Abstract
Systemic sclerosis (SSc) is a chronic autoimmune disease with multi-organ involvement. Historically, SSc classification has focused on the type of skin involvement (limited versus diffuse); however, a growing evidence of organ-specific variability suggests the presence of more than two distinct subtypes. We propose a semi-supervised generative deep learning framework leveraging expert-driven definitions of organ-specific involvement and severity. We model SSc disease trajectories in the European Scleroderma Trials and Research (EUSTAR) database, containing 14,000 patients across 67,000 medical visits, and identify clinically meaningful subtypes to enhance patient stratification and prognosis. We systematically evaluate the model’s predictive accuracy, robustness to missing data, and clinical interpretability. We identified five patient clusters, separating patients based on the degree of organ involvement. Notably, a subset with limited skin involvement still showed high risks of lung and heart complications, underscoring the importance of data-driven methods and multi-organ models to complement established insights from clinical practice.
Similar content being viewed by others
Introduction
Systemic sclerosis (SSc) is a chronic autoimmune disease marked by progressive fibrosis and vascular abnormalities in the skin and multiple internal organs such as the lungs, heart, kidneys, and gastrointestinal tract (GT)1. These multi-organ manifestations vary widely among patients in terms of frequency, onset, and severity, leading to significant morbidity and mortality2. Despite known clinical markers, such as skin involvement (limited cutaneous vs. diffuse cutaneous) and autoantibodies (e.g., anti-centromere, anti-topoisomerase I), it remains unclear which organs will become affected over time and how these manifestations might influence subsequent disease progression3. Early detection of at-risk individuals is therefore crucial for managing disease severity and potentially slowing progression4.
Traditional classification of SSc relies primarily on the extent of skin involvement: limited cutaneous SSc (lcSSc) is characterized by restricted areas of skin thickening, whereas diffuse cutaneous SSc (dcSSc) involves more widespread skin changes and often correlates with a higher risk of internal organ complications1. Specific autoantibodies also serve as important biomarkers for SSc diagnosis, organ involvement, and disease progression5,6. While anti-centromere antibodies (ACA) are predominantly linked with lcSSc and a higher likelihood of pulmonary arterial hypertension (PAH), anti-topoisomerase I antibodies (ATA) are often associated with dcSSc and an increased risk of interstitial lung disease (ILD), and anti-RNA polymerase III antibodies (ARA) are associated with rapid skin thickening, and increased risk of renal crisis7. However, because SSc involves complex, overlapping pathologies in multiple organs, subtyping remains a challenge; many crucial aspects of disease progression are not captured by skin-based classification alone8.
Recent work has leveraged artificial intelligence (AI), particularly deep learning (DL), to address the complexity of diseases with heterogeneous and longitudinal clinical data9 and identify patient subgroups with similar disease evolution10,11. Fully unsupervised models detect latent (i.e. unobserved) patterns without any labels12, while supervised approaches rely heavily on labeled outcomes. Neither paradigm alone is ideal for SSc, where labels (e.g., organ-specific damage) may be incomplete or imprecise, yet expert knowledge exists regarding clinically relevant markers and trajectories. Consequently, semi-supervised or hybrid methods have emerged as a promising alternative, combining partial labels and domain knowledge to guide latent representation learning13,14. Most prior ML-based research for SSc has focused on single-organ complications, such as ILD15, or is limited by sample sizes16. A multi-organ model is needed to capture the true disease complexity and identify subtle, high-risk patient subgroups that might otherwise be overlooked4.
In this work, we propose a semi-supervised deep learning framework for analyzing and clustering multi-organ trajectories in SSc, leveraging the largest global SSc registry from the European Scleroderma Trials and Research (EUSTAR) group17. We build on a previously developed temporal variational autoencoder-based model12,14,18 tailoring it to SSc and incorporating novel expert-guided definitions for two key dynamics, organ involvement and severity, each validated in a prior clinical study19. We model eight organs commonly affected by SSc: the skin, digital ulcers (DU), joints, muscles, lungs, heart, kidneys, and gastrointestinal tract (GT), and learn interpretable representations of patient disease trajectories. We then cluster these learned embeddings to identify clinically meaningful subtypes that may transcend conventional skin-based classification schemes. Figure 1 summarizes our approach.
1. Variable selection process and database preprocessing. 1. A We first screened the medical literature to identify clinical definitions of involvement and severity for each studied organ, and extracted the relevant variables X from the database. 1. B Next, a steering committee of 10 rheumatologists reached an expert consensus to select the most relevant clinical definitions, yielding a more restricted subset of variables G ⊆ X. 1. C Patient data is collected from various EUSTAR-affiliated centers and aggregated by the EUSTAR group. The database is preprocessed and is randomly split into an 85% training set, used for model development and hyperparameter tuning, and a 15% test set for hold-out evaluation. 2. Semi-supervised model architecture. The encoder network processes longitudinal clinical measurements, x1:t up to a time-point t, concatenated with the corresponding missingness indicator mask m1:t, and static patient demographic information s. It learns the distribution of the full latent trajectory z1:T, where T is the time of the last available visit in the registry. 2. A The guidance decoders, each assigned to a specific variable in G, take as input a predefined allocated subset of the dimensions from a sampled z1:T (one allocated subset per organ) and predict the distribution of the corresponding medical variables. 2. B The unsupervised decoder takes a sampled z1:T (all dimensions) and is trained to reconstruct the input x1:t. 3. Hierarchical clustering for disease subtyping in the learned latent space. Our method first divides the cohort into two main clusters—mild and severe trajectories—then further subdivides the mild cluster into two subtypes and the severe cluster into three subtypes. Abbreviations: Long Short-Term Memory Network (LSTM), Multilayer Perceptron (MLP).
