Abstract
Rare gynecological tumors (RGTs) present major clinical challenges due to their low incidence and heterogeneity. The lack of clear guidelines leads to suboptimal management and poor prognosis. Molecular tumor boards accelerate access to effective therapies by tailoring treatment based on biomarkers, beyond cancer type. Unstructured data that requires manual curation hinders efficient use of biomarker profiling for therapy matching. This study explores the use of large language models (LLMs) to construct digital twins for precision medicine in RGTs. Our proof-of-concept digital twin system integrates clinical and biomarker data from institutional and published cases (n = 21) and literature-derived data (n = 655 publications) to create tailored treatment plans for metastatic uterine carcinosarcoma, identifying options potentially missed by traditional, single-source analysis. LLM-enabled digital twins efficiently model individual patient trajectories. Shifting to a biology-based rather than organ-based tumor definition enables personalized care that could advance RGT management and thus enhance patient outcomes.
Similar content being viewed by others
Introduction
Rare Gynecological Tumors (RGTs), comprising over 30 distinct histological subtypes, such as sex cord stromal tumors, and uterine or ovarian carcinosarcomas, account for more than 50% of gynecologic malignancies, presenting a major clinical challenge1. With an incidence rate below six per 100,000 individuals, RGTs are difficult to study through large-scale randomized trials, leading to reliance on less standardized treatment approaches such as retrospective studies, case reports, and expert opinions. This lack of robust clinical guidelines has contributed to persistently poor prognosis for patients with RGTs2.
Technological advancements in cancer diagnostics have enabled the identification of biomarkers as therapeutic targets. Biomarker-guided treatments promise to accelerate the development of precision therapeutics across tumor types, reducing the relevance of organ-based classification3. The prevailing organ-centric approach to clinical trial design hinders the development of effective treatments for rare cancers with shared biomarkers4. This obstacle extends beyond rare cancers: The premature dismissal of olaparib in ovarian cancer and the seven to ten year delay in addressing Programmed Cell Death Ligand 1 (PD-L1) expressing breast and gynecological cancers with PD-L1 inhibition illustrate the need for biomarker-driven stratification for cancer treatment5,6.
Molecular tumor boards (MTBs) are essential for interpreting biomarker profile results and matching cancer patients with appropriate therapies. This includes identifying suitable investigational drugs7. The manual interpretation of multiple, co-occurring molecular alterations requires an in-depth understanding of their functional implications and correlations with treatment sensitivity or resistance. The rapid growth of biomedical literature and the fragmented nature of data sources make manual curation a bottleneck in efficiently translating genomic data into actionable treatment strategies7.
The data produced by MTBs is often stored in unstructured formats within electronic health records (EHRs) or other repositories, hindering their reusability for similar patients. Evaluating the effectiveness of MTB-guided treatments requires extracting follow-up data from EHRs. Unstructured text within EHRs, coupled with the lack of interoperability across healthcare institutions—particularly when MTB patients receive treatment at external facilities—renders the process labor-intensive, error-prone, and time-consuming8. Consequently, applying MTB insights to future patients is hindered.
Advances in data capture and analysis, alongside decreasing costs in genome sequencing, are paving the way for innovative tools to manage rare or refractory cancers more effectively9. Digital twin technology constructs virtual representations of physical entities with dynamic, bidirectional interfaces10. Initially applied in industrial engineering, digital twins can also represent the human body in healthcare. By modeling physiological processes and predicting biomarker-specific responses to treatments, digital twins can address the challenges of patient variability and the limitations of traditional one-size-fits-all approaches11. In the case of RGT, the standard carboplatin and paclitaxel regimen, followed by chemotherapy monotreatments for subsequent lines, may not be the most effective approach12. Digital twins could help stratify RGT patients based on their unique biomarker profiles, enabling more tailored treatments and potentially improving outcomes, even in heavily pretreated cases.
Despite their potential, the adoption of digital twins in clinical practice is constrained by the challenges associated with integrating the diverse and complex data required for their development13. Unlike conventional computational models that often rely on static, population-level assumptions, digital twins are dynamic and personalized—virtual representations of individual patients that evolve in real time with incoming clinical data. This adaptability offers potential for precision medicine, particularly in complex or rare diseases. However, building such models requires synthesizing heterogeneous, often unstructured, information. Traditional natural language processing (NLP) methods are limited in this regard. Recent advances in large language models (LLMs), such as transformer-based architectures, enable more context-aware, scalable, and flexible extraction of clinical insights. Their in-context learning capabilities make them especially suited to support the creation of responsive and individualized digital twins, as explored in this study14.
In this study, we demonstrate the application of an LLM-enabled workflow for constructing digital twins for patients with RGT, specifically metastatic uterine carcinosarcoma (UCS).
The research question was inspired by a real-world UCS case presented to a major German cancer center for evaluation of third-line treatment options. According to a consensus statement by Bogani et al., third-line monotherapy in UCS typically results in a median progression-free survival (PFS) of 1.8 months and a response rate of 5.5%, highlighting the urgent need for novel therapeutic strategies12. The patient presented with a proficient mismatch repair (pMMR) carcinosarcoma with intermediate Tumor Mutational Burden (TMB) and high PD-L1 expression. Although PD-L1 positivity is common in UCS15,16 and has been suggested as an independent prognostic factor17, it has not been validated as a target for immunotherapy12.
Given the potential therapeutic importance of PD-L1, we investigated outcomes in similar patients. We identified cases with high PD-L1 expression, pMMR status and low to intermediate TMB from the institutional MTB database, including non-gynecological cancers, and to expand the pool of UCS cases, from the literature. The unstructured nature of EHR and academic publications posed challenges for immediate analysis. We utilized a local LLM to extract and structure data from EHRs and a cloud-based LLM for literature data. These datasets were integrated into a unified local database, forming the foundation of an RGT Digital Twin system. This system enabled the generation of virtual representations of individual patients, allowing for the simulation of personalized treatment strategies.
