited examples. By leveraging meta-learning approaches or prototype-based classifiers, FSL allows the global model to learn transferable representations that adapt more effectively to new cohorts with minimal labeled data. This is particularly valuable in fetal US imaging, where rare conditions or specific demographic groups may have significantly fewer samples available for training51. A way to incorporate new information as it becomes available is to use continuous learning instead, which keeps the models up-to-date. This means that as new research findings come out or as clinical guidelines evolve, the models can adapt. This ongoing update process helps ensure that the models remain accurate and relevant, reflecting the latest understanding and practices in fetal US imaging. However, working with datasets coming from various centers with different vending machines can introduce variations in data distribution, a common challenge in medical imaging research, including fetal US. These variations, if not addressed, can compromise the performance and reliability of diagnostic models. Domain adaptation techniques, therefore, become invaluable in this context. For instance, differing US system setups, such as brightness or intensity adjustments in B-mode imaging, all of which can produce dataset distribution differences or out-of-distribution effects that impact model learning and performance. These can be mitigated through domain adaptation techniques, such as histogram equalization, CLAHE, GANs, or autoencoders, which enable models to recognize and adjust to differences between datasets collected using diverse equipment or from different sources despite their variability52. This adaptability is particularly beneficial, allowing models to maintain high performance across varied datasets without the need for extensive retraining. Such capability is essential in the medical field, where data might be sourced from numerous hospitals or clinics, each equipped with different machinery and adhering to unique protocols.
Generative AI provides another powerful short-term solution to tackle biases, particularly the lack of diversity in fetal US datasets. By generating synthetic pathological cases, which are often difficult to obtain, it can enhance the variety and depth of training data for developing robust diagnostic models. The scarcity of certain pathological cases in fetal US datasets can lead to biases in diagnostic models, as they may not learn to recognize less common conditions effectively. Generative AI can mitigate this issue by creating realistic, synthetic images of rare fetal conditions, thereby enriching the dataset with diverse pathological examples, faced with the informativeness bias problem. Although these pathologies are inherently more challenging to acquire, recent advancements in generative AI53, including applications in fetal imaging54, have demonstrated the ability to generate clinically significant samples55. While their application to rare conditions such as TTTS remains limited, they offer a promising approach to overcoming data scarcity. By expanding the available dataset, these methods can enhance the model’s diagnostic capabilities and contribute to a more balanced and robust representation of various fetal conditions56. Additionally, generative AI can employ image-to-image translation techniques to enhance the quality of images acquired in low-resource settings57,58. In scenarios where the available US equipment might not produce high-quality images, these advanced AI techniques can transform lower-quality images into higher-quality equivalents, mimicking those captured by more advanced equipment. This application is particularly valuable in democratizing access to high-quality diagnostic imaging, enabling healthcare providers in resource-constrained environments to benefit from advanced diagnostic models. However, it is essential to recognize that training on synthetic data can also amplify biases through what is known as model collapse and disparity amplification. This occurs when synthetic outputs from models are used as new training data, leading to the entrenchment and amplification of existing biases over successive generations of models. Research has shown that these model-induced distribution shifts can encode and perpetuate biases, resulting in degraded performance and fairness59. To mitigate these issues, one promising approach is algorithmic reparation (AR), which aims to address historical discrimination and improve fairness by intentionally curating training batches to represent marginalized groups better. By focusing on creating a more equitable distribution of data and incorporating intersectional interventions at each stage of the model generation process, AR can help maintain balanced representation and performance across different demographics.
However, addressing biases requires more than just technical solutions—it demands integrating ethical considerations throughout the entire AI development process. This principle, known as ”ethics by design,” involves embedding ethical guidelines into every phase of the design process, beginning with data collection. By thoughtfully designing data collection methodologies, developers can better understand the target population and prospective users, thereby avoiding population target bias. Population target bias occurs when there is an asymmetry between the collected data and the actual demographics or characteristics of the prospective users. To mitigate this, it is essential to collect data that accurately represents the diversity of the intended user base, thus preventing the reproduction or exacerbation of existing socio-economic inequalities and power imbalances. Ethical data collection also necessitates training biomedical engineers, clinicians, and other involved personnel to recognize and mitigate intrinsic cognitive biases while understanding the ethical implications of their data collection practices. This involves making informed decisions about the sources and scope of data collection, for instance, whether to integrate temporal information and implement strategies to address potential biases in the data, such as cohort imbalances, missing data patterns, and underrepresentation of minority groups. In addition to designing with ethics in mind, continuous ethical audits by AI ethics experts are necessary to ensure ongoing oversight and bias control. An ethical audit involves regular reviews and assessments of AI systems to identify and address emerging biases or ethical concerns throughout the life cycle of the AI application. This continuous monitoring helps to maintain ethical standards and adapt to new ethical challenges as they arise, ensuring that the AI system remains fair and unbiased in practice.
