Abstract
In order to cope with the complexity and variability of the terrestrial environment, amphibians have developed a wide range of reproductive and parental behaviors. Nest building occurs in some anuran species as parental care. Species of the Music frog genus Nidirana are known for their unique courtship behavior and mud nesting in several congeners. However, the evolution of these frogs and their nidification behavior has yet to be studied. With phylogenomic and phylogeographic analyses based on a wide sampling of the genus, we find that Nidirana originated from central-southwestern China and the nidification behavior initially evolved at ca 19.3 Ma but subsequently lost in several descendants. Further population genomic analyses suggest that the nidification species have an older diversification and colonization history, while N. adenopleura complex congeners that do not exhibit nidification behavior have experienced a recent rapid radiation. The presence and loss of the nidification behavior in the Music frogs may be associated with paleoclimatic factors such as temperature and precipitation. This study highlights the nidification behavior as a key evolutionary innovation that has contributed to the diversification of an amphibian group under past climate changes.
Similar content being viewed by others
Introduction
Understanding how traits facilitate species diversification and adaptive radiation is a fundamental goal in evolutionary biology. The appearances of specific morphological characteristics or behaviors in organisms are usually related to adaptations to specific abiotic environments during evolutionary history and thus act as key innovations contributing to the diversification of life1,2. Nonetheless, most studies focus on understanding morphological or physiological traits that facilitate a species’ survival abilities or exploitation on new resources, such as photosynthesis in cyanobacteria3,4, wings in insects5,6, and jaws in fishes7,8. Much less is known about key innovations that facilitate organisms’ reproduction and parental care, which in turn enhance their ability to adapt to climate change.
Amphibians, being the intermediate form of vertebrates that move from aquatic to terrestrial environments, play an important role in the evolution of tetrapods. The occurrences of four limbs and lungs enable the amphibian species to inhabit a wide variety of terrestrial habitats. However, their inability to produce amniotic eggs and their requirement for metamorphosis restrict them to close proximity with the freshwater environment, particularly during their vital reproductive periods. Thus, amphibians are vulnerable to environmental changes, and hence any traits that facilitate their resilience to climate change or harsh environments, especially during breeding in terrestrial environments, would promote their diversification9,10.
To overcome such environmental challenges and to safeguard their descendants during the fragile egg and larva stages, varied and even seemingly odd modes of incubating behaviors have evolved within anuran amphibians, such as gastric-brooding (Rheobatrachus spp.), mouthbrooding (Rhinoderma darwinii), and dorsal-brooding (Pipa pipa, Gastrotheca spp., etc.)11,12,13,14. Besides these, nest building (also known as nidification) is also regarded as an important strategy of parental care for anuran eggs, tadpoles, and juveniles15,16,17. Studies of nidification behavior usually focus on the most common foam nest type that occurs in many arboreal species (Limnodynastes peronii, Rhacophorus spp., etc.)18,19, while mud nests are relatively overlooked as it has only been documented in a few lentic species, such as Boana faber20, Limnonectes limborgi21, and some species of Nidirana22.
The Music frogs of the genus Nidirana are widespread in subtropical eastern and southeastern Asia, from the southernmost of the Ryukyu Archipelago, throughout southern China including Taiwan and Hainan islands, expanding to the northern Indochina Peninsula. They are known for their mud nest construction behavior which several congeners exhibit. This behavior can even be found in the generic name, as Nidirana is composed of Latin nīdus (“nest”) and rāna (“frog”)22. Among all of the known Nidirana species, ten congeners are reported to build mud nests for reproduction23,24,25,26,27,28. The male frogs will construct a retuse mud-based burrow as their nest, call within it to attract females, and subsequently engage in mating within the nest. The eggs and larvae are, thus, protected in the mud nest from predators and external environmental changes (Fig. 1a, b). Compared with the arboreal foam nests, mud nests are usually constructed at the bank of lentic regions that face the complex challenges from both aquatic and terrestrial environments, and such nidification behavior may serve as an innovative adaptation to enhance their resilience. Meanwhile, other congeners of Nidirana do not possess such complicated behavior and just court and mate in standing water surfaces like most of other lentic frogs (Fig. 1c, d).
a A male N. guangxiensis built a mud-based burrow as its nest and was calling inside it. b Eggs of N. daunchina in a mud nest. c A male N. mangveni called at the standing water surface. d Eggs of N. guangdongensis in the standing water surface. e Map showing sampling localities in this work. Numbers for localities are corresponding to the information in Supplementary Table S1. Locality circles for species exhibiting nidification behavior, lacking nidification behavior, and nidification behavior remaining unknown, are colored with red, blue, and white, respectively.
Considered as a defense mechanism in response to complicated habitats14,29, we hypothesized that the presence and absence of mud nesting may be closely linked to environmental changes. In the distributional range of Nidirana across subtropical eastern and southeastern Asia, the emergence of a predominant monsoonal climate during the Cenozoic profoundly shaped terrestrial habitats and biogeography30,31. The East Asian Monsoon reached its peak intensity during the mid-Miocene, followed by a period of weakening until a re-strengthening during the Pliocene. Such fluctuations led to notable changes in the precipitation and temperature during this time frame32,33,34, which might have played important roles in the evolution of nidification behavior in these lentic Music frogs. Furthermore, our previous studies and observations found that within Nidirana, species that display nidification behaviors are often highly endemic to narrow geographical areas, whereas congeners lacking nidification tend to be more widely distributed35,36. Collectively, these pieces of evidence indicate that the nidification behavior of these species may be correlated with habitat specialties and unique colonization histories.
To understand the evolutionary history of the Music frogs and their unique mud nest construction behavior, we carried out a series of phylogenomic and population genomic analyses based on an extensive sampling from 18 of 20 described species of the genus. We hypothesized that the presence and absence of nidification behavior are associated with ancient climate changes, and that this behavior further acts as a key innovation contributing to the diversification and colonization of these frogs. Firstly, we employed genome-wide single-nucleotide polymorphisms (SNPs) to investigate their time-calibrated phylogeny and reconstruct their ancestral distributions. Secondly, to uncover the speciation of related species, we performed analyses on population genetics and demography for the congener subset with or without nidification behavior. Lastly, we examined the correlation between the nidification behavior and various associated bioclimatic factors.