Our key contributions include:
-
Deep multi-organ SSc model: Development of a semi-supervised generative deep learning approach to model eight clinically relevant organs, capturing both involvement and severity over time, while merging data-driven discovery with expert clinical insights.
-
Deep SSc subtyping: Development of a hierarchical clustering approach for patient trajectories that highlights under-recognized high-risk subgroups and goes beyond traditional SSc subtyping.
-
Large-scale evaluation: Demonstrating predictive accuracy and generalizability through comprehensive training and evaluation on over 14,000 patients and 67,000 visits from the EUSTAR registry.
-
Clinical decision support: Demonstrating how additional features of our framework, such as patient similarity and predictive clustering, can support clinical decision-making and personalized medicine.
Results
As detailed in section "Training", we performed five-fold cross-validation (CV). We then trained a final model for each of the five folds, resulting in 5 final models. We specifically analyzed the model trained on the first fold and used the remaining models to assess result stability, particularly in terms of performance on unseen test data to evaluate generalizability. We support our disease subtyping approach with several analyses: (1) We first evaluate the model’s ability to reconstruct or predict the organ-related variables G (defined in section "Model overview and notations")and its robustness to missing data. (2) Next, we examine how different features and labels shape the structure of the latent space, (3) followed by an in-depth analysis of the identified disease subtypes through hierarchical clustering. We conclude by discussing how various model components can support clinical decision-making.
Predictive performance
We compared our approach against several baselines, including both ML and non-ML approaches in predicting the organ variables in G:
-
Ours – without feature masking: Uses the same architecture as our final approach but does not explicitly train for missing data imputation, unlike our main model, which uses feature masking (i.e. masks 20% randomly during training) and learns to reconstruct missing data (see subsection "Handling missing data"). As a result, this model is optimized purely for prediction rather than also for learning missing variables, and we expect it to perform slightly better on complete datasets.
-
Multilayer Perceptron (MLP): A non-temporal model using only the most recent clinical measurements (unlike our model, which considers the full patient history). It is optimized purely for prediction, and does not learn latent trajectories.
-
Non-ML baselines: Distribution-based predictions/heuristics are included to provide a benchmark for the general capabilities of ML models.
-
Patient-specific: Predicts the future value of a variable based on its current value
-
Cohort mean: Uses the cohort mean of the feature as prediction.
-
Table 1 presents the Mean Absolute Error (MAE) for continuous variables and weighted F1 score for categorical variables for each model, averaged across five CV folds. Our final model and the variant without feature masking (i.e. missingness training) perform similarly and slightly outperform the MLP model. All ML models strongly outperform the non-ML baselines. Moreover, in Supplementary Table 4, we show that our approach outperforms all other models in terms of robustness to missing data.
Latent space analysis: ground truth vs. reconstructed values
As detailed in section “Model Architecture”, our model is trained to project raw patient trajectories into a latent (i.e. unobserved) space. In this section, we examine and interpret these latent representations. To facilitate the analysis, we computed the 2-dimensional UMAP20 decomposition for each time point in the latent trajectories, providing a visualization aid for the latent space. In the resulting UMAP plots (for instance Fig. 2), each point corresponds to a patient at a specific time. By overlaying the UMAP plots with color-coded clinical measurement values, labels, or clusters, we can intuitively visualize patient trajectories, cluster patterns, and feature/label distributions within the latent space.
UMAP decompositions of the latent space are overlaid, respectively, with ground truth values (left) and model-reconstructed values (right) for lung fibrosis features. Plotted data points correspond to values that were masked (not provided to the model), demonstrating its ability to impute missing information.
As discussed in section “Predictive Performance”, we train our model to infer values for missing variables. Fig. 2 shows a side-by-side UMAP visualization comparing the ground truth for masked values (i.e. not provided as input to the model) and the corresponding model reconstructions for two features related to lung fibrosis. The close alignment between ground truth and reconstructed values illustrates that the model reliably imputes missing data. Notably, this applies to all variables, whether available or not, thereby enriching the latent embeddings beyond what is present in the raw inputs. Supplementary Fig. 5 further demonstrates how the model learns to “fill in"gaps in the latent space.