Another way to address biases could be the incorporation of generalist medical AI into fetal health assessments. By leveraging a broader spectrum of data beyond the conventional selective focus of many datasets, generalist AI can help mitigate potential biases while enhancing the robustness and inclusivity of fetal US technology60. Generalist medical AI’s ability to learn from diverse, large-scale datasets allows for a more comprehensive understanding of fetal health, potentially leading to more accurate and holistic assessments. This shift could help in developing models that are not just narrowly accurate in predefined tasks but are adaptable and insightful across a wider range of fetal health conditions, contributing to a more inclusive and equitable healthcare landscape. However, as highlighted in ref. 61, there are inherent risks with this generalist approach, particularly concerning bias and fairness. These models, trained on vast and varied datasets, may inadvertently learn and propagate biases present in the data as well. To mitigate these risks, it is crucial to implement robust fairness quantification and bias mitigation techniques from the outset and continuously monitor and adjust models as they evolve. By addressing these potential pitfalls proactively, we can harness the power of generalist medical AI while ensuring it contributes to equitable and just healthcare outcomes.
However, to ensure equitable outcomes, it is essential to employ robust fairness quantification methods in the development of AI models used in fetal health assessments. Techniques such as—(i) statistical parity analysis, which ensures that AI outcomes are independent of protected attributes like race or sex. It is crucial to recognize that race and sex can be correlated with different pathologies, and this correlation must be carefully considered based on the specific application. For example, certain genetic conditions may be more prevalent in specific racial groups, as well as pregnancy-related complications can vary by fetal sex. Therefore, while statistical parity is important, it should be balanced with medical accuracy to provide effective diagnoses and treatments. (ii) Individual fairness methods which aim to provide similar predictions for individuals who are similar in relevant aspects. (iii) Group fairness measures which ensure that different groups have equal positive rates, enhancing the overall fairness of the system. Incorporating these techniques can help in developing AI models that not only excel in diverse tasks but also uphold and advance the key AI ethics principle of fairness in healthcare AI for prenatal care. Such advancements could significantly enhance prenatal care by making it more personalized and effective, ensuring it addresses the needs of all population segments inclusively and fairly. This inclusive approach could facilitate the development of prenatal diagnostics and treatment in a manner that is unprecedented in the history of medicine, making healthcare more just and accessible. This is not just a scientific advancement, but a societal gain, promoting a healthier future for all communities.
While we have proposed a framework for evaluating bias in fetal US datasets, along with targeted strategies to mitigate these biases, it is equally important to recognize the limitations of this study. These limitations underscore areas requiring further exploration to support the development of more robust and equitable ML models in the future. Our study is exclusively focused on publicly available dataset analysis, without incorporating a comparative analysis on private datasets. Including private datasets could have provided valuable insights into whether significant disparities exist between the quality and structure of publicly accessible datasets and those retained in private settings. While public datasets play a crucial role in fostering collaboration and transparency, they may lack the same level of curation, diversity, or completeness typically associated with private datasets. Such disparities could have further implications for fairness, potentially highlighting gaps in the representativeness and reliability of public datasets. However, analyzing private datasets is not feasible within the scope of this study due to restricted access. Nevertheless, our analysis of public datasets provides a basis for independent evaluations of bias presence in private datasets. Another limitation of our work is that, while it identifies and categorizes biases, it does not quantify their extent or compare their levels across datasets. Developing a standardized quantitative metric or ranking system could enable a more systematic evaluation, allowing for more informed and objective decisions regarding dataset selection and utilization. Bias assessment at the dataset level remains an open research challenge, with only a few studies addressing it. For instance62, examines biases in dermatology imaging datasets through an ethical lens, focusing on aspects such as skin tone and demographic representation. Similarly, in our domain, future research could focus on developing inclusion metrics to assess feature representation, ensuring both the presence of diverse subgroups (e.g., demographic diversity and anatomical region coverage) and their equitable distribution across the dataset (diversity metrics). This approach would help prevent the marginalization of underrepresented groups while avoiding an imbalance that skews model generalizability.
