Abstract
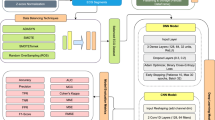
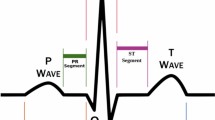
The clinical deployment of artificial intelligence (AI) solutions for assessing cardiovascular disease (CVD) risk in 12-lead electrocardiography (ECG) is hindered by limitations in interpretability and explainability. To address this, we present xGNN4MI, an open-source framework for graph neural networks (GNNs) in ECG modeling for interpretable CVD prediction. Our framework facilitates modeling clinically relevant spatial relationships between ECG leads and their temporal dynamics. We integrated explainable AI (XAI) and developed a task-specific XAI evaluation and visualization workflow to identify ECG leads crucial to the model’s decision-making process, enabling a systematic comparison with established clinical knowledge. We evaluated xGNN4MI on two challenging tasks: diagnostic superclass classification and localization of myocardial infarction. Our findings show that the interpretable ECG-GNN models demonstrate good performance across the tasks. XAI analysis revealed clinically meaningful training effects, such as differentiating between anteroseptal and inferior myocardial infarction. Our work demonstrates the potential of ECG-GNNs for providing trustworthy and interpretable AI-based CVD diagnosis.
Similar content being viewed by others
Data availability
The data of the SHIP study cannot be made publicly available due to the informed consent of the study participants, but it can be accessed through a data application form available at https://fvcm.med.uni-greifswald.de/ for researchers who meet the criteria for access to confidential data.
Code availability
The data analysis code will be freely available on GitHub following publication of the paper (https://github.com/HauschildLab/xGNN4MI). Please contact Miriam Cindy Maurer at miriamcindy.maurer@med.uni-goettingen.de.
References
WHO. World Health Organization https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (1948).
Xie, L., Li, Z., Zhou, Y., He, Y. & Zhu, J. Computational diagnostic techniques for electrocardiogram signal analysis. Sensors 20, 6318 (2020).
de Jager, J., Wallis, L. & Maritz, D. ECG interpretation skills of South African emergency medicine residents. Int. J. Emerg. Med. 3, 309–14 (2010).
Thygesen, K. et al. Third universal definition of myocardial infarction. Circulation 126, 2020–2035 (2012).
Sachdeva, P. et al. Advancements in myocardial infarction management: exploring novel approaches and strategies. Cureus 15, e45578 (2023).
Mechanic, O. J., Gavin, M. & Grossman, S. A. Acute Myocardial Infarction (updated 2023 Sep 3) (StatPearls Publishing, 2023); https://www.ncbi.nlm.nih.gov/books/NBK459269/.
Ribeiro, A. H. et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 11, 1760 (2020).
Raghunath, S. et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat. Med. 26, 886–891 (2020).
Lima, E. M. et al. Deep neural network-estimated electrocardiographic age as a mortality predictor. Nat. Commun. 12, 5117 (2021).
Maurer, M. C. et al. Explainable artificial intelligence on biosignals for clinical decision support. In KDD 24: The 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, 6597–6604 (KDD, 2024).
Ribeiro, M. T., Singh, S. & Guestrin, C. “Why should I trust you?” Explaining the predictions of any classifier. In Proc. of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 1135–1144 (ACM, 2016).
Guidotti, R. et al. A survey of methods for explaining black box models. ACM Comput. Surv. 51, 1–42 (2019).
EU AI Act. European Union Artificial Intelligence Act https://artificialintelligenceact.eu/ (2025).
Goettling, M., Hammer, A., Malberg, H. & Schmidt, M. xECGArch: a trustworthy deep learning architecture for interpretable ECG analysis considering short-term and long-term features. Sci. Rep. 14, 13122 (2024).
Bender, T. et al. Analysis of a deep learning model for 12-lead ECG classification reveals learned features similar to diagnostic criteria. IEEE J. Biomed. Health Inform. https://doi.org/10.1109/JBHI.2023.3271858 (2023).