The RGT Digital Twin system facilitated the identification of additional therapeutic options, which were subsequently evaluated by MTB members. By integrating data from institutional sources (including non-gynecological cancers) and literature sources (to expand the pool of UCS cases), this approach provided novel insights that were not apparent from either data source alone. This integration has the potential to guide more effective treatment strategies for RGT patients and supports a shift towards a biology-based rather than organ-based definition of tumors. LLM technology enabled us to streamline the extraction, structuring, and analysis of EHR and web data, making it readily accessible for MTB evaluation. This is especially valuable in resource-limited settings like MTBs, where results can occasionally arrive too late to guide timely treatment decisions18.
Results
Patient cohort
A retrospective analysis of 1821 cases discussed at the institutional MTB between September 2017 and July 2024 was conducted. Among these, 132 cases exhibited high PD-L1 expression (CPS ≥ 40), encompassing 28 different tumor entities. The analysis was restricted to patients with TMB < 15 mutations/megabase and pMMR status with either gynecological cancers or carcinosarcoma/sarcomatoid carcinoma, resulting in a cohort of nine patients. Of these, seven patients received ICI therapy and were included in the study. The cohort comprised six females and one male aged 32 to 83 years at MTB presentation. Given that the similarity analysis focused on biomarker profiles and cancer morphology, the male patient’s inclusion was appropriate. His sarcomatoid carcinoma aligned with the other inclusion criteria, regardless of his gender or cancer type.
Data extraction
89 clinical reports were extracted for the patient cohort (median: 11, range: 9-21). Documents had a median of two pages and 4340 characters. Documents contained 70 data points for the selected attributes. Experts reviewed all extractions in the sample. The local LLM achieved accuracy of 0.76, precision of 0.85, recall of 0.87 and F1 of 0.86. The highest accuracy was achieved for ‘diagnosis’ and ‘ICI treatment’ (1.00). Low accuracy occurred in ‘previous treatment’ (0.29) and ‘PFS’ (0.14), mainly due to parsing errors in order and dates of previous treatments. See Table 1 for full results of the analysis.
Document analysis revealed that primary tumor sites included metastatic UCS (n = 1), metastatic cervical cancer (n = 4; three squamous cell carcinoma, one adenocarcinoma), metastatic uterine serous carcinoma (n = 1), and metastatic, undifferentiated sarcomatoid carcinoma of the pancreas (n = 1). Patients exhibited high PD-L1 expression with a median CPS of 75 (range: 40–95) and a median TMB of 5.5 (range: 0–11). Median follow-up duration was 48 months (range: 15–132 months). Detailed baseline clinical characteristics are presented in Supplementary Table 3.
The LLM-based systematic literature research yielded a dataset of 663 scientific documents. Files had a median of seven pages and 27,995 characters, with a maximum of 934,513 characters. The LLM extracted 7956 attributes from scientific documents. Attribute extraction was reviewed with a random sample of n = 352 (Z = 1.96, N = 7956, e = 0.05, P = 0.5). The cloud-based LLM achieved accuracy of 0.97, precision of 0.97, recall of 0.99, and F1 of 0.98. Lowest precision was observed in PFS (0.77).
The LLM system identified 15 studies reporting ICI treatment in UCS, encompassing a total of 215 patients. While seven of the studies did not exclusively enroll UCS patients, four provided stratified outcomes for UCS cases. Phase II studies that provided stratified analysis for UCS patients showed objective response rates between 0 and 10% for ICI treatment. None of the seven studies allowed for individual patient-level data extraction to create digital twins (see details on these seven studies in Supplementary Table 4).
PD-L1 status was reported in 10 of the 215 literature-derived UCS cases treated with immunotherapy, with three cases exhibiting PD-L1 positivity. This limited sample size precluded stratified analysis. Notably, two of the PD-L1-positive UCS patients harbored dMMR and one had a high TMB, both of which are known to influence ICI treatment response.
Further analysis of the 15 identified studies yielded eight studies with individual patient follow-up data, comprising a total of 14 cases. The median age of these literature-derived patients was 63 years (range: 55–68 years).
Treatment response outcomes for 21 individual patients
In the institutional cohort, seven patients received ICI therapy: five with pembrolizumab monotherapy, one with pembrolizumab plus lenvatinib, and one with ipilimumab plus nivolumab. ICI therapy was initiated on average in the third line (range: 2–4). Median PFS was 6 months (range: 1–48). One patient remained disease-free after 45 months, two continued to respond, one received a subsequent treatment line, and three had died.
Treatments in the 14 cases of the literature-derived cohort consisted of pembrolizumab (n = 4), pembrolizumab plus lenvatinib (n = 7), pembrolizumab plus lenvatinib plus letrozole (n = 1), PD-1/Cytotoxic T-lymphocyte associated protein 4 inhibitors (n = 1), and avelumab plus axitinib (n = 1). ICI was typically given in the third line (range: 2–5). Median PFS was 4 months (range: 0.9–15), and median OS was 9.9 months (range: 2.1–48). At data cut-off, six patients were alive, seven had died, and one had unknown status.
Table 2 provides a summary of ICI treatment response outcomes for all 21 cases.
RGT digital twins enable predictive modeling of individualized patient treatment strategies
To inform personalized treatment planning for the UCS patient (case 1), digital twins were created based on 21 evaluable patients. Treatment outcomes were predicted based on a database of additional 404,265 cases derived from scientific papers (n = 655). Potential treatment strategies were predicted for a patient with UCS with disease progression following third-line pembrolizumab monotherapy.
Supplementary Figure 1 presents treatment-relevant biomarkers after progression on standard-of-care combination treatment with carboplatin and paclitaxel identified by the digital twin system.