Additionally, while the datasets used in this study do not include metadata that could reveal patient identities, future considerations involving the acquisition of new datasets must account for the potential impact of metadata, such as patient information stored in DICOM files or incorporated in the US frames. Metadata could introduce other biases if utilized as additional features, potentially affecting model generalization and fairness. Exploring these challenges lies outside the scope of this study but represents an important direction for future research. While beyond the scope of our work, it is worth noting that many other biases can arise during the training and evaluation of DL models. For example, the use of pre-training on natural image datasets like ImageNet, which may exhibit poor generalization to the medical field due to the significant differences between natural and medical images, can impact model performance and generalization. These are critical challenges that merit future exploration in relation to the development of fair DL models.
To conclude, this study revealed various biases in publicly available fetal US imaging datasets, which are crucial for training DL algorithms in prenatal diagnostics and follow-up. These biases include an under-representation of demographic groups, a limited range of clinical conditions, variability in US imaging, and a lack of alignment with current clinical practices. These issues can distort DL model performance and lead to healthcare inequities.
To address these challenges, we proposed several strategies that aim to enhance representativeness. These include—improved data collection practices, the use of generative AI, the implementation of generalist medical AI, and federated and continuous learning approaches. Furthermore, we recommend an ethics-by-design approach, embedding ethical principles from the start, and continuous ethical audits by AI ethics experts to maintain ongoing oversight and bias control. Robust fairness quantification methods, such as statistical parity analysis, individual fairness methods, and group fairness measures, are essential to ensure that AI outcomes are equitable and independent of protected attributes, such as race and sex. By implementing these strategies, we can enhance the diversity, accuracy, and fairness of fetal US datasets and DL models, ensuring more equitable health outcomes for all.
Methods
Dataset bias in imaging
The significance of datasets in ML and DL research cannot be overstated, as datasets are often seen as the primary constraint on algorithm advancement and scientific progress. Key benchmarks, such as ImageNet63, for visual object recognition. Although ML systems have recently surpassed human performance on these benchmarks, recent studies have revealed limitations in these datasets as measures of human-like reasoning and raised societal concerns regarding dataset practices64. Extensive research highlights the importance of well-structured data collection and the implications of bias in datasets. For instance, the work in ref. 64 identifies key concerns about dataset practices, including collection, construction, labeling, and sharing processes. These practices can lead to models based on spurious correlations and faulty heuristics, undermining the reliability and validity of ML advancements. Similarly, the work in ref. 65 discusses recent technical advances aimed at making the data-for-AI pipeline more scalable and rigorous, addressing many of the aforementioned concerns. In ref. 66, social bias, measurement bias, representation bias, and label bias are identified, contextualizing them within a real-world case study and additionally illustrating effective mitigation strategies.
In healthcare datasets, the presence of biases can result in the generation of models that perpetuate or exacerbate existing inequalities in healthcare delivery and outcomes26. Different studies address the ethical concerns and risks posed by data bias in clinical settings, from ECG/EEG time series20, sex imbalance in X-ray imaging analysis67, or on a broader level26,68,69,70. Ensuring unbiased datasets requires the inclusion of underrepresented patient subgroups and high-quality annotations to prevent algorithmic bias and ensure reliable model performance71. Additionally, ethical guidelines for algorithm development, transparency in training and validation processes, and thorough documentation of hyperparameters are necessary to mitigate bias and enhance reproducibility. In ref. 69, the issue of non-uniform representation of different populations in medical imaging datasets is emphasized. Non-uniform representation in medical imaging datasets is a significant issue, manifesting as demographic disparities, unequal representation of pathological data, site-specific differences, and technical variations. A recent study highlights that these data biases often arise from established tools and data generation processes, with specific concerns including representation bias, and aggregation bias, which occurs when incorrect assumptions about individuals or subgroups are made based on observations of the entire population72. Additionally, population bias occurs when the demographics and statistical profiles of the dataset population differ significantly from those of the intended patient population.
Although the concept of fairness and data biases has been widely investigated, also in healthcare, no studies have specifically examined these issues focusing on the context of fetal US imaging. This gap is critical, as these biases can significantly affect the evaluation of both fetal and maternal well-being during pregnancy. Furthermore, such biases are often context-sensitive and need to be contextualized to be detected and mitigated adequately. To address this gap, we draw on insights from recent studies such as refs. 25,26,27, which are closely related to our context, as they provide a high-level map of benchmark data biases in the context of healthcare AI and specifically for medical imaging.
Fetal ultrasound imaging
To establish a solid foundation for bias evaluation, an in-depth investigation was conducted to analyze the most active research areas of fetal US image analysis using DL. This section provides a definition of fetal US imaging and an overview of the major tasks for which DL algorithms are developed. This allows a deeper understanding of the key aspects typically evaluated in fetal US datasets, enabling an accurate contextualization of bias evaluation.