Turbé, H., Bjelogrlic, M., Lovis, C. & Mengaldo, G. Evaluation of post-hoc interpretability methods in time-series classification. Nat. Mach. Intell. 5, 250–260 (2023).
Kutluana, G. & Türker, I. Classification of cardiac disorders using weighted visibility graph features from ECG signals. Biomed. Signal Process. Control 87, 105420 (2024).
Dathe, H., Krefting, D. & Spicher, N. Completing the cabrera circle: deriving adaptable leads from ecg limb leads by combining constraints with a correction factor. Physiol. Meas. 44, 105005 (2023).
Scarselli, F., Gori, M., Tsoi, A. C., Hagenbuchner, M. & Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw. 20, 61–80 (2009).
Wu, Z. et al. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 32, 4–24 (2021).
Li, R. et al. Graph signal processing, graph neural network and graph learning on biological data: a systematic review. IEEE Rev. Biomed. Eng. 16, 109–135 (2023).
Chereda, H., Leha, A. & Beißbarth, T. Stable feature selection utilizing graph convolutional neural network and layer-wise relevance propagation for biomarker discovery in breast cancer. Artif. Intell. Med. 151, 102840 (2024).
Zhang, H. et al. ST-ReGE: a novel spatial-temporal residual graph convolutional network for CVD. IEEE J. Biomed. Health Inform. https://ieeexplore.ieee.org/document/10292830/ (2023).
Qiang, Y. et al. Conv-rgnn: an efficient convolutional residual graph neural network for ECG classification. Comput. Methods Prog. Biomed. 257, 108406 (2024).
Zhao, X., Liu, Z., Han, L. & Peng, S. ECGNN: enhancing abnormal recognition in 12-Lead ECG with graph neural network. In: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 1411–1416 (IEEE, 2022).
Guo, L., Wu, Y., Ma, N. & An, Y. KGD-GNN: a knowledge-guided graph neural network for myocardial infarction localization via 12-lead ECG. In: ICASSP 2025—2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 1–5 (IEEE, 2025).
Kan, C., Ye, Z., Zhou, H. & Cheruku, S. R. DG-ECG: multi-stream deep graph learning for the recognition of disease-altered patterns in electrocardiogram. Biomed. Signal Process. Control 80, 104388 (2023).
Mueller, T. T. et al. Differentially private graph neural networks for whole-graph classification. IEEE Trans. Pattern Anal. Mach. Intell. 45, 7308–7318 (2023).
Wagner, P. et al. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 7, 154 (2020).
Vólzke, H. et al. Cohort profile update: the study of health in pomerania (SHIP). Int. J. Epidemiol. 51, e372–e383 (2022).
Ying, Z., Bourgeois, D., You, J., Zitnik, M. & Leskovec, J. Gnnexplainer: Generating explanations for graph neural networks. In Advances in neural information processing systems, 9244−9255 (Curran Associates Inc., 2019).
Ganschow, U. EKG-Kurs (KVM, Berlin, 2016).
Ruiz-Mateos, B. et al. Elevation of ST-segment in aVR is predictive of cardiogenic shock but not of multivessel disease in inferior myocardial infarction. J. Electrocardiol. 58, 63–67 (2020).
Vu, M. & Thai, M.T. PGM-Explainer: Probabilistic graphical model explanations for graph neural networks, In Advances In Neural Information Processing Systems, Vol. 33, (eds Larochelle H., Ranzato M., Hadsell R., Balcan M., Lin H.) pp. 12225–12235 https://dl.acm.org/doi/abs/10.5555/3495724.3496749 (Curran Associates, Inc., 2020).
Chen, R., Stewart, W. F., Sun, J., Ng, K. & Yan, X. Recurrent neural networks for early detection of heart failure from longitudinal electronic health record data: implications for temporal modeling with respect to time before diagnosis, data density, data quantity, and data type. Circ. Cardiovasc. Qual. Outcomes 12, e005114 (2019).
Zhang, X.-M., Liang, L., Liu, L. & Tang, M.-J. Graph neural networks and their current applications in bioinformatics. Front. Genet. 12, 690049 (2021).