Latent space regions
We observe that patients with different disease manifestations are mapped to distinct regions within the latent space. By overlaying the UMAP plots with specific feature values, we can identify the areas corresponding to different patient types, and gain insight into which features most strongly influence the latent space separation. In Fig. 3, the latent space is color-coded based on feature values inferred by our model, revealing a clear separation concerning “HRCT: Lung fibrosis” (true vs false) and the “Cutaneous SSc” (limited vs diffuse). Additionally, a subset of the patients with Digital Ulcers is mapped closely together, and we can distinctly identify regions associated with Esophageal symptoms. Additional plots and discussion for other variables are provided in Supplementary Note 5.
Hierarchical disease subtyping: first hierarchy of clusters
To perform disease subtyping, we followed the hierarchical clustering approach described in section “Trajectory Clustering”. We identified two primary clusters, and then further subdivided each of these into more granular subtypes. The first hierarchy of clusters distinguishes between patients with milder and more severe disease trajectories. The second level divides the mild group into two subtypes and the more severe group into three subtypes (Fig. 1 Panel 3.). In the following, we provide a detailed description of each cluster, followed by a discussion on the differences between clusters, highlighting the key variables driving cluster separation. For every organ, we plotted the empirical involvement and severity curves by averaging the model-inferred probabilities across all patient visits belonging to a given cluster at each follow-up visit.
In the first hierarchy of clusters, patients are split into two clusters (Fig. 4): a mild cluster (green) and a severe cluster (purple).
-
Mild Cluster (green): Patients have moderate to high probabilities of GT, heart and skin involvement, and an increasing likelihood of DU. They have a low risk of severe symptoms across all organs.
-
Severe Cluster (purple): Compared to the mild clusters, patients have a higher likelihood of lung involvement, and exhibit high severity of skin symptoms. Severity is additionally elevated for both heart and lung symptoms.
These observations align with established SSc subtypes based on skin severity (limited vs. diffuse/severe)21 and previous findings linking severe skin involvement with earlier, more frequent internal-organ complications22,23 as well as more pronounced ILD24. Supplementary Fig. 11 compares the average feature values over time in both clusters. Overall, patients in the severe cluster exhibit higher modified rodnan skin scores (mRSS), more dyspnea, increased lung fibrosis (on HRCT and X-ray) and lower forced vital capacity (FVC) compared to those in the mild cluster.
Second hierarchy of clusters
This hierarchy further subdivides the clusters: the mild disease trajectory cluster is split into two subtypes (pale and dark green, Fig. 5A.), while the severe disease trajectory cluster is divided into three subtypes (pale blue, dark blue, and red, Fig. 5B.).
A Mild Disease Subtypes. Patients with milder disease trajectories are further divided into two subtypes. The dark green cluster shows slightly higher probabilities of skin, heart, and GT involvement compared to the pale green cluster. B Severe Disease Subtypes. Patients with severe disease trajectories are subdivided into three subtypes: pale blue, dark blue, and red. The pale blue cluster is marked by severe skin involvement; the dark blue cluster by pronounced heart and lung involvement; and the red cluster by combined skin, GT, lung, and heart involvement.
Figure 5A. shows the average label values over time for the patients categorized in the two mild disease subtypes. In particular, the clusters have the following characteristics:
-
Pale Green Cluster: Patients in this cluster have a high likelihood of skin involvement (non-severe). They have moderate probabilities of heart and GT involvement and experience an increasing probability of DU involvement over time. The probability of severe involvement remains low for all organs.
-
Dark Green Cluster: Patients have a comparatively higher likelihood of heart but particularly GT involvement. Additionally, there is comparatively faster rise in kidney involvement. Symptom severity remains low across organs.
In summary, patients in the pale green cluster generally experience the mildest disease, while those in the dark green cluster exhibit slightly increased risks—particularly for GT and heart involvement. These patterns suggest that even among patients with limited (i.e. non-severe) skin involvement, a subgroup exists with higher probabilities of GT and cardiac issues24. The dark green cluster shows an increasing trend in dyspnea, lower eGFR, and more frequent esophageal symptoms and recurrent DU (Supplementary Fig. 12). Figure 5B. shows the average label values over time for the patients categorized in the more severe disease subtypes. In particular, the clusters have the following characteristics:
-
Pale blue cluster: Patients in this cluster experience high probabilities of severe skin involvement, with slightly increased severity of lung symptoms. Given overall high organ involvement, these patients show prototypical characteristics of diffuse cutaneous SSc, with elevated risks for heart, ILD, GT, and DU24.
-
Red cluster: Compared to the pale blue cluster, patients experience elevated but slightly lower skin severity, with higher severity of heart, lung, GT and DU symptoms. These diffuse cutaneous SSc patients are at high risk for multi-organ complications.
-
Dark blue cluster: Patients in this cluster have even lower skin severity, while still experiencing elevated levels of heart and lung symptoms. Importantly, using the current disease classification criteria based on skin severity, these patients may be overlooked despite facing a high risk of multi-organ complications25,26.
In summary, while all three severe subtypes show high probabilities of skin involvement, only the pale blue and red clusters exhibit severe skin manifestations. Importantly, patients in the dark blue cluster may be overlooked due to their limited skin manifestations, even though they face high mortality risk from ILD and heart complications25,26. Feature comparisons (Supplementary Fig. 12) show that the dark blue cluster has a higher likelihood of lung fibrosis on HRCT or X-ray, while the red cluster is more prone to esophageal or stomach symptoms. Both the red and dark blue clusters experience increasing dyspnea over time, and the pale blue cluster maintains higher eGFR levels compared to the other two groups.
