Abstract
The global biodiversity crisis is generated by the combined effects of human-induced climate change and land conversion. Madagascar is one of the World’s most renewed hotspots of biodiversity. Yet, its rich variety of plant and animal species is threatened by deforestation and climate change. Predicting the future of Madagascar’s chameleons, in particular, is complicated by their ecological rarity, making it hard to tell which factor is the most menacing to their survival. By applying an extension of the ENphylo species distribution model algorithm to work with extremely rare species, we find that Madagascar chameleons will face intense species loss in the north-western sector of the island. Land conversion by humans will drive most of the loss, and will intersect in a complex, nonlinear manner with climate change. We find that some 30% of the Madagascar’s chameleons may lose in the future nearly all their habitats, critically jeopardizing their chance for survival.
Similar content being viewed by others
Introduction
The current biodiversity crisis is elicited by the combined effects of climate change and the other negative consequences deriving from human economic activities1,2,3. These effects are so penetrating that most scholars agree a sixth mass extinction is in fact underway4,5. The intensity of the ongoing extinction wave varies across taxonomic groups and geography, being ostensibly more severe among both the largest and smallest-bodied terrestrial vertebrates4,6,7 and in tropical and subtropical regions5,8. With its extraordinarily diverse fauna exposed to intense human disturbance (mainly via land use change and deforestation) Madagascar stands out as one of the worst-hit regions in the world9,10,11,12. Predicting the future of biodiversity on this large island is complicated. The rapidly expanding network of protected areas may buffer extinction risk in unpredictable ways, depending on the availability of space and corridors left by humans to the species tracking their habitats in the wake of climate change9,13,14. The effects of climate change and land use conversion are not necessarily additive, as some species may even expand their habitats under the anticipated warmer temperatures, whereas the impact of land use conversion is mostly expected to be detrimental15,16. Moreover, producing solid bioclimatic models that can be projected to the future to assess where the habitats will be suitable for rare species is difficult16,17,18. The problem with modelling uncommon taxa is particularly sensitive in regions with high levels of endemism and narrow-ranged species like Madagascar9,12,15. Although a number of solutions were provided in the past to address the problem of rarity in modelling19,20 at least 10 datapoints per species are usually required under such approaches, which translates to geographic range sizes no less than 100 km2 even with a fine-grained, 10 × 10 km gridded cell study design. This figure is larger, for instance, than the geographic range size of one fifth of Madagascar reptiles21 and means that particularly rare species are usually excluded from studies addressing the future distribution of the biota (e.g. refs. 17,19,22,23,24,25). Unsurprisingly, the 10 datapoints limit was applied in modelling the expected future distribution of Madagascar’s plants16 and reptiles17 and an even larger (20) datapoint limit was used to model typically narrow-ranged Madagascar’s chameleons23. To solve this ‘rare species modelling paradox’26 we recently proposed a new algorithm, named ENphylo27, which takes advantage of the strong phylogenetic signal in species climatic preferences28 to infer the presumed climatic niche of rare species, and then provides habitat suitability models for them by intersecting the phylogenetically-imputed niche with the few observational datapoints available. ENphylo proved to be particularly accurate in modelling species for which as few as 10 to 20 observational datapoints where available, and outcompeted similar algorithms within this range27. Here, by using Madagascar’s chameleons as a model, we expanded ENphylo to work with even rarer species, pushing modelling down to two observational datapoints only. The study system is ideal, at 95 taxonomically defined, mostly endemic species21,23, Madagascar’s chameleons are exceptionally diverse, represent a long and complex biogeographic history9,29,30 and yet they are mostly infrequent, with some 30 species having range size smaller than 1000 km2 21. This implies predicting their future distribution is problematic and further complicated by the uncertainty derived by the complex dynamics of Madagascar’s landscapes and socioeconomic growth9,29,31. Albeit Madagascar’s protected areas are expanding13 and include at least in part the geographic range of some 97% of the threatened vertebrate species9, the island is martyrised by decades of deforestation, which erased nearly one half of its forest cover32 and still proceeds at a rate >80 kha/year33. In the 2000 to 2019 interval, the fraction of ecosystem services value generated by Madagascar forests decreased of some 3% while its total values increased by 7%, because of land use conversion to agriculture mainly at the expense of the forests31, where most of the chameleon biodiversity occurrs23,34. To assess how the combined effects of future climate change and land use change on Madagascar’s chameleon diversity, we modelled the potential future distribution of Madagascar’s chameleons with ENphylo, including most of the rarest, under different scenarios of global warming and land use change, and tested how these factors will interact with each other in influencing their survival and distribution. We modelled the effects between climate and land use changes in terms of additive, agonistic or synergistic interactions, testing whether and where their expected contributions are expected to impinge on Madagascar’s chameleon fauna.