US imaging has become a widely adopted modality for the diagnosis, screening, and treatment of various medical conditions. This popularity can be attributed to its inherent advantages, including portability, cost-effectiveness, and non-invasiveness73,74. Over the years, US imaging has emerged as the preferred method for prenatal examinations, routinely employed to evaluate fetal growth and development and to monitor pregnancies, particularly when clinical concerns arise75.
A fetal US examination is usually conducted by a proficient sonographer or a qualified healthcare practitioner. This procedure entails the utilization of a US probe to scan the mother’s abdomen to identify key anatomical features of the fetus for assessment. The specific evaluation taken by the clinician can vary depending on the trimester of pregnancy. During early pregnancy, it is crucial to confirm viability, accurately determine gestational age, establish the number of fetuses, and, in the case of a multiple pregnancy, assess chorionicity and amnionicity76. In contrast, during the second and third trimesters of pregnancy, the procedure is employed for fetal measurements to promptly detect any growth abnormalities that may arise later in pregnancy and to identify congenital malformations77. Although the evaluations conducted throughout the trimesters of pregnancy may differ, the process of assessing fetal well-being consists of three common steps, as illustrated in Fig. 2: SPD, AAS, and BPE.
Regarding SPD, the ISUOG guidelines78 highlight that using standardized acquisition planes improves the reproducibility of both fetal biometry measurements and overall fetal assessment.
In the first trimester of gestation, when the fetus is relatively small, the evaluation includes different standard fetal planes. The mid-sagittal plane is used for assessing nuchal translucency, crown-rump length (CRL), and overall fetal morphology. The transverse plane of the head is utilized to observe brain structures such as the forebrain, midbrain, and hindbrain. The coronal plane of the trunk and the abdomen is important for evaluating internal organs such as the liver and stomach. The sagittal plane of the abdomen is useful for visualizing the stomach, the heart, and the major vessels such as the aorta and the inferior vena cava. Finally, the transverse plane of the heart is employed to assess the position and structure of the heart, including the four chambers and heart valves. These planes are essential for a comprehensive US evaluation of the fetus in the first trimester, helping to identify early developmental anomalies79.
In mid-trimester fetal US scans, a comprehensive assessment of the central nervous system (CNS) is of paramount importance due to the high incidence of CNS malformations. The diagnostic protocol typically involves evaluating the fetal brain’s integrity using at least two axial planes—TV and TC—and often includes a third, the TT plane, for additional biometric data. A particular focus is given to key anatomical features, including the lateral ventricles, cerebellum, cisterna magna, and cavum septi pellucidi, as well as to the head’s morphology and the brain’s textural characteristics within these planes. A longitudinal spinal section is also essential, as it can reveal specific spinal abnormalities, providing a comprehensive view of the CNS.
In addition to CNS evaluation, the acquisition of the fetal abdominal standard plane (FASP) is critically important for examining the development and positioning of essential organs such as the stomach, liver, kidneys, and intestines. In a clinical setting, the successful identification of FASP requires the radiologist’s ability to simultaneously visualize three key anatomical structures—stomach bubble, umbilical vein, and spine, while maneuvering the US probe across the maternal abdomen. To effectively screen for congenital heart disease (CHD)—a leading cause of infant mortality—during midgestation, a careful cardiac evaluation is also important. This comprehensive assessment should incorporate both the four-chamber (4CH) and outflow tract planes. Within the context of the 4CH view, a focused examination is required to meet specific anatomical criteria: the two atria should be discernible at the top, with the two ventricles situated at the bottom, separated by a septum. Additionally, cardiac valves ought to be clearly visible, and the apex of the heart should point to the left at an angle of 45 ± 20°. When it comes to examining the outflow tract views, it is critical to include observations of both the left and right ventricular outflow tracts (LVOT and RVOT) as essential components of fetal cardiac screening. Furthermore, as part of a seamless progression beginning with the RVOT, there are additional cross-sectional perspectives that reveal various facets of the great vessels and their adjacent structures. These perspectives encompass the three-vessel view and the three vessels along with the trachea view. Fetal evaluation also requires the acquisition of the fetal facial standard plane, which includes axial, coronal, and sagittal planes.