Results
Prevalent discordance between mitochondrial and SNPs’ phylogeny
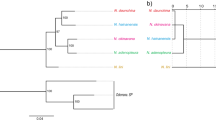
Both ML and BI analyses for mitochondrial phylogeny generated nearly identical topologies (Supplementary Fig. S1a). The genus Nidirana was monophyletic, and four major clades A–D were revealed with strong support (BS ≥ 90, BPP = 1.00, respectively). The relationship among these four clades was (A, (B, (C, D))), even though the support for (C, D) were slightly weak in the ML tree. Each of the clades was further comprised of single or multiple lineages with strong support values (BS ≥ 99, BPP = 1.00, respectively). In total 18 lineages can be recognized, and 17 of them corresponded to 17 described species. The remaining unnamed lineage of the “Poyang population”, which was previously recorded as N. adenopleura, was placed as a sister taxon to N. mangveni, thus this lineage was treated independently in the subsequent analyses. Furthermore, samples of N. “guibeiensis” clustered within N. leishanensis, indicating they are conspecific.
The phylogenetic result based on SNPs data (Fig. 2a; Supplementary Fig. S1b) showed a topology with several incongruences to the ones from the mitochondrial loci. The genus Nidirana was also recovered as monophyletic, but the interspecific relationships were not completely consistent. Clades A and B revealed from the mtDNA data were also supported by the SNPs data (BS = 100, respectively). However, taxa from mitochondrial clades C and D were intermixed and placed together in the phylogeny constructed from SNPs data (labeled as clade “C + D”, BS = 100). This group can be further divided into four clades N1, N2, N3, and N4 (BS = 100, respectively). The relationship among these six clades in the SNPs phylogeny was ((A, B), (N1, (N2, (N3, N4)))). At the species level, most of the species, including the synonymy of N. “guibeiensis” with N. leishanensis, were well recognized by SNPs data (BS = 100, respectively). Yet there were two exceptions: species within clade N4 (the N. adenopleura complex) and clade A (the N. pleuraden complex). As illustrated by plots of the PCA result of SNPs data (Fig. 2b), all sampled individuals were primarily grouped into six clusters, corresponding to the above six clades. Congeners that possess nidification behavior (clades N1 and N2) were close to each other in phylogeny and PCA plots, despite the unknown nidification status for N. yeae.
a Phylogeny of the genus Nidirana, using maximum likelihood based on SNPs data. Values of bootstrap supports (BS) larger than 90 are shown. Within the N. adenopleura complex, different populations are labelled with solid circles of different colors: N. mangveni, the Poyang population, N. guangdongensis, and four populations of N. adenopleura: Taiwan, north, center, and west. b Two-dimensional PCA plot for genetic variation in the genus Nidirana.
Spatiotemporal reconstruction of diversification patterns
The divergence time estimation from PAML indicated that the split between Nidirana and its sister taxon Babina was at ca 27.2 (95% CI 22.2–32.3) Ma (Supplementary Fig. S2). The Music frogs had the most recent common ancestor (MRCA) at ca 23.5 (95% CI 18.4–28.8) Ma. The divergence time for nidification species within clades N1 and N2 mainly occurred in the period of ca 10.0–15.4 Ma, while the divergence time for congeners without nidification behavior in clades A, N3, and N4 mainly occurred in the period of ca 2.1–9.3 Ma.
As inferred by the ancestral distribution reconstruction, it is highly likely that the MRCA of Nidirana inhabited the region encompassing the Yunnan-Guizhou Plateau (Area C) and Luoxiao Mountain Range and nonboring river basin (Area E), and then began to migrate westward or eastward as two groups (Fig. 3a, b; Supplementary Fig. S3). The first group (clades A and B) mostly moved westward across the Yunnan-Guizhou Plateau and entered into the western Yunnan longitudinal valleys (clade A) and tropical hills in the northern Indochinese Peninsula (clade B). The second group (clade “C + D”) primarily migrated eastward and subsequently split into three subgroups. One subgroup (clade N1) spread into different localities in Sichuan, Guangxi, Hainan, and Vietnam, where are close to the birthplace of the genus. Another subgroup (clade N2) migrated southeastward to Guangdong, with one population eventually dispersing to the Ryukyu Archipelago. The final subgroup (clades N3 and N4) evolved into the populations found in regions of central China (clade N3), as well as eastern mainland China and Taiwan Island (clade N4). Analyses on range shifts through time showed that the Music frogs had a major episode of dispersal events since Pliocene (Fig. 3c) and a minor episode of cladogenetic changes events around the mid-Miocene (Fig. 3d).
a Ten geographical areas delineated for the genus Nidirana. Characters for geographical areas and dashed lines for barriers are corresponding to the information in Supplementary Table S2. b Ancestral distributions reconstructed by Dispersal-Extinction-Cladogenesis model. Areas with the highest probabilities at nodes are shown with the same characters in Fig. 3a. Within the N. adenopleura complex, populations are labelled with solid circles of different colors: N. mangveni (MV), the Poyang population (PY), N. guangdongensis (GD), and four populations of N. adenopleura: Taiwan (At), north (An), center (Ac), and west (Aw). c Dispersal events along branches through time. d Cladogenetic change events through time. e Development of East Asian Monsoon from Miocene with the fluctuations of fluvial mass accumulation rate representing level of precipitation and carbon isotopes representing level of temperature30,34,45.
Population structure and differentiation
The PCA result of the nidification species suggested six clusters (Fig. 4a), corresponding to six recognized species. Among them, clusters of Nidirana nankunensis and N. okinavana were relatively closer and forming a larger cluster, when compared with the other larger cluster including N. chapaensis, N. hainanensis, N. yaoica, and N. daunchina. This was consistent with their phylogenetic relationships (Fig. 2), and also reflected their different dispersal routes (Fig. 3b), which shaped their current distribution patterns (Supplementary Fig. S4a).
a PCA plot of PC1 and PC2 for nidification species. b PCA plot of PC1 and PC2 for the Nidirana adenopleura complex. c Admixture plot for nidification species with K = 6. d Admixture plot for the N. adenopleura complex with K = 4. e Demographic trajectories for different populations for five nidification species. Nidirana okinavana was not included in this analysis due to a limited sample size. f Demographic trajectories for different populations for seven populations of the N. adenopleura complex. Labels for different populations: N. chapaensis (CP), N. hainanensis (HN), N. yaoica (Y), N. daunchina (D), N. nankunensis (N), N. okinavana (OK), N. mangveni (MV), the Poyang population (PY), N. guangdongensis (GD), and four populations of N. adenopleura: Taiwan (At), north (An), center (Ac), and west (Aw).