Results
We modelled 134 chamaeleonid species under different SDM algorithms. In particular, 25 out 56 Madagascar’ chameleons were modelled under ENphylo. Fifteen out of 56 were modelled via ESM, and 16 more were eventually modelled by the model ensemble of MaxEnt, RF and GLM algorithms. Overall, we built 45 model predictions for each Chamaeleonidae species combining the three binarization thresholds, the three GCMs, and the two mild and severe scenarios for both climate and LULC, for a total of 1620 (45 maps × 56 species) predictive maps.
We assessed the effect of spatial autocorrelation by quantifying its amount in ensemble model residuals by means of Moran’s I correlograms. According to this analysis, the average Moran’s I value among all the species is equal to −0.13 (sd = 0.06), with only 14% of significant replicates. These results indicate an overall negligible effect of spatial autocorrelation on models.
Since evaluation metrics can provide misleading results when quantifying the accuracy of models calibrated on sample size <10 presences35, we reported the ENphylo performances without considering the models related to the species with <10 occurrences. Under this filtering, ENphylo achieved fair-to-excellent predictive performances with an AUC value averaged among the modelled species equal to 0.918 (sd = 0.106), an average TSS equal to 0.703 (sd = 0.166), and an average Boyce index equal to 0.394 (sd = 0.122). It is worth nothing, though, that model selection (i.e., phylogeny selection) was implemented by selecting the replicate with the highest AUC value in ENphylo, meaning that TSS and Boyce values might be underestimated.
The other approaches led to equally robust model performances: averaged AUC = 0.953 (sd = 0.033), averaged TSS = 0.871 (sd = 0.086), and average Boyce index = 0.827 (sd = 0.067) for the ESMs, while averaged AUC = 0.964 (sd = 0.033), averaged TSS = 0.864 (sd = 0.100), and averaged Boyce index = 0.965 (sd = 0.04) for traditional SDMs.
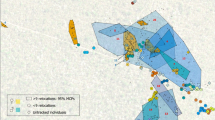
Future projections showed large loss and gain areas for Chamaeleonidae species in different regions of Madagascar (Fig. 1, Supplementary Figs. 1–2). The magnitude of the species loss/gain estimates increased/decreased passing from the mild to severe future scenarios (Table 1). The impact of future LULC change for both loss and gain dynamics seems relevant at large spatial scale, whereas future climate predictions suggest an important role at local scale (Fig. 1, Supplementary Figs. 1–2). As for the interaction type, the “only LULC” effect has the highest percentage for both gain and loss dynamics, whereas the synergic effect has the lowest value. The role of antagonistic interaction is relevant for species loss only (Fig. 2, Table 2).