AAS using fetal US, instead, serves multiple critical functions during the first and mid-trimesters of pregnancy, each corresponding to different clinical needs and developmental stages. During the first trimester, the primary objective is often the early identification of gross anatomical anomalies78, the assessment of nuchal translucency, and the establishment of basic fetal viability. This early-stage analysis facilitates timely genetic testing, offers reassurance to mothers considered at risk, and allows for the option of elective pregnancy termination if severe abnormalities are detected. Transitioning to the mid-trimester, the anatomical examination becomes more detailed, covering structures such as the brain, spine, heart, kidneys, and limbs. The focus of the examination shifts toward the identification of more subtle morphological abnormalities, the evaluation of fetal growth patterns, and the preparation for any necessary interventions, either prenatally or immediately after birth. Additionally, mid-trimester anatomical analysis can provide insights into placental health and positioning, thereby aiding in the prediction and prevention of complications such as placental abruption or preterm birth. A detailed description of this anatomical assessment is provided in Table 3.
Another critical aspect is the evaluation of fetal size and gestational age, in conjunction with the identification of deviations in fetal growth, whose primary medical investigation employed for this purpose is the BPE.
Before the 14th week of pregnancy, gestational age and fetal size are determined by measuring the CRL, as illustrated in Fig. 2. This measurement calculates the distance from the top of the fetus’s head to the bottom of its torso. Once the CRL exceeds a predetermined threshold (typically at 14 weeks), standard measurements encompass HC, biparietal diameter, occipito-frontal diameter, trans-cerebellar diameter, lateral ventricles, AC, and femur diaphysis length. These fetal biometric measurements are used to assess the trajectory of fetal growth and to ensure the fetus’s normal development when assessed at different stages of pregnancy (trimesters). Furthermore, the cardio-thoracic ratio and cardiac axis biometrics are employed for the diagnosis of CHD.
Search strategy
While exploring relevant research on DL for fetal US, we embarked on a quest to uncover datasets pertinent to this domain. Our exploration spanned various databases and online repositories, including Kaggle, GitHub, Mendeley Data, MICCAI Challenges, and Zenodo.
To streamline our search and ensure relevance:
-
Keywords played a central role and our primary guiding terms encompassed “fetal US”, “US image datasets”, and “fetal anomalies”, further expanded upon with their various related terms to cast a wider net.
-
We applied explicit inclusion criteria to refine our selection: (i) Datasets had to be publicly available and specific to fetal US images, deliberately excluding general US datasets and any datasets from pre- or intra-partum stages, or those using fetal phantoms. Public datasets were prioritized as they serve as benchmarks for the research community, facilitating results replication and comparison across studies, while non-public datasets were excluded because their inaccessibility undermines fairness in evaluations. (ii) With an inclination towards DL applications, datasets with a large number of samples were favored. (iii) Only datasets in standard formats (e.g., PNG, JPEG, BMP) were considered; datasets containing DICOM files or patient-identifiable information in file names or overlaid on US frames were excluded to ensure anonymization.
Data availability
The authors used only publicly available datasets: FETAL_PLANES_DB (https://zenodo.org/record/3904280), Maternal-fetal US planes in African countries (https://zenodo.org/records/7540448), Fetal Abdominal Structures Segmentation Dataset (https://data.mendeley.com/datasets/4gcpm9dsc3/1) and HC18 (https://hc18.grand-challenge.org/).
References
Liu, S. et al. Deep learning in medical ultrasound analysis: a review. Engineering 5, 261–275 (2019).
Fernandez-Quilez, A. Deep learning in radiology: ethics of data and on the value of algorithm transparency, interpretability and explainability. AI Ethics 3, 257–265 (2023).
Fiorentino, M. C., Villani, F. P., Di Cosmo, M., Frontoni, E. & Moccia, S. A review on deep-learning algorithms for fetal ultrasound-image analysis. Med. Image Anal. 83,102629 (2023).
Alzubaidi, M. et al. Toward deep observation: a systematic survey on artificial intelligence techniques to monitor fetus via ultrasound images. Iscience 25, 104713 (2022).
Torres, H. R. et al. A review of image processing methods for fetal head and brain analysis in ultrasound images. Comput. Methods Prog. Biomed. 215, 106629 (2022).
Ramirez Zegarra, R. & Ghi, T. Use of artificial intelligence and deep learning in fetal ultrasound imaging. Ultrasound Obstet. Gynecol. 62, 185–194 (2023).
Sendra-Balcells, C. et al. Generalisability of fetal ultrasound deep learning models to low-resource imaging settings in five African countries. Sci. Rep. 13, 2728 (2023).
Fabbrizzi, S., Papadopoulos, S., Ntoutsi, E. & Kompatsiaris, I. A survey on bias in visual datasets. Comput. Vis. Image Underst. 223, 103552 (2022).
Rädsch, T. et al. Labelling instructions matter in biomedical image analysis. Nat. Mach. Intell. 5, 273–283 (2023).
Chlap, P. et al. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 65, 545–563 (2021).