The result of the Admixture analysis suggested that the nidification species was comprised of six genetic populations (K = 6) (Fig. 4c). Almost each of them was distinct without genetic mixtures from other populations, except Nidirana chapaensis that was slightly intermixed with N. hainanensis. Genetic differentiations among populations of the nidification species were at a high level, with the lowest differentiation being observed between N. yaoica and N. daunchina (FST = 0.329, dxy = 0.00193, respectively; Supplementary Fig. S4c).
For the Nidirana adenopleura complex without nidification behavior, the PCA plot showed that individuals were grouped into four clusters (Fig. 4b). Among them, samples of N. guangdongensis (GD), N. mangveni (MV), and the Poyang population (PY) were roughly separated from each other and formed three clusters. Surprisingly, samples of N. adenopleura were not gathered together but instead split into four populations, corresponding to their geographical distributions, respectively (Supplementary Fig. S4b): the samples from Taiwan Island (At) formed an independent cluster which was distinctly separated from others; samples from southern Zhejiang and northeastern Jiangxi (An) were gathered with MV; samples from northern Fujian and southeastern Jiangxi (Ac) were gathered with PY; and samples from southwestern Jiangxi (Aw) were gathered with GD. Therefore, the N. adenopleura complex was treated as seven populations in four clusters (At, An+MV, Ac+PY, and Aw+GD) in the subsequent analyses. Particularly, the rough relationship among clusters An+MV, Ac+PY, and Aw+GD in PCA result suggests their gradient of differentiation and admixture.
The Admixture analysis revealed that the Nidirana adenopleura complex was comprised of four genetic populations (K = 4) (Fig. 4d), consistent with the four clusters in the above PCA result. The insular population At was pure without genetic mixtures from other populations, while all of the other mainland populations were found to be somewhat intermixed. Samples within cluster An+MV had similar genetic mixture despite different proportions among individuals, and similar patterns also occurred within the other two clusters Ac+PY and Aw+GD. High genetic differentiation was found between population At and other populations (Supplementary Fig. S4d), which were more distinct in At vs. Aw (FST = 0.265, dxy = 0.00168) and At vs. An (FST = 0.177, dxy = 0.00148). Low genetic differentiation occurred within the clusters An+MV, Ac+PY, and Aw+GD (FST < 0.044, dxy < 0.00107, respectively).
Demographic trajectories and dynamics
The nidification species have a relatively plain demographic history, as can be seen from the demographic trajectories depicted at ca 5–10 Ma with the effective population size showing a distinct decrease (Fig. 4e). After this single dramatic decrease, no other distinct fluctuation could be observed for the effective population size of each of the nidification species until the present. Nidirana okinavana was not included in this analysis due to a limited sample size. By contrast, in the analysis of demographic trajectories from ca 5 Ma, the effective population size gradually decreased in each of the seven populations within the N. adenopleura complex (Fig. 4f). Even though the degrees of decrease and processes varied among different populations, no distinct bottlenecks or population expansion events were inferred. For demographic dynamics analysis for the Nidirana adenopleura complex, we selected three pairs of populations based on the upstream results (Figs. 2, 4; Supplementary Fig. 1): (1) An vs. MV, representing the populations that showed distant relationships in mtDNA tree but were in one cluster using SNPs data; (2) An vs. Ac, representing the populations that have been clustered into a single lineage in mtDNA tree but were separated distinctly in SNPs data; (3) At vs. MV, representing the populations that were placed distinctly using both datasets. The results revealed that reciprocal migrations were present between the populations pairs of An and MV, nonreciprocal migration was present from population Ac to population An, and migration was absent between the populations of At and MV (Supplementary Table S3).
Environmental correlation of nidification behavior
The GLMMs analysis indicated that the presence/absence of nidification behavior of Music frogs is markedly associated with 12 bioclimatic variables (P-values < 0.05) and especially three variables with P values < 0.01 (Supplementary Table S4). The six major variables were annual temperature range (Bio 7), temperature seasonality (Bio 4), precipitation seasonality (Bio 15), precipitation of warmest quarter (Bio 18), precipitation of coldest quarter (Bio 19), and minimum temperature of coldest month (Bio 6) (Fig. 5). As distinctly indicated in the boxplots and scatterplots, Nidirana species with nidification behavior are more likely to inhabit environments with smaller fluctuations in temperature (Bio7 and Bio4), warmer weather in the coldest month (Bio6), and higher precipitation of the warmest quarter but lower precipitation of the coldest quarter (Bio18, Bio 15, and Bio19), while the species without nidification behavior were adapted to the opposite conditions.
a Boxplots showing the correlation for absence/presence of nidification behavior with Bio 7 (annual temperature range), Bio 4 (temperature seasonality), Bio 6 (minimum temperature of coldest month), Bio 18 (precipitation of warmest quarter), Bio 15 (precipitation seasonality), and Bio19 (precipitation of coldest quarter). b Scatterplot showing the associations for absence/presence of nidification behavior with Bio 7 and Bio 18, with Bio4 and Bio15, and with Bio6 and Bio19.
Discussion
Single origin of nidification behavior
Previous phylogenetic studies using mitochondrial markers suggested the nidification behavior may evolve multiple times independently22,24. In this work, the mitochondrial phylogenetics with complete sample coverage of genus yielded the same result as previous studies. However, when a larger dataset of SNPs was used, the phylogenomic analyses revealed that the Nidirana species with nidification behavior essentially form two paraphyletic clades N1 and N2, and these two clades and another two clades N3 and N4, which are species without the nidification behavior, have a MRCA to form the clade “C + D” (Fig. 2). Populations within this “C + D” clade have a similar spatiotemporal history that represent the eastward colonizers (Fig. 3b).