The digital twin system tailored treatment recommendations based on the patient’s specific tumor characteristics, treatment history, and geographic location. Considering the patient’s ongoing pembrolizumab therapy, the system suggested testing for Folate Receptor Alpha (FRα) to assess potential eligibility for an off-label treatment regimen currently under clinical investigation. This trial investigated the combination of mirvetuximab soravtansine and pembrolizumab in FRα-positive UCS, with eligibility criteria including pMMR status and prior pembrolizumab progression. However, the trial was no longer recruiting participants and was limited to the United States19. Therefore, the digital twin system suggested considering off-label use of this regimen for the patient. A previous evaluation (2021) identified HER2 amplification in the patient’s tumor, a biomarker linked to high objective response rates to trastuzumab deruxtecan20. Due to the potential for HER2 status to evolve, the system recommended confirming this finding through a new biopsy21. Additionally, the digital twin system suggested evaluating Melanoma-Associated Antigen A4 (MAGE-A4) and Preferentially Expressed Antigen in Melanoma (PRAME), biomarkers frequently expressed in UCS22,23. Ongoing research explores targeted therapies for these markers. Three relevant clinical trials were accessible within the patient’s geographic area. To monitor disease progression, the system recommended continued tracking of serum Cancer Antigen-125 (CA-125) levels24 based on its established correlation with disease progression identified in the patient’s 2021 EHR data.
Potential treatment trajectories for treatment line four derived from the Digital Twin pipeline are demonstrated in Table 3.
Quantifying LLM throughput
For local patient data processed on a standard PC (Macbook Pro M4), LLM extraction took a total of 61 min, averaging 0.68 min (41 s) per file. We estimate that this could be reduced to around 10 s per file when using a clustered computing environment. Manual review of the LLM output added an average of 8 min per file, resulting in a total time of 8.69 min per file for the combined LLM-assisted and human-reviewed approach. In comparison, manual extraction of the same data took an average of 55 min per file, representing a 6.3-fold reduction in time.
When processing a larger volume of literature with a cloud-based LLM, extraction took 83 minutes, averaging 0.125 min (7.5 s) per document. Manual validation added another 6 minutes per document, for a total of 6.125 min per document. In contrast, manual extraction from literature averaged 17 min per document, resulting in a 2.8-fold time saving with the LLM-assisted method.
Discussion
Extracting meaningful data from unstructured medical text is a prerequisite for precision medicine. In this study, we implemented an LLM-based extraction pipeline to systematically retrieve, structure, and analyze data from real-world EHRs and online sources to support and evaluate diagnostic and targeted therapeutic strategies for constructing patient-specific digital twins for metastatic UCS.
The limited data for this rare cancer led us to strategically use in-context learning rather than fine-tuning, which risks overfitting on small datasets and poor generalization. In-context learning leverages the pre-trained LLMs’ broad knowledge through prompt examples for effective data extraction without extensive retraining.
The LLM-based extraction pipeline facilitated timely and accurate synthesis of all relevant full-text scientific publications available through institutional access up to August 15, 2024.
Quantifying LLM throughput highlights the considerable efficiency gains achieved through LLM-assisted data extraction, even when incorporating necessary manual review. These improvements demonstrate the practical viability of integrating LLMs into clinical workflows, particularly for accelerating data curation from unstructured sources.
The cloud-based LLM achieved precision of 0.97 on a complex corpus of medical literature, close to the 0.96 observed by other researchers25. This enabled the generation of evidence-based recommendations and predictive insights grounded in the latest research. Key gaps were observed in extraction of complex data structures, with precision of ‘PFS’ (0.77) and ‘OS’ (0.94) below the overall precision of 0.97. This was due to the fragmented and unstructured way of reporting PFS and OS. Sentences such as “Patient survived for 14 months with the residual tumor post-relapse,”26 make it challenging to accurately determine PFS, as it requires estimation based on prior treatments and the number of treatment cycles. However, estimated PFS may not be accurate if treatment cycles were prolonged. This highlights the challenge of extracting precise outcome data when the primary source lacks comprehensive reporting. We strongly advocate for standards in reporting treatment outcomes, e.g., by clearly stating PFS in months and not date ranges.
In institutional data, unstructured EHR impeded the extraction of key clinical information. This limitation delayed the integration of institutional patient data for informing the management of similar cases. Phase II trials neglecting biomarker-stratification in patients with UCS yielded low objective response rates to ICI therapy, ranging from only zero to ten percent27,28. For our UCS patient, this bottleneck might have precluded ICIs based on high PD-L1 expression, despite the fact that pembrolizumab proved highly efficacious with no adverse effects in this patient. The local LLM system was able to extract structured follow-up data from EHRs across a diverse and complex set of medical documents. While it achieved lower precision than the cloud-based model at 0.85, this is in line with the performance of similar models on complex EHR29. Notably, precision for biomarkers was rated the second-lowest at 0.57. The LLM achieved full recall for all biomarkers given as examples for in-context learning, but did not recognize biomarkers that were not explicitly mentioned (e.g., BRAF for case ID 4). The LLM again achieved lowest precision for PFS at 0.14. This is due to the highly unstructured and fragmented way of reporting PFS, often across multiple documents.
Error analysis of the LLM-based extraction highlighted two challenges: Errors were found in extracting PFS from literature and EHRs, likely due to inconsistent formats and the need for temporal reasoning across multiple paragraphs. Similarly, identifying the number and type of prior treatments from free-text clinical notes led to incorrect extractions. In contrast, the LLMs performed reliably when extracting clearly defined and consistently reported data points. Notably, errors were either presented as missing or incomplete data points, rather than the fabrication of information. The use of in-context learning improved the model’s grounding for well-structured tasks but was less effective in addressing the ambiguities and contextual dependencies inherent in more complex extractions.The European Society for Medical Oncology Precision Medicine Working Group recently established criteria for evaluating the tumor-agnostic potential of molecularly guided therapies, mandating an ORR of ≥20% in at least one of five patients across at least four investigated tumor types, with a minimum of five evaluable patients per type3. Our institutional MTB database identified six analogous cases involving four additional tumor types, most of which exhibited durable responses to ICIs. While the limited number of evaluable patients per tumor type in our single-institution cohort restricted the statistical power, an LLM-driven literature review highlighted an underreporting of PD-L1 expression in UCS in studies conducted to date, despite the known high prevalence of PD-L1 positivity in this malignancy15,16. This underreporting impeded our ability to assess the predictive value of PD-L1 to guide ICI treatment in UCS. Despite the limited sample size of our institutional cohort, the promising outcomes observed suggest that targeting PD-L1 expression in RGT may be a viable therapeutic strategy. The inclusion of diverse tumor types in our institutional cohort further strengthens the role of PD-L1 inhibition in both gynecological and non-gynecological cancers, making it a potential tumor-agnostic marker.