Chaudhari, A. S. et al. Prospective deployment of deep learning in MRI: a framework for important considerations, challenges, and recommendations for best practices. J. Magn. Reson. Imaging 54, 357–371 (2021).
Abràmoff, M. D. et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Investig. Ophthalmol. Vis. Sci. 57, 5200–5206 (2016).
Liu, G. et al. Cx22: a new publicly available dataset for deep learning-based segmentation of cervical cytology images. Comput. Biol. Med. 150, 106194 (2022).
Maier-Hein, L. et al. Bias: transparent reporting of biomedical image analysis challenges. Med. Image Anal. 66, 101796 (2020).
Maier-Hein, L. et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 9, 5217 (2018).
Eisenmann, M. et al. Why is the winner the best? In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 19955–19966 (IEEE, 2023).
Drukker, L., Droste, R., Chatelain, P., Noble, J. A. & Papageorghiou, A. T. Expected-value bias in routine third-trimester growth scans. Ultrasound Obstet. Gynecol. 55, 375–382 (2020).
Drukker, L. et al. Transforming obstetric ultrasound into data science using eye tracking, voice recording, transducer motion and ultrasound video. Sci. Rep. 11, 14109 (2021).
Drukker, L. et al. How often do we identify fetal abnormalities during routine third-trimester ultrasound? a systematic review and meta-analysis. BJOG: Int. J. Obstet. Gynaecol. 128, 259–269 (2021).
Dakshit, S., Dakshit, S., Khargonkar, N. & Prabhakaran, B. Bias analysis in healthcare time series (BAHT) decision support systems from meta data. J. Healthc. Inform. Res. 7, 225–253 (2023).
Organization, W. H. et al. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance: Executive Summary (World Health Organization, 2021).
Ganapathi, S. et al. Tackling bias in AI health datasets through the standing together initiative. Nat. Med. 28, 2232–2233 (2022).
Migliorelli, L. et al. Accountable deep-learning-based vision systems for preterm infant monitoring. Computer 56, 84–93 (2023).
Stanley, E. A., Wilms, M. & Forkert, N. D. A flexible framework for simulating and evaluating biases in deep learning-based medical image analysis. In Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention 489–499 (Springer, 2023).
Chen, R. J. et al. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat. Biomed. Eng. 7, 719–742 (2023).
Giovanola, B. & Tiribelli, S. Beyond bias and discrimination: redefining the AI ethics principle of fairness in healthcare machine-learning algorithms. AI Soc. 38, 549–563 (2023).
Abràmoff, M. D. et al. Considerations for addressing bias in artificial intelligence for health equity. NPJ Digit. Med. 6, 170 (2023).
Ogasawara, K. K. Variation in fetal ultrasound biometry based on differences in fetal ethnicity. Am. J. Obstet. Gynecol. 200, 676–e1 (2009).
Sletner, L. et al. Ethnic differences in fetal size and growth in a multi-ethnic population. Early Hum. Dev. 91, 547–554 (2015).
Jacquemyn, Y., Sys, S. U. & Verdonk, P. Fetal biometry in different ethnic groups. Early Hum. Dev. 57, 1–13 (2000).
Hanley, G. E. & Janssen, P. A. Ethnicity-specific birthweight distributions improve identification of term newborns at risk for short-term morbidity. Am. J. Obstet. Gynecol. 209, 428.e1–428.e6 (2013).
Kiserud, T. et al. The World Health Organization fetal growth charts: a multinational longitudinal study of ultrasound biometric measurements and estimated fetal weight. PLoS Med. 14, e1002220 (2017).
Mujugira, A., Osoti, A., Deya, R., Hawes, S. E. & Phipps, A. I. Fetal head circumference, operative delivery, and fetal outcomes: a multi-ethnic population-based cohort study. BMC Pregnancy Childbirth 13, 1–6 (2013).
Casella, A. et al. A shape-constraint adversarial framework with instance-normalized spatio-temporal features for inter-fetal membrane segmentation. Med. Image Anal. 70, 102008 (2021).
Stagnati, V. et al. Early prediction of twin-to-twin transfusion syndrome: systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 49, 573–582 (2017).
Kontopoulos, E., Chmait, R. H. & Quintero, R. A. Twin-to-twin transfusion syndrome: definition, staging, and ultrasound assessment. Twin Res. Hum. Genet. 19, 175–183 (2016).
Gaglioti, P., Oberto, M. & Todros, T. The significance of fetal ventriculomegaly: etiology, short- and long-term outcomes. Prenat. Diagn. 29, 381–388 (2009).
Cohen Jr, M. M. Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res. Part A 76, 658–673 (2006).