Nidification is an obviously complicated behavior as it may be involved in the courtship and the mating site, and consists of different steps: building the burrow, emitting calls, and mating15,37,38, which can be reasonably expected to be of a single origin rather than evolving multiple times independently during their evolutionary history39,40. Furthermore, the above-mentioned steps of nidification are primarily initiated and performed by the male individuals, thus maternal mitochondrial data might take bias for the origin of this behavior. Hence, we consider that the behavior of nest construction is much more likely to have initially evolved at the MRCA of clade “C + D” (ca 19.3 Ma), and was subsequently lost at the MRCA of clades N3 and N4 (ca 11.4 Ma).
Different evolutionary histories of the congeners with different behavior
The nidification species have relatively consistent phylogenetic relationships in both analyses. Population genomics suggest that no sign of admixture and high levels of genetic differentiations among them. Demographic analyses further indicate their relatively stable population size and much plainer demographic history. As revealed from the spatiotemporal reconstruction, divergence time for these species mainly concentrated around the mid-Miocene, accompanying with a minor episode of cladogenetic changes of range evolution in this period. Thus, the nidification species are considered to have an older history of diversification and colonization.
By contrast, the Nidirana adenopleura complex without nidification behavior may have experienced a recent rapid radiation. There is discordance in the topologies derived from mtDNA and SNPs data for the complex, with four clusters revealed by SNPs data being different from the four lineages identified in the mitochondrial phylogeny. Several factors may drive such patterns, such as incomplete lineage sorting from ancestral polymorphism and introgressive hybridization between taxa41,42,43. Combining the results of demographic analyses, it is most likely that incomplete lineage sorting from ancestral polymorphism has driven these incongruent phylogenetic topologies. The decreasing effective population sizes without bottlenecks and population expansion for different populations indicate their isolations from each other, and secondary contacts are unlikely to exist. Besides, the migrations mostly occurred between populations with a closer SNPs relationship (An vs. MV), and became fewer (An vs. Ac) or even absent (At vs. MV) as the divergence increased, suggesting low likelihood of introgressive hybridization.
Moreover, similar to the condition of the Nidirana adenopleura complex, conflicting topologies are also observed in terminal lineages within clade A (namely N. pleuraden complex), and complicated intraspecific relationships are found within clade N3 (N. leishanensis and N. xiangica) (Fig. 2; Supplementary Fig. S1). All of these taxa are without nidification behavior, and their major divergent time is relatively close from each other (Supplementary Fig. S2). A recent rapid radiative evolution similar to that in the N. adenopleura complex could be also expected in these species, which is closely associated with the major episode of dispersal events since Pliocene.
Paleoclimate changes benefit Nidirana congeners with different behaviors
Environmental correlation analysis illustrates that nidification species tend to inhabit environments where the temperature is relatively stable and warmer and the precipitation is mainly concentrated in the warmest quarter, which are the typical characteristics of strengthened monsoon climate. However, the congeners without nidification behavior are adapted to the opposite conditions. Such contrasting tendencies suggest that the presence or absence of nidification behaviors are associated with variations in temperature and precipitation due to climate changes.
Through the ancient development stages of monsoons in eastern Asia (Fig. 3e), the East Asian Monsoon intensified to a maximum during the mid-Miocene. This resulted in an environment characterized by extremely high precipitation and stable temperature in southern China, which in turn facilitated the diversification of Music frogs exhibiting nidification. Since Pliocene, a recent rapid radiation occurred in Nidirana, generating the diversity of extant congeners lacking nidification behavior. During this time period, the East Asian Monsoon was weakening, resulting in a dryer and cooler climate.
The nidification behavior is revealed to occur before the mid-Miocene and the maximum of the East Asian Monsoon (Fig. 3b). During the intensification of monsoons in southern China, the constantly higher temperatures and higher levels of precipitation caused an increase of overland floods (Fig. 3e)44,45. For frogs inhabiting open lentic ecosystems such as ponds, swamps, and paddy fields23,26,28, constructing nests for mating and incubating eggs would act as a means to mitigate the impact caused by the perennial high temperatures, heavy rainfall, and frequent floods. Such nests would also be an advantage against desiccation after flood waters recede, especially during their breeding season in summer. Therefore, the frog group benefited from the nidification behavior in such climate and underwent species diversification. Since ca 8 Ma, the temperature cooled down and the East Asian Monsoon substantially weakened, resulting in decreased precipitation and overland floods (Fig. 3e)32. Consequently, lacustrine environments were well developed, forming abundant lentic ecosystems46. As a result, the Music frog congeners without nidification behavior presumably started their rapid radiation from that time onwards till the present since they were not constrained by the need for nesting during mating.
Nidirana possesses adaptative flexibility under climate changes
A nest is an animal architecture that is usually elaborate and painstakingly built to protect offspring from predation and environmental variables47,48. While nests are primarily associated with birds of various types47, such architecture can be found across vertebrate taxa, including the little-known nests constructed by anuran amphibians16,49.
Our work investigates the evolutionary history of the Music frogs and their unique mud nest construction behavior, which has received less attention in previous studies. These findings indicate that mud nests likely evolve as a key evolutionary innovation during the paleoclimate characterized by perennial high temperatures and heavy rainfall. However, it was subsequently lost in certain descendants as the environment transitioned to a dryer and colder state. In contrast to the nesting behavior observed in Music frogs, the foam nest in the Treefrog family Rhacophoridae has been found to be well-adapted for cold and dry settings18,50. Foam nests are usually built in trees and are regarded as one of the adaptative mechanisms for arboreal life, being a shelter against the dry air that is uneasy to be replaced18,50. On the contrary, the mud nests built on the bank of lentic regions serve as a residence during intense rainfall and floods and can be easily discarded when no longer needed. Therefore, the construction behavior of mud nests in Nidirana would exhibit greater evolutionary flexibility in response to environmental variations. Climate models have predicted a significant increase in temperature in the near future, accompanied with intensified heavy rainfall and flooding51,52. Given their evolutionary history, it is plausible that the nesting behavior could resurface in Nidirana as a strategy to endure the increasingly hot and humid climate and flourish in the future. However, it is claimed that climate changes during the Anthropocene are characterized by greater variability and an increase in extreme weather events53,54. Thus, the extent of Music frogs’ adaptability in the face of unpredictable future climate conditions remains undetermined.