To inform treatment strategies in the event of disease progression, we constructed 21 individualized digital twins, including 7 from our institutional database and 14 from the literature, and queried an LLM-derived database containing 404,265 patient cases. Although our systematic PubMed search was specifically limited to the terms “uterine carcinosarcoma” and “endometrial carcinosarcoma,” the resulting sample also included other uterine and ovarian malignancies. This is because UCS is frequently reported within the broader context of clinical trials involving more common gynecologic cancers.
The RGT digital twin system generated individualized trajectory predictions for various targeted therapies within a secure local environment that respects patient data privacy, offering guidance on further diagnostics, potential treatment options and continued treatment monitoring with serum CA-125. Additionally, since our real-world cohort comprised only White patients, being able to extract data on patients of other races from a vast corpus of literature helped us validate the generalizability of our treatment recommendations30.
This study successfully demonstrated the utility of RGT digital twins for individualized treatment prediction and response modeling. The digital twin not only provided generalized recommendations for additional diagnostic testing but also incorporated specific clinical details from the patient’s treatment history—such as prior pembrolizumab administration—to refine eligibility assessments for targeted therapies. The extraction and analysis of follow-up data revealed that, following the MTB recommendation, the patient received pembrolizumab due to high PD-L1 expression and exhibited a sustained partial response for over 30 months. Subsequent to pembrolizumab initiation, CA-125 levels normalized within three months and have remained stable in the context of ongoing stable disease.
This study had several limitations. Firstly, despite a large dataset, the combination of stringent similarity criteria, a limited institutional cohort, and underreported PD-L1 status in published UCS cases prevented us from stratifying patients by PD-L1 status. Efficacy of ICI treatment in PD-L1–positive UCS remains uncertain, and current trials lack PD-L1 as a stratification factor. Bogani et al. listed nine clinical trials currently exploring ICI treatment in UCS, many of which are nearing completion12. None of these trials included PD-L1 expression as a stratification factor. Our findings could inform the design of future trials that specifically evaluate ICI efficacy in pMMR UCS with high PD-L1 expression and low to intermediate TMB. Secondly, only somatic biomarkers were included, potentially underestimating clinical actionability by excluding germline mutations, such as BRCA1/2, which are predictive of PARP inhibitor response31. Thirdly, a local LLM was used for data extraction, impacting extraction performance due to its smaller size and no fine-tuning on German medical texts32. Lack of a German-language equivalent to the English-language MIMIC labeled medical record dataset33 precluded fine-tuning our own model. Finally, the German Network for Personalized Medicine (DNPM) data model is under revision34, necessitating the use of a custom data model for this study and highlighting the importance of future validation for compatibility with DNPM v2.
National and international collaborative initiatives, such as the DNPM Data Integration Platform35 and the Molecular Tumor Board Portal by Cancer Core Europe36, aim to enhance MTB decision-making by standardizing and harmonizing data collection across institutions. These platforms stand to benefit significantly from the integration of LLM-based extraction pipelines, which could facilitate the automated extraction of both baseline and follow-up data, thereby enabling the real-time utilization of MTB data across different healthcare systems. Once the DNPM database is fully operational, clinical narratives derived from EHR data could be transformed into HL7 Fast Healthcare Interoperability Resources (FHIR), streamlining interoperability and reducing the biases and costs associated with manual documentation37. Such automation would enable the analysis of larger patient cohorts, thereby providing the statistical power necessary for accurate treatment predictions in rare cancers, a critical step towards advancing personalized oncology. These outcome data offer a valuable resource for fine-tuning LLMs and for guiding both preclinical research and stratified clinical trial design.
At present, LLM knowledge synthesis forms the core of the system’s predictive capabilities. The future incorporation of auxiliary machine learning models could offer complementary approaches for enhanced predictive analytics.
Beyond the specific limitations of our study, the broader applicability of generating digital twins necessitates careful consideration of local environments and ethical implications. Our framework is designed to prioritize local data processing within institutional settings, ensuring sensitive patient data remains secure, as reflected in our accompanying GitHub code. For the 27b model used, local processing is feasible with standard computer hardware. This is paired with a cloud-based approach for processing publicly available literature, allowing us to leverage external resources while maintaining local control over sensitive information, thereby addressing key ethical and privacy concerns. Future work should continue to explore strategies for secure and ethical deployment across diverse institutional infrastructures and data governance models.
Our LLM-enabled precision oncology approach can inform more effective treatment strategies for RGT patients and suggests the potential for incorporating biomarkers, such as PD-L1 expression, to broaden therapeutic options and inform more personalized strategies that transcend traditional organ-based boundaries, particularly in the context of cancers with limited standard treatments.
Given the increasing volume and complexity of precision oncology data from MTBs, and the limited availability of precision oncologists to translate this abundance of information into clinically meaningful actions7, there is an urgent need for advanced digital tools to facilitate the extraction, structuring, and analysis of large datasets18. Our proof-of-concept study demonstrates the potential of LLMs to efficiently synthesize relevant information for MTB evaluation.
While this study focused on RGTs, the LLM-enabled digital twin approach holds potential for a wide range of refractory cancers. By accurately predicting individual patient trajectories, these digital twins can inform personalized diagnostics and treatment strategies in a timely and cost-effective manner, potentially improving patient outcomes.
Methods
Study setup
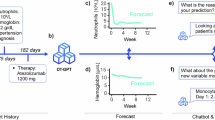
We employed an RGT Digital Twin system to create personalized treatment suggestions for UCS. A real-world patient case, along with molecular profiling data, was analyzed using the RGT Digital Twin system. The findings were then compared to analogous cases drawn from institutional and public databases. Treatment options were discussed at the MTB to inform individualized care decisions. Post-treatment outcomes were documented in the patient’s EHR and updated for the individual RGT Digital Twin to improve future predictions. The RGT Digital Twin provided rationale for cost coverage requests and supported study inclusion decisions. Refer to Fig. 1 for an overview of the study process.