Kipf, T. N. & Welling, M., Semi-Supervised Classification with Graph Convolutional Networks, In 5th International Conference on Learning Representations, Toulon, France, Conference Track Proceedings', OpenReview.net, (ICLR, 2017) https://doi.org/10.48550/arXiv.1609.02907.
He, K., Zhang, X., Ren, S. & Sun, J., Deep Residual Learning for Image Recognition, IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770−778, (Las Vegas, NV, USA, 2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q., Densely Connected Convolutional Networks, 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2261−2269, (Honolulu, HI, USA, 2017).
Yuan, H., Yu, H., Wang, J., Li, K. & Ji, S. On explainability of graph neural networks via subgraph explorations. In Proc. 38th International Conference on Machine Learning, Vol. 139, 12241−12252 https://doi.org/10.48550/arXiv.2102.05152 (2021).
Longo, L. et al. Explainable artificial intelligence (XAI) 2.0: A manifesto of open challenges and interdisciplinary research directions. Inf. Fusion 106, 102301 (2024).
Metsch, J. M. & Hauschild, A.-C. Benchxai: comprehensive benchmarking of post-hoc explainable ai methods on multi-modal biomedical data. Comput. Biol. Med. 191, 110124 (2025).
Klein, L., Lüth, C., Schlegel, U. & Bungert, T. El-Assady M., and Jäger P. Navigating the maze of explainable AI: a systematic approach to evaluating methods and metrics. In Proc. 38th International Conference on Neural Information Processing Systems (NIPS '24), Vol. 37, 67106–67146 (Curran Associates Inc., Red Hook, NY, USA, 2024).
Bender, T. et al. Analysis of a deep learning model for 12-Lead ECG classification reveals learned features similar to diagnostic criteria. IEEE J. Biomed. Health Inform. https://ieeexplore.ieee.org/document/10113187/ (2023).
Fey, M. & Lenssen, J. E., Fast graph representation learning with pytorch geometric, In ICLR 2019 Workshop on Representation Learning on Graphs and Manifolds. https://doi.org/10.48550/arXiv.1903.02428 (2019).
Acknowledgements
We gratefully acknowledge the computing time granted by the Resource Allocation Board and provided on the supercomputer Emmy at NHR@Göttingen as part of the NHR infrastructure, under the project nib00044. This work is supported in part by the German Federal Ministry of Research, Technology and Space (BMFTR) under grant agreement no. 01KD2208A and no. 01KD2414A (FAIrPaCT), grant no. 01ZZ2324A and no. 01ZZ2324C (Somnolink), by the Innovation Committee at the Federal Joint Committee no. 01VSF20014 (KI-Thrust), by the Lower Saxony “Vorab” of the Volkswagen Foundation and the Ministry for Science and Culture of Lower Saxony, grant no. 76211-12-1/21 and by the ACRIBiS project of the Medical Informatics Initiative (FK: 01ZZ2317B). SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-Western Pomerania, and the network “Greifswald Approach to Individualized Medicine (GANI_MED)” funded by the Federal Ministry of Education and Research (grant no. 03IS2061A).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.C.M.: conceptualization, methodology, software, validation, formal analysis, investigation, data curation, visualization, and writing—original draft, review, and editing. P.H: validation, software, and writing—review and editing. K.E.S.: investigation and writing—review and editing. H.C.: methodology and writing—review and editing. M.V.: resources and writing—review and editing. D.K.: supervision, funding acquisition, and writing—review and editing. N.S.: investigation, visualization, supervision, project administration, and writing—review and editing. A.-C.H.: investigation, funding acquisition, supervision, project administration, and writing—review and editing. All authors have read and approved the final manuscript
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maurer, M.C., Hempel, P., Steinhaus, K.E. et al. xGNN4MI: explainability of graph neural networks in 12-lead electrocardiography for cardiovascular disease classification. npj Digit. Med. (2026). https://doi.org/10.1038/s41746-026-02367-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-026-02367-1