In summary, cluster separation is primarily driven by lung, skin, heart, and gastrointestinal involvement. For mild trajectories, two clusters emerged—both with low probabilities of severe organ involvement, though one exhibits slightly higher overall organ involvement. Three subtypes of severe trajectories were identified: one cluster shows a high likelihood of severe skin involvement with minimal severe involvement elsewhere, while the other two present increased probabilities of severe lung and heart complications. Notably, we identified a high-risk cluster (dark blue) with limited skin severity.
Cluster stability
As described in subsection “Handling missing data”, we performed 5-fold CV, producing five models each trained on different subsets of the training data. We also reserved a hold-out test set—not included in the CV process—for an independent clinical evaluation of the clustering results. To assess how consistently the clusters formed across these models, we examined which features most strongly contributed to cluster separation. Specifically, for each cluster and each model, we computed the average value (or class probability) of every feature and then calculated the standard deviation of these averages across the clusters. A higher standard deviation indicates a greater influence on cluster separation. Ranking the features by this standard deviation revealed that the same subset of features consistently drove clustering across models. The bar charts in Fig. 6 illustrate the standard deviation of feature values, with larger bars indicating more pronounced variability across clusters and error bars capturing variation among the five models. Notably, the error bars are generally small, suggesting strong consistency in feature ranking across the models. These findings also confirm the trends discussed in section “Hierarchical Disease Subtyping: First Hierarchy of Clusters”, where skin- and lung-related features are the primary drivers of cluster separation.
Clinical decision support system
Using our trained model, we can build a clinical decision support system that enables predicting future patient latent trajectories and early identification of disease subtypes. By comparing predicted cluster assignments at different stages of a patient’s journey to the final cluster assignment—after all medical visits have been encoded—we can anticipate the most likely disease subtype early in the disease course. Figure 7 illustrates these capabilities within a CDSS for a sample patient:
-
Panel A: Predicted (in blue) versus final (in red) latent trajectory, with corresponding cluster assignments.
-
Panel B: Final trajectory alongside nearest neighbors.
-
Panel C: Trajectories of key clinical variables for the patient and nearest neighbors.
-
Panel D: Trajectories of selected medical labels for the patient and nearest neighbors.
A The model predicts future latent trajectories and assigns patients to likely severity subtypes. For an index patient, it visualizes their latent trajectory and predicted disease progression (start at the X). B Similar trajectories to the index patient can be identified using k-nearest neighbors (start at the X). C Medical feature trajectories of the retrieved similar patients can be visualized and compared. D Organ involvement trajectories of these similar patients can also be visualized and compared.
For this patient, the CDSS suggests they likely belong to the purple subtype, characterized by a high risk of severe skin involvement (Fig. 7A.). Similar patients are located in regions with likely lung fibrosis and esophageal symptoms (Fig. 7B.). Moreover, as shown in Supplementary Fig. 15b, predicting cluster assignment at various stages of a patient’s journey to the final cluster yields a high F1 score (around 0.8), demonstrating the model’s effectiveness in early severity stratification. This capability allows clinicians to intervene sooner, potentially mitigating organ involvement. Furthermore, following the procedure in section “Trajectory Clustering”, our model identifies the top-k similar patient trajectories (here, k = 3) to any given patient from the test set. Clinicians can leverage this feature to compare disease progressions, offering insights into a patient’s likely trajectory.
Discussion
In this work, we introduced a semi-supervised generative deep learning model that leverages expert-defined disease criteria to capture the complexity of systemic sclerosis across eight organs. Our approach uncovered five distinct hierarchical SSc subtypes spanning a mild-to-severe spectrum (Fig. 5). Among the two “mild” subtypes, one cluster showed only little involvement, whereas the other displayed higher tendencies for GT and heart issues. In the “severe” subtypes, we found one cluster aligned with a classic diffuse disease profile and elevated multi-organ involvement, another marked by pronounced multi-organ severity, and a particularly noteworthy cluster with limited skin involvement yet elevated risks of lung and heart complications. This highlights the shortcomings of relying on skin phenotypes alone.
These findings underscore the clinical utility of combining expert-guided label definitions with data-driven representation learning. By leveraging even partially labeled information, the model aligned learned trajectories with known clinical patterns, while also revealing less apparent subtypes that may carry significant morbidity risk. Overall, our approach moves beyond skin-based distinctions, offering a framework for translating complex patient data into interpretable, actionable insights to support personalized clinical decision support.
The primary limitation of our approach stems from the challenge of modeling highly imbalanced and sparse datasets. We observed that organ dynamics with highly imbalanced data tended to have less impact on subtyping, suggesting the need to investigate techniques like re-weighting minority classes during training. Alternatively, a more targeted model could be developed, focusing only on specific labels rather than the holistic approach used in this study.