Rare Gynecological Tumor (RGT) Digital Twin is a dynamic system that can integrate diverse data sources to predict individual patient trajectories. Molecular profiling identifies patient biomarkers. Large Language Model (LLM) capabilities support clinical interpretation of molecular profiles, patient matching to clinical trials, reasoning for cost-coverage requests, medical documentation, and data preparation for advanced computing. Advanced computing techniques such as classification and regression algorithms enable the creation and exploration of Digital Twin models. By adjusting parameters such as biomarker expression or previous treatment strategies, clinicians can model potential patient outcomes and determine suitable treatment strategies. The RGT Digital Twin then integrates outcome data back into the RGT Patient Electronic Health Record (EHR), supporting a continuous learning process. RGT Rare Gynecological Tumor, DB Database, LLM Large Language Model Fig. 1 uses free icons obtained from Flaticon.com, in accordance with their licensing terms.
Patient description and research question
The patient is a 77-year-old woman with metastatic UCS, initially diagnosed with FIGO IIIC2 UCS at age 66. Six years after surgery and adjuvant chemotherapy with carboplatin and paclitaxel, the patient experienced a recurrence in the cervical lymph nodes and pelvis, which was also associated with an increase in CA-125 levels. A cervical lymph node biopsy confirmed the recurrence, and the patient underwent the same chemotherapy regimen followed by MTB presentation in 2021 (see Supplementary Table 1 for detailed results). Genomic profiling was conducted using the TruSight Oncology 500 (TSO 500) and TruSight Tumor 170 (TST 170) panels. By analyzing a wide range of cancer-related genes, these panels facilitate the discovery of potential therapeutic targets. Molecular profiling revealed an intermediate TMB of 6.3 mutations/megabase, high PD-L1 expression (Combined Positive Score, CPS: 41), and a pMMR status. The patient’s high PD-L1 expression prompted us to investigate the potential efficacy of PD-L1 inhibitors in metastatic tumors, regardless of primary tumor site or regional drug approval status. To this end, we searched our institutional MTB database for analogous cases, not restricted to gynecological cancers, and expanded our cohort with additional UCS cases identified in the literature.
Data collection
EHR data obtained from Technical University of Munich (TUM) University Hospital was the primary data source. Data downloaded from web-based repositories through institutional access extended the dataset. These repositories included PubMed, ClinicalTrials.gov, and clinical practice guidelines from the National Comprehensive Cancer Network (NCCN) and the German Cancer Society (DKG), which ensured adherence to up-to-date clinical standards.
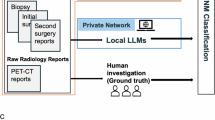
A two-stage approach was employed for extracting structured, actionable data from source files. First, a locally deployed LLM system extracted relevant data from institutional EHRs. Second, a cloud-based LLM system processed documents from web-based repositories. Afterwards, the extracted structured dataset was made available to clinicians and the locally deployed LLM for further analysis. This process is shown in Fig. 3.
Identification of analogous institutional cases
We utilized a locally deployed large language model to systematically analyze pathology reports from MTB cases discussed at TUM University Hospital between September 2017 and July 2024. PD-L1 expression, TMB and MMR status were documented in a total of 1821 cases. Eligibility criteria included high PD-L1 expression (CPS ≥ 40) and the availability of both MMR and TMB status. Patients with high TMB ( ≥ 15 mutations/megabase) or deficient MMR were excluded to avoid bias, as these features are already established FDA-approved biomarkers for Immune Checkpoint Inhibitor (ICI) therapy responsiveness.
The LLM-assisted screening process applied predefined inclusion criteria and stratified cases based on similarity to the presented UCS patient. Similarity was determined by either medical specialty (gynecological oncology) or histopathological features, specifically carcinosarcoma or sarcomatoid carcinoma morphology, independent of gender or anatomical origin. This strategy was based on the recognized molecular overlap between UCS and high-grade serous ovarian or endometrial carcinomas12. Given the limited clinical value of distinguishing between carcinosarcoma and sarcomatoid carcinoma in practice, the institutional MTB members grouped these under the unified category of “morphology.”
As visualized in the CONSORT diagram (Fig. 2), 9 cases met all inclusion criteria: CPS ≥ 40, pMMR, TMB < 15 mutations/megabase, and concordance with at least one similarity parameter. Of these, 7 patients received ICI treatment and were included in the final LLM-driven analysis, which focused on extracting clinical and treatment outcomes, and evaluating case-level comparability to the index UCS patient.
Flow diagram depicting patient selection for analysis from the institutional Molecular Tumor Board (MTB), including those eligible for and treated with immune checkpoint inhibitors (ICI). Due to the limited number of analogous digital twins within the institutional MTB cohort (n = 7), additional UCS cases receiving ICI treatment were identified through a targeted literature review and included to supplement the Rare Gynecological Tumor (RGT) Digital Twin (DT) dataset. MTB Molecular Tumor Board, PD-L1 Programmed Cell Death-Ligand 1, MMR Mismatch Repair, TMB Tumor Mutational Burden, CPS Combined Positive Score, pMMR proficient Mismatch Repair, Mut/Mb Mutations/Megabase, UCS Uterine Carcinosarcoma, RGT Rare Gynecological Tumor. DT Digital Twin.
Figure 2 provides an exemplary execution of the system for institutional data enhanced with external literature due to the limited availability of analogous digital twins in EHR.
Institutional patient data extraction pipeline
EHR of selected patients were processed in a secure hospital environment. Documents varied in format, from (handwritten) medical notes to obituaries. Ten attributes were extracted from documents for each patient to form the RGT Digital Twin. Supplementary Table 2 shows the full list of attributes. Optical Character Recognition (OCR) was performed using Tesseract38. Raw text was processed with a locally deployed version of pre-trained LLM gemma-2-27b-it, chosen for its ability to run locally while maintaining strong performance on medical texts39. This privacy-preserving architecture ensured that patient data would not leave the local clinic environment. In-context learning was used to adapt the LLM to the task at hand. This involves providing the LLMs with extensive instructions in their prompts, including example input and output (e.g., “age”: “63”, “gender”: “female”, “diagnosis”: “Uterine Carcinosarcoma”). The publicly available GitHub repository provides the full prompt templates and examples. This improves their recall and precision40.