Discussion
Madagascar is a hotspot of biodiversity, hosting a great variety of terrestrial tetrapods and plants unique to the island12. This rich and diversified biota is facing serious threats posed by humans via overexploitation of the natural landscape and species, and by human-driven climate change9. These activities are prolonging a pattern of extinction initiated by the first human colonizers who eradicated the island’s megafauna36,37,38. Despite protected areas now cover 10.4% of the country and encompass, at least in part, the distributional range of virtually all threatened vertebrates9 rapid deforestation has taken its toll on Madagascar biodiversity for almost fifty years now32,33. Besides direct human activities, climate change poses another obvious threat to the island’s biota. Albeit climate change is anticipated to exert a negative effect on Madagascar’s forested biomes39, projected range maps suggest a mixed blessing, with most range contraction but also sizeable range expansion being forecasted15,16,39. Consequently, human disturbance is usually perceived as posing a greater threat to the survival of Madagascar’s species compared to the potential effects of climate change9. Whereas vegetational cover has been readily modelled, though, the future distribution of the tetrapod fauna is less well-understood and new species are still being described12, possibly because of the restricted geographic range of several, small-bodied endemic species23. Thus, the future of the small vertebrates is surrounded by greater uncertainty than with plants, which imperils the proper design of protected areas and corridors management to save them from extinction13,14. Here, we applied species distribution modelling to 56 Madagascar’s chameleon species, including many of the rarest. The list of uncommon species comprise leaf chameleons Brookesia (B. desperata, B. karchei, B. micra, and B. tristis), several species in the genus Calumma (C. guibei, C. furcifer, C. guillaumeti, C. marojezense) and Furcifer (F. timoni, F. bifidus) whose range is mostly restricted to small areas in the North, like Montagne d’Ambre and Marojejy National Parks and Nosy Hara archipelago. Unfortunately, we found that for some 10–20 species the predicted geographic range change will exceed 90% loss, depending on the LULC and climate change future scenario (Supplementary Data 3), meaning they will be in grave danger of extinction. This set of imperilled taxa mostly occupy dry deciduous, subhumid and lowland forest habitats such as canopy chameleon Furcifer willsii, Decary’s leaf chameleon B. decaryi and B. brunoi, flat-casqued chameleon Calumma globifer, and the abovementioned B. desperata, B.karchei, B. micra, and B. tristis, additional species within Calumma (C. amber, C. guibei, C. ambreense, C. nasutum, C. fallax, C. peltierorum and C. boettgeri) and Furcifer (F. petteri). In contrast, geographic range change predictions are positive (meaning they are expected to gain range) for widespread species, some of which occur in multiple habitats such as giant and rhinoceros chameleon Furcifer oustaleti and F. rhinoceratus, Parson’s, blue-legged, short-horned and O’Shaughnessy’s chameleons (C. parsonii, C. crypticum, C. brevicorne, and C. oshaughnessyi) and the stump-tailed chameleon B. superciliaris. Overall, though, there is no clear relationship between commonness and the predicted percentage of habitat loss, especially for the species occurring in the dry deciduous forest habitat, such as Calumma ambreense, C. amber, and Brookesia tuberculata, all predicted to be losing >90% of their habitat despite their commonness.
At comparing the effects of climate change and land use conversion, and their intersection, on species habitat suitability in the future, we found that the effects of LULC were preponderant. We predict that almost one chameleon species per cell in Madagascar will find unsuitable habitats in the future (Table 1). This result does not change by considering different land use future scenarios whereby the intensification of the climatic and socioeconomic factors will only affect the maximum of species loss ranging from 8 (mild) to 11 (severe) (Table 1, Supplementary Fig. 2). This is mostly due to changes in LULC, which are predicted to override the effects of climate change and the interaction terms of both sets of variables, regardless of which climate change scenario, or SDM parameters are used (Fig. 2, Table 2). The highest chameleon biodiversity loss is expected to occur in the dry deciduous forest in the western and north-western sectors of the island (Fig. 1, Supplementary Fig.1 and Supplementary Data 4). In contrast, most potential species losses and gains (hence higher turnover) are concentrated in the lowland forest in the eastern sector of the island (Fig.1). The true nature of species gains, though, is hard to tell. Chameleons are not particularly good at dispersal, and we consequently bounded dispersal distance to around 1 km yr−1 40. While this figure is not particularly high by vertebrate standards, it is liberal in assuming the habitat corridors can in fact be trespassed. This will critically depend on their existence and functioning and ignores lack of connectivity between suitable habitats. This implies the potential gains, in the absence of human-mediated translocation, are hypothetical at best.