Next, we plan to leverage the learned latent trajectories to answer questions specific to particular patient subsets, for instance, patients who develop ILD early in the disease course. By pretraining our model on the full dataset and subsequently clustering only within the ILD cohort, we can uncover ILD-specific subtypes.
Furthermore, our choice of five clusters, although guided by both mathematical and clinical validation, should not be interpreted as a definitive “ground truth”. For more fine-grained results, a similar hierarchical strategy could be extended through further sub-clustering, potentially revealing additional patterns in sparser organ dynamics.
Finally, the present study is purely retrospective, relying on observational patient data. A key limitation is the absence of a healthy-control reference: the EUSTAR registry does not enroll unaffected individuals, and no external cohort provides longitudinal, organ-specific assessments of comparable granularity. As a result, our analysis is confined to delineating phenotypic heterogeneity within the SSc population rather than benchmarking these trajectories against normative patterns. A possible next step would be to conduct a silent prospective evaluation in clinical practice to assess how well the model supports rheumatologists’ decision-making in real-time.
Methods
Analyzing and comparing raw longitudinal patient trajectories presents significant challenges due to heterogeneity, temporality, missingness, and biases9. To overcome these issues, we propose a two-stage approach. First, we develop a deep learning model to transform raw, heterogeneous data into smoother temporal patient representations. These refined representations are then used for disease subtyping through temporal clustering. Supplementary Note 11 summarizes the key machine-learning concepts referenced in this work.
Cohort description
We use SSc patient data from the European Scleroderma Trials and Research group (EUSTAR) registry (database export from June 1, 2022), a comprehensive dataset detailed in refs. 17,27. This study was conducted in accordance with the Declaration of Helsinki and was approved by the local ethical committees of the participating EUSTAR centers. All patients provided written informed consent for their data to be used for research purposes as required by the local ethics committees for this study. The project was approved by the EUSTAR board (project number: CP125).
After preprocessing, the database comprises 14, 060 patients and 67, 894 medical visits, averaging approximately 4.8 medical visits per patient, see Supplementary Fig. 2 for the distribution of the number of patient visits. We included demographic variables such as gender and age, along with temporal variables measuring the disease progression across different organs, following the variable selection approach detailed in section “Variable selection for organ-specific definitions”. Moreover, Supplementary Note 2 provides additional details about the database, such as feature distribution plots (Supplementary Figs. 3 and 4 and Supplementary Tables 2 and 3) and a list of variable names with brief descriptions (Supplementary Table 1). To facilitate comparison with other EUSTAR studies, we retained the original variable names from the EUSTAR database when they were sufficiently clear.
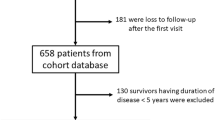
We excluded patients with fewer than two or 15 and more medical visits and removed outliers. Additionally, all patients included in the analysis were 18 years or older. Patients with at least 15 medical visits were excluded to avoid biasing the model towards a few heavily sampled trajectories. A consort diagram describing patient inclusion during the different steps of our analysis is shown in Supplementary Fig. 1. Prior to model training or application, continuous variables were standardized, and categorical variables were one-hot encoded.
Variable selection for organ-specific definitions
For each organ, we model two dynamics: (a) involvement and (b) severity stage (if applicable), representing organ-specific outcome labels. These labels are computed based on clinical definitions (i.e. list of criteria) applied to a set of organ-specific variables recorded in the dataset.
More specifically, to create these labels, (1) we first reviewed the literature to compile all clinical definitions for each organ, usually ending up with multiple definitions per label (i.e. definitions for organ involvement and organ severity). (2) We then identified the relevant clinical variables available in the EUSTAR database (list of variables per definition), resulting in an extensive set of input variables X to describe organ dynamics. (3) In the second stage, a steering committee of ten SSc experts from various EUSTAR centers selected the most clinically relevant definition for each organ and label19. The final definitions are provided in Supplementary Note 3, and this process yielded a refined subset of EUSTAR variables G ⊆ X, derived from the final definitions. A complete list of variables in X and G is available in Supplementary Note 2. Panel 1 in Fig. 1 illustrates the variable selection process of our study. Note that autoantibody profiles were intentionally omitted, as their prognostic value in SSc is already well-documented, and our objective was to derive patient subtypes exclusively from longitudinal organ-specific trajectories.
Model overview and notations
For each patient, our model learns to summarize raw medical measurements into organ-specific representations that encode both the presence and severity of organ involvement. A sequence of these representations yields a longitudinal trajectory for every patient, and clustering those trajectories uncovers five distinct SSc phenotypes, each with a characteristic pattern of multi-organ disease. Following standard ML practice, we develop and tune the model on a training partition of the data and reserve an independent test set for final evaluation, confirming that the identified phenotypes generalize to previously unseen patients. See Supplementary Note 11 for an overview of the key ML concepts.