Literature extraction pipeline
To extend the limited sample of analogous digital twins available in institutional EHR, a systematic literature search was conducted on PubMed using the terms ‘uterine carcinosarcoma’ and ‘endometrial carcinosarcoma’. Studies and case reports that included individual clinical follow-up data on patients with UCS treated with ICI were considered for inclusion in the analysis. Potential alternative treatment options and therapeutic targets were identified through a comprehensive review of national (DKG) and international (NCCN) oncological guidelines, PubMed-indexed publications, as well as the ClinicalTrials.gov database.
Data points were extracted in the structure shown in Supplementary Table 2. The sample size was captured as an additional data point. Additionally, the LLM was instructed to extract the main treatment recommendation based on the patient profile in the paper. General purpose LLM Google-Gemini-1.5-Pro was selected for this task due to its large context window, which enabled it to process all files in the sample without splitting them into smaller chunks41. Since no institutional patient data was processed in this step, use of public cloud resources was permitted. The LLM was instructed to return results in the form of a JSON object. The LLM processed all documents sequentially, with each document processed in-context. Outputs were exported to a Pandas dataframe on the local machine in the secure hospital environment for convenient analysis by clinicians.
Ethics statement
Patients from the institutional MTB were included in a clinical registry that allowed for retrospective analyses of clinical and molecular anonymized data in accordance with the Declaration of Helsinki. The retrospective analysis was approved by the Ethics committee of the Medical Faculty of the Technical University of Munich (Reference No. 2023-486-S-SB).
All web-based research procedures were conducted exclusively on publicly accessible, anonymized patient data and in accordance with the Declaration of Helsinki, maintaining all relevant ethical standards.
Construction of LLM-enabled digital twin system
Next, extracted data points were stored in a database in the secure hospital environment that constituted the patient’s digital twin. Clinicians were able to model potential outcomes for their patients and determine suitable treatment strategies by reviewing treatment outcomes from patients with similar biomarkers and treatment history. Additionally, they were able to employ the local LLM to combine treatment strategies identified from web sources with the patient’s digital twin, creating personalized treatment recommendations.
The digital twin system provided a comprehensive overview of all potential recommendations relevant to the patient’s digital representation. Suitable recommendations were determined based on matching similar cases based on disease, prior treatments and biomarkers extracted from institutional data, oncological guidelines, clinical trials and literature. If the patient had received a specific treatment before, it was not suggested by the system. If relevant biomarkers were not specified in the patient’s digital representation, the digital twin system proactively suggested their analysis to inform treatment decisions. The recommendations represented the complete set of applicable and viable options for that consultation. As new patient follow-up data and relevant literature were added to the local database, the LLM’s accessible knowledge base expanded. This growing repository enabled the system to generate increasingly informed and updated insights for subsequent patients, enhancing its ability to provide personalized and temporally relevant recommendations.
After selecting a treatment strategy, the database served as evidence for MTB evaluation, clinical trial matching, and creation of cost coverage requests with health insurance providers.
The developed pipeline is illustrated in Fig. 3.
To obtain institutional patient data and matching patient profiles from literature, we first filtered institutional records and public data sources (e.g., PubMed) by biomarker profiles and patient disease. We then extracted structured patient data from EHR using a locally deployed, privacy-preserving LLM, and extracted similar data from published literature using a cloud-based LLM. By utilizing a broader patient population than what is available in institutional data, the RGT Digital Twin system generated personalized treatment plans for MTB evaluation. This method revealed additional treatment options that might have been missed when considering each data source alone. EHR Electronic Health Record, LLM Large Language Model. Figure 3 uses free icons obtained from Flaticon.com, in accordance with their licensing terms.
Analysis
Clinical characteristics, treatment regimens, duration of therapy, treatment responses, PFS, and overall survival (OS) were systematically collected from patient records and reports. Treatment response was captured from radiology reports and categorized as complete response (CR), partial response (PR), stable disease (SD), mixed response (MR), or progressive disease (PD). For additional treatment strategies, outcomes were summarized for each therapeutic approach. Cases were sequentially numbered, starting with those retrieved from the institutional MTB database, followed by cases identified from the literature.
Formal statistical analysis to evaluate the accuracy of LLM data retrieval was performed by experts. Due to the large amount of data processed by the Digital Twin pipeline, we adopted human-in-the-loop reviews, an important aspect of machine learning42. To ensure that no information was missed during extraction, a sample-based review of LLM output was performed according to machine learning leading practice43. For institutional data, experts reviewed all attributes extracted from EHR by the LLM for correctness. For public research data, experts reviewed a random sample of attributes extracted from scientific studies for correctness. Additionally, all attributes that were used by the LLM to construct the literature-derived digital twins were manually reviewed. Afterwards, accuracy, precision, recall, and F1 scores of LLM extraction were calculated. Finally, all treatment recommendations generated by the LLM were manually reviewed and corrected by human experts. The data extraction review panel included two bioinformaticians and two gynecological oncologists with five and 16 years of clinical experience.
A panel of five MTB members, including three clinicians, one pathologist, and one biologist, along with a senior gynecological oncologist, evaluated the personalized treatment recommendations generated by the RGT Digital Twin system.
All statistical analyses were conducted using Pandas and SciPy libraries in Python (Version 3.10.12). The full code and documentation is available on GitHub.
Data availability
The datasets analysed during the study are not publicly available due to institutional patient privacy regulations and the inclusion of full EHR, but are available from the corresponding author on reasonable request and pending appropriate ethical approval. All notebooks and prompts used in this study are publicly available at GitHub (https://github.com/LammertJ/RGT-Digital-Twin/).
Code availability
The underlying code for this study is available on GitHub and can be accessed via this link.
References
Gatta, G. et al. Rare cancers are not so rare: The rare cancer burden in Europe. Eur. J. Cancer 47, 2493–2511 (2011).