Prefumo, F. & Izzi, C. Fetal abdominal wall defects. Best Pract. Res. Clin. Obstet. Gynaecol. 28, 391–402 (2014).
Nicolaides, K., Snijders, R., Cheng, H. & Gosden, C. Fetal gastro-intestinal and abdominal wall defects: associated malformations and chromosomal abnormalities. Fetal Diagn. Ther. 7, 102–115 (1992).
Illescas, T. et al. Prenatal diagnosis of fetal skeletal dysplasias in a tertiary hospital in Spain. Eur. J. Obstet. Gynecol. Reprod. Biol. 250, 209–215 (2020).
Herbst, M. K., Tafti, D. & Shanahan, M. M. Obstetric ultrasound (StatPearls Publishing, 2017).
Prevedello, L. M. et al. Challenges related to artificial intelligence research in medical imaging and the importance of image analysis competitions. Radiology 1, e180031 (2019).
Yan, Y., Rosales, R., Fung, G., Subramanian, R. & Dy, J. Learning from multiple annotators with varying expertise. Mach. Learn. 95, 291–327 (2014).
Arora, A. et al. The value of standards for health datasets in artificial intelligence-based applications. Nat. Med. 29, 2929–2938 (2023).
Norori, N., Hu, Q., Aellen, F. M., Faraci, F. D. & Tzovara, A. Addressing bias in big data and AI for health care: a call for open science. Patterns 2, 100347 (2021).
Migliorelli, G. et al. On the use of contrastive learning for standard-plane classification in fetal ultrasound imaging. Comput. Biol. Med. 174, 108430 (2024).
Alzubaidi, M. et al. Large-scale annotation dataset for fetal head biometry in ultrasound images. Data Br. 51, 109708 (2023).
Chen, Y., Huang, W. & Ye, M. Fair federated learning under domain skew with local consistency and domain diversity. In Proc. of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12077–12086 (2024).
Li, Z., Lin, T., Shang, X. & Wu, C. Revisiting weighted aggregation in federated learning with neural networks. In International Conference on Machine Learning, 19767–19788 (PMLR, 2023).
Pachetti, E. & Colantonio, S. A systematic review of few-shot learning in medical imaging. Artif. Intell. Med. 156, 102949 (2024).
Huang, L. et al. Standardization of ultrasound images across various centers: M2o-diffgan bridging the gaps among unpaired multi-domain ultrasound images. Med. Image Anal. 95, 103187 (2024).
Ruiz, N. et al. Hyperdreambooth: hypernetworks for fast personalization of text-to-image models. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 6527–6536 (IEEE, 2024).
Lasala, A., Fiorentino, M. C., Bandini, A. & Moccia, S. FetalBrainAwareNet: bridging GANs with anatomical insight for fetal ultrasound brain plane synthesis. Comput. Med. Imaging Graph 116,102405 (2024).
Wang, J. et al. Self-improving generative foundation model for synthetic medical image generation and clinical applications. Nat. Med. 31, 609–617 (2025).
Gowal, S. et al. Improving robustness using generated data. Adv. Neural Inf. Process Syst. 34, 4218–4233 (2021).
Zhou, Z., Wang, Y., Guo, Y., Qi, Y. & Yu, J. Image quality improvement of hand-held ultrasound devices with a two-stage generative adversarial network. IEEE Trans. Biomed. Eng. 67, 298–311 (2019).
Liu, J. et al. Speckle noise reduction for medical ultrasound images based on cycle-consistent generative adversarial network. Biomed. Signal Process Control 86, 105150 (2023).
Wyllie, S., Shumailov, I. & Papernot, N. Fairness feedback loops: training on synthetic data amplifies bias. In Proc. of the 2024 ACM Conference on Fairness, Accountability, and Transparency, pp. 2113–2147 (2024).
Moor, M. et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023).
Bommasani, R. et al. On the opportunities and risks of foundation models. arXiv preprint arXiv:2108.07258 (2021).
Mittal, S. et al. On responsible machine learning datasets emphasizing fairness, privacy and regulatory norms with examples in biometrics and healthcare. Nat. Mach. Intellig. 6, 936–949 (2024).
Russakovsky, O. et al. Imagenet large-scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015).
Paullada, A., Raji, I. D., Bender, E. M., Denton, E. & Hanna, A. Data and its (dis) contents: a survey of dataset development and use in machine learning research. Patterns 2, 100336 (2021).
Liang, W. et al. Advances, challenges and opportunities in creating data for trustworthy AI. Nat. Mach. Intell. 4, 669–677 (2022).