As outlined in section “Variable selection for organ-specific definitions”, the temporal input variables set X comprises a broad range of clinical measurements related to organ dynamics. Furthermore, a more refined subset of these variables, G ⊆ X, reflects the latest medical knowledge on organ impact in SSc. These variables are continuous, binary, or categorical, with all categorical variables being ordinal. For each patient, let x ≔ x1:T ∈ X and g ≔ g1:T ∈ G, where \(x\in {{\mathbb{R}}}^{D\times T}\) and \(g\in {{\mathbb{R}}}^{P\times T}\) represent the temporal clinical measurements, T is the index of the most recent measurement (i.e. last available in the database), and D and P are the number of variables in X and G respectively. Additionally, we define m ≔ m1:T ∈ M, where \(m\in {{\mathbb{R}}}^{D\times T}\) is a boolean mask indicating the availability of clinical variables. We also incorporate N static demographic variables s ∈ S, \(s\in {{\mathbb{R}}}^{N}\). Our goal is to model the distribution of L latent, i.e. unobserved, variables z ≔ z1:T ∈ Z, where \(z\in {{\mathbb{R}}}^{L\times T}\), that generate the observed X and G conditioned on S. These latent variables should contain the key information necessary to reconstruct X and predict G.
Model architecture
We adopt a probabilistic approach leveraging and adapting the well-established variational autoencoder (VAE) framework12 to learn interpretable latent (unobserved) temporal organ-specific representations. Our method is designed to model organ behaviors in SSc by learning from the entire dataset while separately modeling each organ, thereby facilitating the analysis of organ-specific dynamics. We build on our prior deep probabilistic model14, in which we designed a temporal VAE-based approach to model the behavior of three organs (lungs, heart, and joints) in SSc to perform online patient monitoring. A key design element is “guiding” distinct latent dimensions for each organ (i.e. non-overlapping subsets of dimensions of the z vector), ensuring each subset of the latent dimension learns specialized organ-specific trajectories. In ref. 14, we used preliminary label definitions to guide these dimensions in a semi-supervised manner, training separate networks to predict all clinical variables. Here, we instead focus on learning predictive latent processes specifically for the organ-related variables G, with final label definitions aimed at improving disease subtyping.
We model eight organs (previously three), adapting the architecture to handle higher dimensionality and missing data. As in ref. 14, we dedicate separate latent dimensions to learn each organ’s dynamics (see Fig. 1). Following the bottleneck principle, the model is trained to reconstruct the variables in X. Additionally, we implement individual multilayer perceptrons (MLPs) as “guidance” networks for each variable in G. These networks receive the organ-specific latent subsets and learn to reconstruct and predict the current and future values of their respective variables. Intuitively, we integrate these organ-specific medical definitions as partial labels to guide the latent space for each organ dimension. We also train our model using an additional mask (denoted feature masking) by randomly dropping 20% of the input features to make the model more robust in reconstructing missing data (see subsection "Handling missing data"). In summary, for each patient, given x1:t, s, m1:t, and g1:t, the model learns the distribution of z1:T and uses a sampled z to reconstruct and predict x1:t and g1:T. The encoder network relies on MLPs and Long Short-Term Memory networks (LSTMs)28, while the decoder and guidance networks are independent MLPs (Fig. 1). A separate neural network models the prior distribution of z (not shown in Fig. 1).
Handling missing data
The model expects a fixed-length input vector, so unobserved measurements are initially filled with cohort means computed on the training split. We then supply an accompanying missingness mask m that flags every imputed entry. The encoder, therefore, sees two channels per variable: its (possibly imputed) value and its missingness mask (i.e. boolean indicator). During training, the reconstruction/prediction loss is computed exclusively on observed values; imputed placeholders are ignored. In addition, we randomly drop 20% of the observed inputs in every mini-batch ("feature masking”). This forces the decoder to learn the joint structure of the data and produces reliable model-based imputations. All analyses, therefore, operate on the reconstructed time series, preventing bias from simple mean imputation even for variables with very high missingness. A detailed ablation showing the resulting robustness is reported in Supplementary Table 4.
Training
We first split the full dataset into training and validation (85%) and test (15%) sets; the training portion was used exclusively for model development and tuning, while the test set remained untouched until final evaluation. We then performed five-fold cross-validation (CV) on the training data: the training set was divided into five equal folds, and in turn, one fold served as a validation set while the model was trained on the other four. Within each training split, we executed a random search over hyperparameter combinations, selecting the configuration that minimized validation loss. This procedure yielded five separate final models, one per fold. To assess the stability and consistency of the results, each of the five models is then evaluated on the independent 15 % hold-out test set that was never seen during training and tuning.
To train our model, we adapted the objective function from refs. 14,18 to our specific setting. We outline the key aspects of the optimization process here and refer the reader to ref. 14 for detailed computational information. Consider observational patient data x1:T, g1:T and s, where T is the index of the most recent clinical measurement. For each time step t = 1, . . . , T, given x1:t, the model is trained to predict the distribution of the full latent trajectory z1:T. Using a sample of this latent distribution, the guidance decoders are then trained to reconstruct and predict gt:T, minimizing the cross entropy loss for binary or categorical variables and the mean squared error (MSE) for continuous variables. Similarly, the decoder is trained to reconstruct x1:t given z1:T, also using cross-entropy or MSE depending on the variable type. The model learns the distribution of the latent space by minimizing the Kullback-Leibler (KL) divergence, a regularization term that aligns the prior assumptions about the latent space with the distribution learned by the encoder. Following the approach in ref. 14, we assume a Gaussian distribution with constant variance for continuous variables, Bernoulli or categorical distributions for binary and categorical variables, and a Gaussian prior distribution for the latent space. During the model training, the parameters of these predefined distributions are learned and optimized.