Lainé, A., Hanvic, B. & Ray-Coquard, I. Importance of guidelines and networking for the management of rare gynecological cancers. Curr. Opin. Oncol. 33, 442–446 (2021).
The ESMO Tumour-Agnostic Classifier and Screener (ETAC-S): A tool for assessing tumour-agnostic potential of molecularly guided therapies and for steering drug development. Ann. Oncol. https://doi.org/10.1016/j.annonc.2024.07.730 (2024).
Horak, P. et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov. 11, 2780–2795 (2021).
Ledermann, J. A. PARP inhibitors in ovarian cancer. Ann. Oncol. 27, i40–i44 (2016).
André, F., Rassy, E., Marabelle, A., Michiels, S. & Besse, B. Forget lung, breast or prostate cancer: why tumour naming needs to change. Nature Publishing Group UK https://doi.org/10.1038/d41586-024-00216-3 (2024).
Tsimberidou, A. M. et al. Molecular tumour boards - current and future considerations for precision oncology. Nat. Rev. Clin. Oncol. 20, 843–863 (2023).
Botsis, T. et al. Precision Oncology Core Data Model to Support Clinical Genomics Decision Making. JCO Clin. Cancer Inf. 7, e2200108 (2023).
Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical AI. Nat. Med. 28, 1773–1784 (2022).
Kamel Boulos, M. N. & Zhang, P. Digital twins: From personalised medicine to precision public health. J. Pers. Med. 11, 745 (2021).
Food, U. S., Administration, D. & Others. Paving the way for personalized medicine: FDA’s role in a new era of medical product development. Silver Spring, MD: US Food and Drug Administration.
Bogani, G. et al. Endometrial carcinosarcoma. Int. J. Gynecol. Cancer 33, 147–174 (2023).
Katsoulakis, E. et al. Digital twins for health: A scoping review. NPJ Digit Med 7, 77 (2024).
Adams, L. C. et al. Leveraging GPT-4 for post hoc transformation of free-text. Radiol. Rep. into Struct. Rep. A Multiling. Feasibility Study Radiol. 307, e230725 (2023).
Hacking, S., Chavarria, H., Jin, C., Perry, A. & Nasim, M. Landscape of immune checkpoint inhibition in carcinosarcoma (MMMT): Analysis of IDO-1, PD-L1 and PD-1. Pathol. Res. Pract. 216, 152847 (2020).
Jenkins, T. M., Cantrell, L. A., Stoler, M. H. & Mills, A. M. PD-L1 and mismatch repair status in uterine carcinosarcomas. Int. J. Gynecol. Pathol. 40, 563–574 (2021).
Kucukgoz Gulec, U. et al. Prognostic significance of programmed death-1 (PD-1) and programmed death-ligand 1 (PD-L1) expression in uterine carcinosarcoma. Eur. J. Obstet. Gynecol. Reprod. Biol. 244, 51–55 (2020).
The Lancet Oncology Incorporating whole-genome sequencing into cancer care. Lancet Oncol. 25, 945 (2024).
ClinicalTrials.Gov. https://clinicaltrials.gov/study/NCT03835819.
Nishikawa, T. et al. Trastuzumab deruxtecan for human epidermal growth factor receptor 2-expressing advanced or recurrent uterine carcinosarcoma (NCCH1615): The STATICE Trial. J. Clin. Oncol. 41, 2789–2799 (2023).
Yoshida, H. et al. Discordances in expression of human epidermal growth factor receptor 2 between primary and metastatic uterine carcinosarcoma: A proposal for HER2-targeted therapy specimen selection. Ann. Diagn. Pathol. 65, 152150 (2023).
Resnick, M. B. et al. Cancer-testis antigen expression in uterine malignancies with an emphasis on carcinosarcomas and papillary serous carcinomas. Int. J. Cancer 101, 190–195 (2002).
Alrohaibani, A. et al. PReferentially expressed antigen in MElanoma expression in uterine and ovarian carcinosarcomas. Int. J. Gynecol. Pathol. 43, 284–289 (2024).
Huang, G. S. et al. Serum CA125 predicts extrauterine disease and survival in uterine carcinosarcoma. Gynecol. Oncol. 107, 513–517 (2007).
Konet, A. et al. Performance of two large language models for data extraction in evidence synthesis. Res. Synth. Methods https://doi.org/10.1002/jrsm.1732 (2024).
Yano, M. et al. Pembrolizumab and radiotherapy for platinum-refractory recurrent uterine carcinosarcoma with an abscopal effect: A case report. Anticancer Res. 40, 4131–4135 (2020).
Rubinstein, M. M. et al. Durvalumab with or without tremelimumab in patients with persistent or recurrent endometrial cancer or endometrial carcinosarcoma: A randomized open-label phase 2 study. Gynecol. Oncol. 169, 64–69 (2023).
Lheureux, S. et al. Translational randomized phase II trial of cabozantinib in combination with nivolumab in advanced, recurrent, or metastatic endometrial cancer. J. Immunother Cancer 10, e004233 (2022).
Saab, K. et al. Capabilities of Gemini Models in Medicine. arXiv [cs.AI] (2024).
Arora, K. et al. Genetic ancestry correlates with somatic differences in a real-world clinical cancer sequencing cohort. Cancer Discov. 12, 2552–2565 (2022).
Mendes Gomes, L. B. et al. Primary peritoneal carcinosarcoma in a breast cancer patient harboring a germline BRCA2 pathogenic variant: Case report. Case Rep. Oncol. 17, 1–9 (2024).
Han, T. et al. MedAlpaca - An open-source collection of medical conversational AI models and training data. arXiv [cs.CL] (2023).
Johnson, A. E. W. et al. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data 10, 1 (2023).
Siebrasse, B. MTB Kerndatensätze_V01 (öffentlich zugänglich) - DNPM AG - MedInf - IBMI-UT Confluence. https://ibmi-ut.atlassian.net/wiki/spaces/DAM/pages/2592722/MTB+Kerndatens+tze_V01+ffentlich+zug+nglich.
Illert, A. L. et al. The German network for personalized medicine to enhance patient care and translational research. Nat. Med. 29, 1298–1301 (2023).