Given its intricate nature, it is imperative to conduct further research to unveil the genetic basis of the nidification behavior. The regulatory mechanisms underlying this complicated behavior at genetic, physiological, and behavioral levels are not well understood, particularly regarding the transition between nidification and non-nidification. Nidirana congeners show great potential for studies in this field. Our present study sheds light on the external environmental forces, and further research will be conducted to clarify and comprehend the adaptative mechanisms when tetrapods evolved from water to the land.
Methods
Sampling and sequencing
Field surveys were conducted from 2014 to 2021, throughout the distribution range of the genus Nidirana (Fig. 1e; Supplementary Table S1). A total of 237 samples of 18 Nidirana from 83 localities were collected. Four additional samples of Babina and Odorrana species were collected as out-groups26. All specimens were fixed in 10% buffered formalin and later transferred to 70% ethanol; muscle or liver samples were obtained from euthanized specimens and then preserved in 95% ethanol and stored at −40 °C in the Museum of Biology, Sun Yat-sen University. All the procedures related animals were performed in accordance with the ethical guidelines and approval of the of Institutional Animal Care and Use Committee of Sun Yat-sen University (2005DKA21403-JK).
Genomic DNA was extracted using a CTAB-extraction protocol with Proteinase K to digest tissue. All the DNA integrities were inspected on a 1% agarose gel to ensure the completion of DNA. All collected samples, except two of Babina species due to the quantity limitation, were amplified and sequenced for two mitochondrion segments (mtDNA), namely partial 16 S ribosomal RNA gene (16 S) and partial cytochrome C oxidase 1 gene (COI). Primers, PCR amplification, and sequencing protocol followed our previous studies55,56,57. In addition, published sequences from 22 individuals (13 with both 16 S and COI data and nine with only 16 S; Supplementary Table S1) of Nidirana and Babina species were downloaded from GenBank and incorporated into our dataset for mitochondrial phylogenetic analysis.
Double-digest restriction site-associated DNA sequencing (ddRAD-seq)58 conducted by Shenzhen RealOmics (Biotech) Co., Ltd. was applied to a subset consisting of 192 samples of 16 Nidirana species and four samples of the out-groups (Supplementary Table S1). Restriction enzymes EcoRI and NlaIII were used for double digests. The enzyme digestion products were ligated to index adaptors for PCR-amplification. Paired-end 150-bp reads were generated on an Illumina Hiseq platform. Sequence data from ddRAD-seq were performed for de novo SNPs calling using Stacks v2.5359. Raw sequences were filtered using process_radtags tool, and reads with Phred scores lower than 20 were discarded and the rest were all trimmed to 130 bp. After that, the reads were assembled and SNPs were called based on the main Stacks pipeline: ustacks, cstacks, sstacks, gstacks, and populations. To ensure sufficient yields of polymorphic loci and exclude erroneous assemblies, we tested different combinations of parameters, and the final parameter set was: minimum depth of coverage required to create a stack (m) = 3, maximum mismatches allowed between stacks (M) = 2, and maximum allowed mismatches between loci when building the catalog (n) = 2. VCFtools v0.1.16 was used for quality filtering60, and the final filtered SNPs dataset of 196 individuals and 1,460,825 polymorphic sites was finally obtained for downstream analysis.
Phylogenetic analysis
For mitochondrial phylogeny, raw sequences of 16 S and COI were aligned respectively by the Clustal W algorithm with default parameters and subsequently refined in MEGA 661,62. The two segments, 1,045 base pairs (bp) of 16 S and 639 bp of COI, were concatenated into a 1,684-bp matrix. The matrix was tested in jmodeltest v2.1.763, resulting in the best-fitting nucleotide substitution model as GTR + I + G based on the Bayesian information criterion. The dataset was analyzed using maximum likelihood (ML) in RAxML v8.064, and Bayesian inference (BI) in MrBayes 3.2.465. In the ML analysis, the bootstrap consensus tree inferred from 1,000 replicates was used to represent the evolutionary history of the taxa analyzed. Two independent runs were conducted for the BI analysis, each for 10,000,000 generations with four Markov Chains Monte Carlo chains and sampled every 1,000 generations, and the first 25% of samples discarded as burn-in. The result was confirmed to be converged based on the average standard deviation of split frequencies below 0.01, as well as effective sample size larger than 200 which was evaluated in Tracer v1.766. ML bootstrap support (BS) larger than 90 and Bayesian posterior probabilities (BPP) larger than 0.95 were considered to be strong supports.
For filtered SNPs data, vcf2phylip (https://github.com/edgardomortiz/vcf2phylip, last accessed November 20, 2021) was used to generate the shared SNPs matrix. The matrix was analyzed using ML in RAxML-NG67. The best-fitting nucleotide substitution model tested by jmodeltest was GTR + I + G based on BIC. Branch supports were evaluated with 1,000 rapid bootstrapping replicates, and BS larger than 90% were considered to be strong supports. Furthermore, the filtered SNPs matrix was extracted by VCFtools to retain 192 Nidirana individuals. PLINK 1.968 was used to calculate linkage disequilibrium and prune the SNPs matrix to those with linkage disequilibrium < 0.2. Principle component analysis (PCA) was conducted in PLINK to find the best low-dimensional representation of each SNP in the genus Nidirana.
Divergence time estimation
The divergence time for the genus Nidirana was estimated using the MCMCTREE program in PAML v4.869. The ML topology based on the SNPs data was used as the reference tree. Due to the lack of fossils for Nidirana and its related genera, an inference time from the previous study was used to calibrate the clock, and the root age was set to 28.63 Ma with the 95% credible interval (CI) as 24.0–33.0 Ma70. Data binning strategy was used to reduce sampling error that divided the SNPs data into five bins71,72. The ML estimates of branch lengths for the bins of alignments and the substitution rate per time unit were obtained by the BASEML program in PAML. In MCMCTREE, the clock model was set as independent rate model. The Markov Chains Monte Carlo run was first executed for 10,000,000 generations as burn-in and then sampled every 150 generations until a total of 100,000 samples were collected. Two independent runs using random seeds were performed to examine whether similar results were obtained.