Importantly, when computing the loss, we only include the observed (non-missing) variables. This ensures that the model is not trained to reconstruct imputed data, reducing potential bias. Furthermore, to enhance the model’s ability to handle missing data, we randomly mask 20% of the available clinical measurements in each batch during each training epoch. We use the Adam29 algorithm with mini-batch processing to optimize the objective function.
Trajectory clustering
For clustering, we used k-means with dynamic time-warping (DTW) distance30 on the learned latent patient trajectories. DTW allows us to align patient trajectories with varying length. After model training, k-means centroids were learned only on the embeddings from the training data. The 15 % hold-out cohort was subsequently projected into the same latent space and assigned to the nearest centroids. Reporting cluster characteristics on this unseen test set, therefore, provides a strictly out-of-sample evaluation of our subtyping approach. To determine the optimal number of clusters, we varied k from 2 to 15, and evaluated the clustering performance by computing the inertia, which measures cluster compactness (Supplementary Fig. 15a), prompting us to set k = 5. Then, for k ∈ [2, 3, 4, 5], we assigned the test embeddings to the nearest cluster centers. We observed a natural hierarchy in the clustering process: as k increased, new clusters were almost perfectly nested within the existing ones (Supplementary Fig. 14). For instance, when k = 2, let \({c}_{1}^{2}\) and \({c}_{2}^{2}\) be the identified clusters. As k increased to 5, \({c}_{1}^{2}\) split into two clusters (\({c}_{1}^{5}\) and \({c}_{2}^{5}\)), while \({c}_{2}^{2}\) divided into three clusters (\({c}_{5}^{3}\), \({c}_{5}^{4}\), and \({c}_{5}^{5}\)). This inherent hierarchy led us to adopt a strict hierarchical clustering approach for the final cluster assignment, resulting in more interpretable and clinically meaningful groupings. Following this procedure, we identified k = 5 main clusters and identified a natural hierarchy among the clusters.
Similarly, we used a k-Nearest Neighbors method to identify similar patients (here k=3), retrieving each test patient’s closest trajectories from the training data (based on the DTW distance).
Data availability
The raw dataset is owned by the EUSTAR group, and may be obtained by request after approval and permission from the EUSTAR board.
Code availability
The code is available at https://github.com/uzh-dqbm-cmi/eustar_npj.
References
Denton, C. P. & Khanna, D. Systemic sclerosis. Lancet 390, 1685–1699 (2017).
Del Galdo, F. et al. Eular recommendations for the treatment of systemic sclerosis: 2023 update. Ann. Rheum. Dis. 84, 29–40 (2025).
Jaeger, V. K. et al. Incidences and risk factors of organ manifestations in the early course of systemic sclerosis: a longitudinal eustar study. PloS one 11, e0163894 (2016).
Hoffmann-Vold, A.-M. et al. Setting the international standard for longitudinal follow-up of patients with systemic sclerosis: a delphi-based expert consensus on core clinical features. RMD open 5, e000826 (2019).
Elhai, M. et al. Stratification in systemic sclerosis according to autoantibody status versus skin involvement: a study of the prospective eustar cohort. Lancet Rheumatol. 4, e785–e794 (2022).
Nihtyanova, S. I. et al. Using autoantibodies and cutaneous subset to develop outcome-based disease classification in systemic sclerosis. Arthritis Rheumatol. 72, 465–476 (2020).
Fretheim, H. et al. Multidimensional tracking of phenotypes and organ involvement in a complete nationwide systemic sclerosis cohort. Rheumatology 59, 2920–2929 (2020).
Petelytska, L. et al. Heterogeneity of determining disease severity, clinical course and outcomes in systemic sclerosis-associated interstitial lung disease: a systematic literature review. RMD open 9, e003426 (2023).
Allam, A., Feuerriegel, S., Rebhan, M. & Krauthammer, M. Analyzing patient trajectories with artificial intelligence. J. Med. internet Res. 23, e29812 (2021).
Lee, C. & Van Der Schaar, M. Temporal phenotyping using deep predictive clustering of disease progression. In International conference on machine learning, 5767–5777 (PMLR, 2020).
Chen, I. Y., Joshi, S., Ghassemi, M. & Ranganath, R. Probabilistic machine learning for healthcare. Annu. Rev. Biomed. data Sci. 4, 393–415 (2021).
Kingma, D. P. & Welling, M. Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114 (2013).
Locatello, F. et al. A sober look at the unsupervised learning of disentangled representations and their evaluation. J. Mach. Learn. Res. 21, 1–62 (2020).
Trottet, C. et al. Semi-Supervised Generative Models for Disease Trajectories: A Case Study on Systemic Sclerosis. Machine Learning for Healthcare Conference. PMLR, 2024.