Tamborero, D. et al. The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology. Nat. Cancer 3, 251–261 (2022).
Li, Y., Wang, H., Yerebakan, H. Z., Shinagawa, Y. & Luo, Y. FHIR-GPT enhances health interoperability with large language models. NEJM AI 1, AIcs2300301 (2024).
Hegghammer, T. OCR with Tesseract, Amazon Textract, and Google Document AI: a benchmarking experiment. J. Computational Soc. Sci. 5, 861–882 (2022).
Gemma Team et al. Gemma 2: Improving Open Language Models at a Practical Size. arXiv [cs.CL] (2024).
Xie, S. M., Raghunathan, A., Liang, P. & Ma, T. An explanation of in-context learning as implicit Bayesian inference. arXiv [cs.CL] (2021).
Gemini Team et al. Gemini 1.5: Unlocking multimodal understanding across millions of tokens of context. arXiv [cs.CL] (2024).
Mosqueira-Rey, E., Hernández-Pereira, E., Alonso-Ríos, D., Bobes-Bascarán, J. & Fernández-Leal, Á Human-in-the-loop machine learning: a state of the art. Artif. Intell. Rev. 56, 3005–3054 (2023).
Marshall, I. J. & Wallace, B. C. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Syst. Rev. 8, 163 (2019).
Jenkins, T. M., Cantrell, L. A., Stoler, M. H. & Mills, A. M. HER2 overexpression and amplification in uterine carcinosarcomas with serous morphology. Am. J. Surg. Pathol. 46, 435–442 (2022).
Nicoletti, R. et al. T-DM1, a novel antibody-drug conjugate, is highly effective against uterine and ovarian carcinosarcomas overexpressing HER2. Clin. Exp. Metastasis 32, 29–38 (2015).
Edmondson, R. J. et al. Phase 2 study of anastrozole in rare cohorts of patients with estrogen receptor/progesterone receptor positive leiomyosarcomas and carcinosarcomas of the uterine corpus: The PARAGON trial (ANZGOG 0903). Gynecol. Oncol. 163, 524–530 (2021).
Soiffer, J. L. et al. Durable partial response to pembrolizumab, lenvatinib, and letrozole in a case of recurrent uterine carcinosarcoma with ESR1 gene amplification. Gynecol. Oncol. Rep. 54, 101426 (2024).
Saito, A. et al. Folate receptor alpha is widely expressed and a potential therapeutic target in uterine and ovarian carcinosarcoma. Gynecol. Oncol. 176, 115–121 (2023).
Porter, R. L. et al. Abstract CT008: A phase 2, two-stage study of mirvetuximab soravtansine (IMGN853) in combination with pembrolizumab in patients with microsatellite stable (MSS) recurrent or persistent endometrial cancer. Cancer Res. 84, CT008–CT008 (2024).
Tymon-Rosario, J. R. et al. Homologous recombination deficiency (HRD) signature-3 in ovarian and uterine carcinosarcomas correlates with preclinical sensitivity to Olaparib, a poly (adenosine diphosphate [ADP]- ribose) polymerase (PARP) inhibitor. Gynecol. Oncol. 166, 117–125 (2022).
Roszik, J. et al. Overexpressed PRAME is a potential immunotherapy target in sarcoma subtypes. Clin. Sarcoma Res. 7, 11 (2017).
Raji, R. et al. Uterine and ovarian carcinosarcomas overexpressing Trop-2 are sensitive to hRS7, a humanized anti-Trop-2 antibody. J. Exp. Clin. Cancer Res. 30, 106 (2011).
Lopez, S. et al. Preclinical activity of sacituzumab govitecan (IMMU-132) in uterine and ovarian carcinosarcomas. Oncotarget 11, 560–570 (2020).
Santin, A. et al. Preliminary results of a phase II trial with sacituzumab govitecan-hziy in patients with recurrent endometrial carcinoma overexpressing Trop-2. J. Clin. Orthod. 41, 5599–5599 (2023).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conceptualisation: J.L., M.T.; project administration: J.L., M.T.; resources: J.L., N.P., K.S. and S.L.; validation: J.L., N.P., L.K., K.S., S.L., U.S., H.B., M.T.; writing—original draft preparation: J.L., M.T.; writing—review & editing: S.M., T.D., L.M., P.M., D.F., J.N.K., D.T., L.C.A., K.K.B., M.B., M.K., U.S., H.B. All authors contributed scientific advice and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.L. received honoraria from the Forum for Continuing Medical Education (FomF), AstraZeneca GmbH (Germany), and Novartis Pharma GmbH (Germany). J.N.K. declares consulting services for Owkin, France; DoMore Diagnostics, Norway; Panakeia, UK, and Scailyte, Basel, Switzerland; furthermore J.N.K. holds shares in Kather Consulting, Dresden, Germany; and StratifAI GmbH, Dresden, Germany, and has received honoraria for lectures and advisory board participation by AstraZeneca, Bayer, Eisai, MSD, BMS, Roche, Pfizer and Fresenius. D.T. received honoraria for lectures by Bayer and holds shares in StratifAI GmbH, Germany. K.K.B. has received honoraria for lectures by GE HealthCare and Canon Medical Systems Corporation. M.K. has received fees from Springer Press, Biermann Press, Celgene, AstraZeneca, Myriad Genetics, TEVA, Eli Lilly, GSK, Seagen, AllergoSan, FomF, Roche, BESINS, Bayer AG. M.K. has received honoraria for consulting and advisory board participation by Myriad Genetics, Bavarian KVB, DKMS Life, BLAEK, TEVA, Exeltis, Roche, BESINS, Bayer AG. M.K. holds shares in AIM GmbH, in-manas GmbH, Therawis Diagnostic GmbH. H.B. has received personal fees from Roche, AstraZeneca, Gilead, and GlaxoSmithKline outside the submitted work. The authors have no additional financial or non-financial conflicts of interest to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lammert, J., Pfarr, N., Kuligin, L. et al. Large language models-enabled digital twins for precision medicine in rare gynecological tumors. npj Digit. Med. 8, 420 (2025). https://doi.org/10.1038/s41746-025-01810-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01810-z