Van Giffen, B., Herhausen, D. & Fahse, T. Overcoming the pitfalls and perils of algorithms: a classification of machine learning biases and mitigation methods. J. Bus. Res. 144, 93–106 (2022).
Larrazabal, A. J., Nieto, N., Peterson, V., Milone, D. H. & Ferrante, E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. USA 117, 12592–12594 (2020).
Pagano, T. P. et al. Bias and unfairness in machine learning models: a systematic review on datasets, tools, fairness metrics, and identification and mitigation methods. Big Data Cogn. Comput. 7, 15 (2023).
Ricci Lara, M. A., Echeveste, R. & Ferrante, E. Addressing fairness in artificial intelligence for medical imaging. Nat. Commun. 13, 4581 (2022).
Vrudhula, A., Kwan, A. C., Ouyang, D. & Cheng, S. Machine learning and bias in medical imaging: opportunities and challenges. Circulation 17, e015495 (2024).
Jha, D. et al. Ensuring trustworthy medical artificial intelligence through ethical and philosophical principles. arXiv preprint arXiv:2304.11530 (2023).
Yang, Y. et al. A survey of recent methods for addressing AI fairness and bias in biomedicine. J. Biomed. Inform. 154, 104646 (2024).
Zaffino, P., Moccia, S., De Momi, E. & Spadea, M. F. A review on advances in intra-operative imaging for surgery and therapy: imagining the operating room of the future. Ann. Biomed. Eng. 48, 2171–2191 (2020).
Fiorentino, M. C., Moccia, S., Cipolletta, E., Filippucci, E. & Frontoni, E. A learning approach for informative-frame selection in US rheumatology images. In Proc. New Trends in Image Analysis and Processing–ICIAP 2019: ICIAP International Workshops, BioFor, PatReCH, e-BADLE, DeepRetail, and Industrial Session, Trento, Italy, September 9–10, 2019, Revised Selected Papers 20 228–236 (Springer, 2019).
Ibrahimi, J. & Mumtaz, Z. Ultrasound imaging and the culture of pregnancy management in low- and middle-income countries: a systematic review. Int. J. Gynecol. Obstet. 165, 76–93 (2023).
Salomon, L. et al. ISUOG practice guidelines: performance of first-trimester fetal ultrasound scan. Ultrasound Obstet. Gynecol. 41, 102–113 (2013).
Salomon, L. J. et al. Practice guidelines for performance of the routine mid-trimester fetal ultrasound scan. Ultrasound Obstet. Gynecol. 37, 116–126 (2011).
Salomon, L. et al. ISUOG practice guidelines: ultrasound assessment of fetal biometry and growth. Ultrasound Obstet. Gynecol. 53, 715–723 (2019).
Bilardo, C. et al. ISUOG practice guidelines (updated): performance of 11–14-week ultrasound scan. Ultrasound Obstet. Gynecol. 61, 127–143 (2023).
Acknowledgments
The authors would like to acknowledge the ERASMUS + KA 220 - Cooperation partnerships in Higher Education Agreement no - 2024-1-IT02-KA220-HED-000249955 EthicAI4CARE Implementing Ethics by Design in AI: A Training Framework for the Healthcare Sector and The National Recovery along with the Resilience Plan (NRRP), Mission 4, Component 2, Investment 1.1, Call for tender No. 104 published on 2.2.2022 by the Italian Ministry of University and Research (MUR), funded by the European Union - NextGenerationEU- Project Title THAI-MIA - CUP B53D23005000006. Grant assignment Decree No. 20227HSE83 adopted on May 29, 2023 by the Italian MUR. The authors would also like to thank Ms. Arianna Fiorentino for providing us with the icons used in this paper.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.C.F., S.M. and S.T. Data curation: M.C.F., M.D.C. and S.T. Formal analysis: M.C.F., M.D.C. and S.T. Investigation: M.C.F. Methodology: M.C.F., S.M. and S.T. Supervision: S.M. and S.T. Project administration: S.M., S.T., B.G and E.F. Visualization: M.C.F., S.M., M.D.C. and S.T. Writing—original draft preparation: M.C.F. and M.D.C. Writing—review and editing: M.C.F, M.D.C., S.M., S.T., B.G. and E.F. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fiorentino, M.C., Moccia, S., Cosmo, M.D. et al. Uncovering ethical biases in publicly available fetal ultrasound datasets. npj Digit. Med. 8, 355 (2025). https://doi.org/10.1038/s41746-025-01739-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01739-3
This article is cited by
-
Benchmarking Vision LLMs in fetal ultrasound interpretation: a five-point expert evaluation of standard vs. custom prompts
BioMedical Engineering OnLine (2025)