Allam, A. et al. Predicting interstitial lung disease progression in patients with systemic sclerosis using attentive neural processes-a eustar study. medRxiv 2024–04 (2024).
Bonomi, F. et al. The use and utility of machine learning in achieving precision medicine in systemic sclerosis: a narrative review. J. Personalized Med. 12, 1198 (2022).
Meier, F. M. et al. Update on the profile of the eustar cohort: an analysis of the eular scleroderma trials and research group database. Ann. Rheum. Dis. 71, 1355–1360 (2012).
Trottet, C., Schürch, M., Mollaysa, A., Allam, A. & Krauthammer, M. Generative time series models with interpretable latent processes for complex disease trajectories. In Deep Generative Models for Health Workshop NeurIPS 2023 (2023).
Hoffmann-Vold, A. et al. Pos0203 evidence-based expert consensus definition of organ involvement in systemic sclerosis–a eustar study (2024).
McInnes, L., Healy, J. & Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 (2018).
Varga, J.Systemic Sclerosis (Scleroderma) and Related Disorders (McGraw-Hill Education, New York, NY, 2018). accessmedicine.mhmedical.com/content.aspx?aid=1179365261.
Herrick, A. L., Assassi, S. & Denton, C. P. Skin involvement in early diffuse cutaneous systemic sclerosis: an unmet clinical need. Nat. Rev. Rheumatol. 18, 276–285 (2022).
Steen, V. D. & Medsger Jr, T. A. Severe organ involvement in systemic sclerosis with diffuse scleroderma. Arthritis Rheumatism: Off. J. Am. Coll. Rheumatol. 43, 2437–2444 (2000).
Adigun, R., Goyal, A. & Hariz, A.Systemic Sclerosis (Scleroderma) (StatPearls Publishing, Treasure Island (FL), 2025), updated 2024 apr 5 edn. Available from: https://www.ncbi.nlm.nih.gov/books/NBK430875/.
Campochiaro, C. & Matucci-Cerinic, M. Interstitial lung disease in limited cutaneous systemic sclerosis patients: never let your guard down (2024).
Zanatta, E. et al. Phenotype of limited cutaneous systemic sclerosis patients with positive anti-topoisomerase i antibodies: data from the eustar cohort. Rheumatology 61, 4786–4796 (2022).
Hoffmann-Vold, A.-M. et al. Progressive interstitial lung disease in patients with systemic sclerosis-associated interstitial lung disease in the eustar database. Ann. Rheum. Dis. 80, 219–227 (2021).
Hochreiter, S. Long short-term memory. Neural Computation MIT-Press (1997).
Kinga, D., Adam, J. B. et al. A method for stochastic optimization. In International conference on learning representations (ICLR), vol. 5, 6 (San Diego, California;, 2015).
Müller, M. Dynamic time warping. Information retrieval for music and motion 69–84 (2007).
Acknowledgements
The authors thank the patients and caregivers who made the study possible, as well as all involved clinicians from the EUSTAR who collected the data. A list of contributing centers can be found at https://eustar.org/centers/. C.T., M.S., A.A., and M.K. received funding from the Swiss National Science Foundation (grant number 201184) for this work.
Author information
Authors and Affiliations
Consortia
Contributions
A.H. and M.K. devised the study. C.T. and M.S. curated and analyzed the data and implemented the algorithms. C.T., M.S., A.A., L.P., O.D., A.H., and M.K. analyzed the results. MK, O.D., and A.H. supervised the project. C.T. wrote the original manuscript draft and prepared the figures. All authors critically reviewed, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
A.H. has/had consultancy relationship with and/or has received research funding from or has served as a speaker for the following companies in the area of potential treatments for systemic sclerosis and its complications in the last 36 months: Abbvie, Avalyn, CallunaPharma, BMS, Boehringer Ingelheim, Genentech, Janssen, Merck Sharp&Dohme, Medscape, Novartis, Pliant therapeutics, Roche and Werfen. A.H. is a CTD-ILD ERS/EULAR convenor and a EULAR study group leader on the lung in rheumatic and musculoskeletal diseases.OD has/had consultancy relationship with and/or has received research funding from or has served as a speaker for the following companies in the area of potential treatments for systemic sclerosis and its complications in the last two years: 4P-Pharma, Abbvie, Acepodia, Aera, AnaMar, Anaveon AG, Argenx, Boehringer Ingelheim, BMS, Calluna, Cantargia AB, Citus AG, CSL Behring, Galderma, Galapagos, Hemetron AG, Innovaderm, Lilly, MSD Merck, Mitsubishi Tanabe; Nkarta Inc., Orion, Pilan, Quell, Scleroderma Research Foundation, EMD Serono, Topadur and UCB. Patent issued “mir-29 for the treatment of systemic sclerosis” (US8247389, EP2331143). OD is a co-founder of CITUS AG. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Trottet, C., Schürch, M., Allam, A. et al. Deep hierarchical subtyping of multi-organ systemic sclerosis trajectories - a EUSTAR study. npj Digit. Med. 8, 563 (2025). https://doi.org/10.1038/s41746-025-01962-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-025-01962-y