Ancestral distribution reconstruction
To infer the ancestral distributions of Nidirana, we applied the Dispersal-Extinction-Cladogenesis model in a Bayesian framework implemented in RevBayes73,74. The trees dataset was estimated using BEAST v2.5 for a total of 10,000 trees75, and the phylogeny with divergence time estimating from PAML was used as the condensed tree, with out-groups removed. For the distributions statement, we divided the distribution of the whole genus into ten areas (A–J) on the basis of topography, which were separate from each other by distinct geographical barriers of mountains, rivers, or straits (Supplementary Table S2). Then tip lineages were assigned areas based on their known occurrences. The dispersal rates between areas were scaled to their relative distances, represented by 10^ the minimum number of boundaries needed to be crossed from one area to the other. We used a time-stratified model with the time split into two epochs by the emergence of Taiwan island (the time bounds of this breakpoint: 9–6 Ma)76, which means that before this break there was no connectivity between area I and other nine areas, whereas after this break all ten areas are allowed to be connected. Two cladogenetic event types, allopatric and subset sympatric speciation were considered in the Dispersal-Extinction-Cladogenesis model, and the maximum number of areas was limited to two, given narrow distributions across extant lineages and limited dispersal ability in this group. We performed the analyses in RevBayes for 100,000 generations, and all other parameters were set to the default following the tutorial. With Markov Chains Monte Carlo samples, we annotated the condensed tree with ancestral states and generated stochastic mapping across the phylogeny to count biogeographic events over time. All parameter posteriors have effective sample sizes well over 1000. Meanwhile, we applied stochastic character mapping to generate simulated histories consistent with the observed data. With 1000 stochastic maps, we discarded the first 25% as burn-in, and parsed sequences of range states along the branches (anagenetic changes) and at nodes (cladogenetic changes) in the phylogeny. We then estimated numbers of range shifts through time by slicing the phylogeny into 1-Ma time windows, mainly focusing on dispersal/range expansion and cladogenetic changes (including allopatric and subset sympatric speciation).
Population and demographic analyses
Detailed population and demographic analyses were performed on the Nidirana congeners with or without nidification behavior, respectively. Nidirana chapaensis, N. hainanensis, N. yaoica, N. daunchina, N. nankunensis, and N. okinavana were used as nidification species. Nidirana adenopleura complex (including populations of N. adenopleura, N. guangdongensis, and N. mangveni) are used as the representatives for the congeners without nest construction behavior.
The SNPs dataset of nidification species including 18 samples and dataset of the Nidirana adenopleura complex including 127 samples, were extracted from the original filtered SNPs matrix by VCFtools and further pruned with linkage disequilibrium < 0.2. PCA was conducted in PLINK for the SNPs dataset of the nidification species and N. adenopleura complex, respectively. The population structure among individuals of the complex was inferred by Admixture 1.377 under different numbers of subgroups (K) setting from 2 to 10. The most likely number of genetic clusters was computed with a 10-fold cross-validation error. To obtain the level of genetic differentiation among species, Wright’s fixation index (FST) and absolute genetic divergences (dxy) statistics were computed using the populations program in Stacks.
To understand effective population size changes through historical time, demographic trajectories for each of the nidification species and the Nidirana adenopleura complex were inferred using Stairway Plot 278. The 1D folded observed site frequency spectrum for each population was calculated on the SNPs matrix using easySFS (https://github.com/isaacovercast/easySFS, last accessed October 28, 2021), using a strategy of maximizing the number of segregating sites. In the Stairway Plot 2 analysis, the mutation rate per site per generation was set as 7.76e-1079, the generation time was set at 1 years, and 200 bootstrap iterations were conducted.
The upstream analysis results showed that species within the Nidirana adenopleura complex have relatively complicated evolutionary histories (see Results above), therefore, we used fastsimcoal v2.7 to further infer the demographic dynamics80. To simplify its demographic histories, we performed four scenarios for three pairs of Nidirana populations selected according to the results of upstream analyses (Supplementary Table S4). The four scenarios were: (1) mig0: the absence of migrations between the two populations; (2) mig12: the presence of nonreciprocal migration from population 1 to population 2; (3) mig21: the presence of nonreciprocal migration from population 2 to population 1; (4) mig2: the presence of reciprocal migrations between the two populations. The population pairs were selected based on the results of upstream analyses. The 2D folded observed site frequency spectrum for population pairs was also calculated using easySFS, and the mutation rate per site per generation was also set as 7.76e-1079. To maximize the likelihood of each model, we ran 100 expectation-conditional maximization cycles with a total of 100,000,000 coalescent simulations. For each model, we obtained the best likelihood values and estimated parameters from 100 optimizations. The best model was determined using the Akaike information criterion weight method81.
Environmental correlation of nidification behavior
To assess the correlation and dependence of the nidification behavior with the environmental factors, we performed Generalized Linear Mixed Models (GLMMs) using the glm function in the lme4 R package with a binomial distribution and a logit link82. For environmental factors of each sampling locality (Supplementary Table S1) of Nidirana species, we extracted 19 bioclimatic variables (Supplementary Table S4) from WorldClim v2.1 at a resolution of 1 × 1 km for each variable83. P-value in the result was adjusted using False Discovery Rate method. Boxplots and scatterplots were visualized with the ggplot2 R package84.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All mitochondrial DNA Sanger sequencing data were uploaded to NCBI Nucleotide Database and the accession numbers are provided in Supplementary Table S1. All raw sequencing data generated by ddRAD-seq were uploaded to NCBI Sequence Read Archive under the BioProject accession number PRJNA1091219. Alignment for phylogeny and supplementary tables and figures are available on Dryad at https://doi.org/10.5061/dryad.wstqjq2rs.
References
Darwin, C. On the origin of species by means of natural selection 1–502 (John Murray, 1859).
Dobzhansky, T. Genetics of the evolutionary process 1–505 (Columbia University Press, 1970).
Kopp, R. E., Kirschvink, J. L., Hilburn, I. A. & Nash, C. Z. The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl Acad. Sci. Usa. 102, 11131–11136 (2005).
Schirrmeister, B. E., de Vos, J. M., Antonelli, A. & Bagheri, H. C. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc. Natl Acad. Sci. Usa. 110, 1791–1796 (2013).
Medved, V. et al. Origin and diversification of wings: Insights from a neopteran insect. Proc. Natl Acad. Sci. USA. 112, 15946–15951 (2015).
Prokop, J. et al. Paleozoic nymphal wing pads support dual model of insect wing origins. Curr. Biol. 27, 263–269 (2017).
Kimmel, C. B., Miller, C. T. & Keynes, R. J. Neural crest patterning and the evolution of the jaw. J. Anat. 199, 105–119 (2001).
Zhu, Y. A. et al. The oldest complete jawed vertebrates from the early Silurian of China. Nature 609, 954–958 (2022).
Buss, N., Swierk, L. & Hua, J. Amphibian breeding phenology influences offspring size and response to a common wetland contaminant. Front. Zool. 18, 11 (2021).
Lertzman-Lepofsky, G. F., Kissel, A. M., Sinervo, B. & Palen, W. J. Water loss and temperature interact to compound amphibian vulnerability to climate change. Glob. Change Biol. 26, 4868–4879 (2020).
Furness, A. I. & Capellini, I. The evolution of parental care diversity in amphibians. Nat. Commun. 10, 4709 (2019).
Nunes-de-Almeida, C. H. L., Haddad, C. F. B. & Toledo, L. F. A revised classification of the amphibian reproductive modes. Salamandra 57, 413–427 (2021).
Schulte, L. M., Ringler, E., Rojas, B. & Stynoski, J. L. Developments in amphibian parental care research: history, present advances, and future perspectives. Herpetol. Monogr. 34, 71–97 (2020).
Vági, B., Végvári, Z., Liker, A., Freckleton, R. P. & Székely, T. Parental care and the evolution of terrestriality in frogs. Proc. R. Soc. B 286, 20182737 (2019).
Cui, J. G., Tang, Y. Z. & Narins, P. M. Real estate ads in Emei music frog vocalizations: female preference for calls emanating from burrows. Biol. Lett. 8, 337–340 (2012).
Fischer, E. K. Form, function, foam: evolutionary ecology of anuran nests and nesting behaviour. Philos. Trans. R. Soc. B 378, 20220141 (2023).
Zhu, B., Sun, X., Zhang, H., Tang, Y. & Cui, J. Circadian Rhythm and Intersexual Differences in Auditory Frequency Sensitivity in Emei Music Frogs. Asian Herpetol. Res. 13, 43–52 (2022).
Ellepola, G., Pie, M. R., Pethiyagoda, R., Hanken, J. & Meegaskumbura, M. The role of climate and islands in species diversification and reproductive-mode evolution of Old World tree frogs. Commun. Biol. 5, 1–14 (2022).
Pereira, E. B. et al. Evolution of the anuran foam nest: trait conservatism and lineage diversification. Biol. J. Linn. Soc. 122, 814–823 (2017).
Luza, A. L., da Silva, E. R., Failace, D. M. & Colombo, P. Nest site selection by Hypsiboas faber (Anura, Hylidae) in southern Brazil. Iheringia Sér. Zool. 105, 453–460 (2015).
Rowley, J. J. & Altig, R. Nidicolous development in Limnonectes limborgi (Anura, Dicroglossidae). Amphib.-Reptilia 33, 145–149 (2012).
Dubois, A. Notes sur la classification des Ranidae (Amphibiens anoures). Bull. Mens. Soc. Linn. Lyon 61, 305–352 (1992).
Fei, L., Ye, C. Y. & Jiang, J. P. Colored atlas of Chinese amphibians and their distributions 1–619 (Sichuan Publishing House of Science & Technology, 2012).
Kuramoto, M. A new frog (genus Rana) from the Yaeyama Group of the Ryukyu Islands. Herpetol 41, 150–158 (1985).
Liu, C. C. Amphibians of western China. Fieldiana.: Zool. Mem. 2, 1–400 (1950).
Lyu, Z. T. et al. Resurrection of genus Nidirana (Anura: Ranidae) and synonymizing N. caldwelli with N. adenopleura, with description of a new species from China. Amphib.-Reptilia 38, 483–502 (2017).
Lyu, Z. T. et al. Description of a new species of Music frogs (Anura, Ranidae, Nidirana) from Mt Dayao, southern China. ZooKeys 858, 109–126 (2019).
Lyu, Z. T. et al. A new species of Music frog (Anura, Ranidae, Nidirana) from Mt Daming, Guangxi, China. ZooKeys 1059, 35–56 (2021).
Gomez-Mestre, I., Pyron, R. A. & Wiens, J. J. Phylogenetic analyses reveal unexpected patterns in the evolution of reproductive modes in frogs. Evolution 66, 3687–3700 (2012).
Clift, P. D., Wan, S. & Blusztajn, J. Reconstructing chemical weathering, physical erosion and monsoon intensity since 25 Ma in the northern South China Sea: a review of competing proxies. Earth-Sci. Rev. 130, 86–102 (2014).
Guo, Z. T. et al. A major reorganization of Asian climate by the early Miocene. Climate 4, 153–174 (2008).
Ding, W. et al. Sedimentary geochemical records of late Miocene-early Pliocene palaeovegetation and palaeoclimate evolution in the Ying-Qiong Basin, South China Sea. Mar. Geol. 445, 106750 (2022).
Farnsworth, A. et al. Past East Asian monsoon evolution controlled by paleogeography, not CO2. Sci. Adv. 5, eaax1697 (2019).
Westerhold, T. et al. An astronomically dated record of Earth’s climate and its predictability over the last 66 million years. Science 369, 1383–1387 (2020).
Lyu, Z. T. et al. A new species of Nidirana from the N. pleuraden group (Anura, Ranidae) from western Yunnan, China. Zootaxa 4861, 43–62 (2020).
Lyu, Z. T. et al. Comprehensive approaches reveal three cryptic species of genus Nidirana (Anura, Ranidae) from China. ZooKeys 914, 127–159 (2020).
Chen, Q., Cui, J., Fang, G., Brauth, S. E. & Tang, Y. Acoustic analysis of the advertisement calls of the music frog, Babina daunchina. J. Herpetol. 45, 406–416 (2011).
Cui, J. et al. Coevolution of male and female response preferences to sexual signals in music frogs. Asian Herpetol. Res. 7, 87–95 (2016).
Churchill, M., Martinez-Caceres, M., de Muizon, C., Mnieckowski, J. & Geisler, J. H. The origin of high-frequency hearing in whales. Curr. Biol. 26, 2144–2149 (2016).
Handrigan, G. R. & Wassersug, R. J. The anuran Bauplan: a review of the adaptive, developmental, and genetic underpinnings of frog and tadpole morphology. Biol. Rev. 82, 1–25 (2007).
Joly, S., McLenachan, P. A. & Lockhart, P. J. A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Naturalist 174, E54–E70 (2009).
Meyer, B. S., Matschiner, M. & Salzburger, W. Disentangling incomplete lineage sorting and introgression to refine species-tree estimates for Lake Tanganyika cichlid fishes. Syst. Biol. 66, 531–550 (2017).
Seehausen, O. Hybridization and adaptive radiation. Trends Eco. Evo. 19, 198–207 (2004).
Clift, P. D. Asian monsoon dynamics and sediment transport in SE Asia. J. Asian Earth Sci. 195, 104352 (2020).
Wan, S., Li, A., Clift, P. D. & Stuut, J. B. W. Development of the East Asian monsoon: mineralogical and sedimentologic records in the northern South China Sea since 20 Ma. Palaeogeogr., Palaeoclimatol., Palaeoecol. 254, 561–582 (2007).
Zheng, L., Huang, X., Ji, X. & Deng, C. Paleoenvironmental implications of molecular and carbon isotopic distributions of n-alkanes in the hominoid-bearing upper Miocene sediments from the Zhaotong Basin, southwestern China. Palaeogeogr., Palaeoclimatol., Palaeoecol. 544, 109627 (2020).
Collias, N. E. & Collias, E. C. Nest building and bird behavior 1–358 (Princeton University Press, 1984).
Hansell, M. H. Animal architecture 1–336 (Oxford University Press, 2005).
Wells, K. The ecology & behavior of amphibians 1–1400 (University of Chicago Press, 2007).
Chen, J. M. et al. An integrative phylogenomic approach illuminates the evolutionary history of Old World tree frogs (Anura: Rhacophoridae). Mol. Phylogenet. Evol. 145, 106724 (2020).
Easterling, D. R. et al. Climate extremes: observations, modeling, and impacts. Science 289, 2068–2075 (2000).
Kotz, M., Lange, S., Wenz, L. & Levermann, A. Constraining the pattern and magnitude of projected extreme precipitation change in a multimodel ensemble. J. Clim. 37, 97–111 (2024).
Fischer, E. M. & Knutti, R. Observed heavy precipitation increase confirms theory and early models. Nat. Clim. Change 6, 986–991 (2016).
Perkins-Kirkpatrick, S. E. et al. On the attribution of the impacts of extreme weather events to anthropogenic climate change. Environ. Res. Lett. 17, 024009 (2022).
Lyu, Z. T. et al. A new species of Amolops (Anura: Ranidae) from China, with taxonomic comments on A. liangshanensis and Chinese populations of A. marmoratus. Zootaxa 4609, 247–268 (2019).
Lyu, Z. T. et al. Generic classification of Asian horned toads (Anura: Megophryidae: Megophryinae) and monograph of Chinese species. Zool. Res. 44, 380–450 (2023).
Lyu, Z. T. et al. Taxonomic clarifications on the floating frogs (Anura: Dicroglossidae: Occidozyga sensu lato) in southeastern China. Vertebr. Zool. 72, 495–512 (2022).
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS one 7, e37135 (2012).
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A. & Cresko, W. A. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140 (2013).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, s13742–015 (2015).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Wu, Y. H. et al. A combined approach of mitochondrial DNA and anchored nuclear phylogenomics sheds light on unrecognized diversity, phylogeny, and historical biogeography of the torrent frogs, genus Amolops (Anura: Ranidae). Mol. Phylogenet. Evol. 148, 106789 (2020).
Bayzid, M. S. B. & Warnow, T. Naive binning improves phylogenomic analyses. Bioinformatics 29, 2277–2284 (2013).
Mirarab, S., Bayzid, M. S., Boussau, B. & Warnow, T. Statistical binning enables an accurate coalescent-based estimation of the avian tree. Science 346, 1250463 (2014).
Höhna, S. et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65, 726–736 (2016).
Ree, R. H. & Smith, S. A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 (2008).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Sibuet, J. C. & Hsu, S. K. How was Taiwan created? Tectonophysics 379, 159–181 (2004).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Liu, X. & Fu, Y. X. Stairway Plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol. 21, 280 (2020).
Sun, Y. B. et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc. Natl Acad. Sci. Usa. 112, E1257–E1262 (2015).
Excoffier, L. et al. fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics 37, 4882–4885 (2021).
Burnham K. P. & Anderson, D. R. Model Selection and Multimodel Inference 1–488 (Springer, 2002).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 67, 1–48 (2015).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, New York, 2016).
Acknowledgements
We thank Jian Wang, Yao Li, Guoling Chen, Bicheng Zhu, Xiao-Yun Wang, Si-Min Lin, Yun-Ming Mo, Song Huang, Zhongxing Wang, Jian Zhao, Hai-Long He, Jian Luo, Jian-Huan Yang, and Ying Liu for their help in the field and lab. We thank Zhiyong Yuan and Jin-Long Ren for providing samples even though they were not used in this study. Richard H. Ree and Michael J. Landis provided guidance on the RevBayes analysis. Isaac Overcast, Mengjie Jin, He Chen, and Xin-Kai Wu provided valuable comments on the initial manuscript. This work was supported by DFGP Project of Fauna of Guangdong-202115 and the Interdisciplinary Innovation Team of the Chinese Academy of Sciences (CAS) “Light of West China” Program (xbzg-zdsys-202207).
Author information
Authors and Affiliations
Contributions
ZTL and YYW conceived the study; ZTL, ZCZ, and HW collected samples and conducted laboratory work; A Tominaga, K Nishikawa, M Matsui, and SZ Li, contributed materials; ZT Lyu, ZC Zeng, Q Li, and ZW Jiang analyzed the data; ZT Lyu, ZC Zeng, Q Li, and YL wrote the manuscript with supports from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Luke R. Grinham. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lyu, ZT., Zeng, ZC., Wan, H. et al. Contrasting nidification behaviors facilitate diversification and colonization of the Music frogs under a changing paleoclimate. Commun Biol 7, 638 (2024). https://doi.org/10.1038/s42003-024-06347-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06347-7
This article is cited by
-
New insight into the origin and evolution of the flora and fauna of Hainan Island, China
Science China Earth Sciences (2025)