Predicting species distribution for extremely rare species cannot usually be done26,41, and they are usually omitted18 or presence datapoints simulated within the extent of occurrence so that up to 10 geographic cells are included35. With SDM validation, statistical issues introduced by rarity are still present, as with rare species common validation metrics such the AUC suffers from variation in the species prevalence and sample size42. In addition, pseudo-presences necessarily introduce some autocorrelation in the data, and can only be supposed to may harbour the species presence. However, it is important to notice that our procedure is not meant to ‘learn’ the niche from the ‘pseudo-presences’. In ENphylo, the niche of a rare species is imputed after modelling the niches of its relatives with ENFA. True presences and ‘pseudo-presences’ are just used to convert the imputed niche into habitat suitability, by computing the Mahalanobis distance between sampled and the ‘mean’ habitat43,44. Since the ‘pseudo-presences’ are sampled exactly according to their similarity to the real presence datapoints, this distance could be safely assumed to be left almost unaltered by our procedure. Of course, this does not mean that five datapoints provide a faithful representation of the species niche, especially because we selected the pseudo-presences as to minimize their dissimilarity from the true presences. This may possibly imply the rare species niches are probably narrow. We believe maintaining this non-random narrowness, though, provides a more genuine representation of the actual niche than producing an artificially broad niche for an otherwise little-known species. As a matter of fact, ENphylo predictive performance was quite high (AUC > 0.9) which testifies that the modelling procedure was robust. By applying ENphylo, and by testing explicitly the combined effects of land-use and climate change under different scenarios, we found that the most serious threat to Madagascar’s future chameleon biodiversity is posed by land conversion to agriculture and other human activities, rather than by climate change.
Materials and methods
Chamaeleonidae occurrences
We downloaded the Chamaeleonidae modern occurrences from the “Global Biodiversity Information Facility” online database (GBIF; www.gbif.org/), including only the data provided with geographical coordinates. Data were further filtered by selecting “Material citation and both Machine and Human observations” as the basis of record categories (GBIF.org (26 March 2024) GBIF Occurrence Download https://doi.org/10.15468/dl.86bqsw). Record accuracy was assessed by including only occurrences given to at least two decimal places (0.01 decimal degrees, corresponding to 1.11 km at the equator) and by removing duplicated or unrealistic records. Then, we excluded the data outside the African continent and the Mediterranean area. Overall, we gathered 17170 occurrences belonging to 151 Chamaeleonidae species.
Environmental variables
To address the potential impact of climate and land use change on the future of Madagascar’s chameleons, we started by considering the 19 bioclimatic variables listed in the CHELSA database version 2.145 (http://chelsa-climate.org/) as environmental predictors. Specifically, we downloaded the high-resolution (1 × 1 km) modern climatic data (1981–2010) as the reference temporal period, while for future scenarios, we referred to projections for the 2071–2100 interval. We took into account both the mild and severe “Shared Socioeconomic Pathways” (SSP), i.e., SSP1-2.6 and SSP5-8.5 scenarios. For each of them, we considered three global circulation models (GCMs) from the Coupled Model Intercomparison Project (CMIP6), namely GFDL-ESM4, MRI-ESM2-0, and IPSL-CM6A-LR. To take into account Madagascar’s land use change scenarios, we considered seven Land Use/Land Cover (LULC) categories (https://www.geosimulation.cn/), as provided in ref. 46. Specifically, we calculated Euclidean distances from each of the seven LULC categories and used them as predictors. Climate and LULC variables were rasterized at 1 km spatial resolution and cropped along Madagascar extent. Subsequently, the variables were checked for multicollinearity by using the “usdm” R package47. After excluding the variables with a high Pearson’s correlation coefficient (using 0.7 as the threshold), we retained 11 predictors: Temperature Seasonality (BIO4), Max Temperature of Warmest Month (BIO5), Temperature Annual Range (BIO7), Precipitation of Wettest Month (BIO13), Precipitation Seasonality (BIO15) and Euclidean distance from Water bodies, Forests, Grasslands, Barren areas, Urban areas, and Croplands.
Species distribution models (SDMs)
The selected predictors were used to feed species distribution models (SDMs) sought to predict the current and future potential distribution of Chamaeleonidae species. We used three different modelling approaches depending on the number of occurrences available per species. Specifically, we adopted the ENphylo modelling algorithm27 for the species with less than 15 occurrences, given the proven ability of this algorithm to outperform ensembles of small models (ESMs20) and “traditional” SDMs when rare species are modelled27. To run ENphylo, for each species we randomly generated 10,000 background points across the ecoregion included within the study area48. In ENphylo, the climatic niche dimensions of rare species are derived estimating the marginality and specialization axes of well-sampled species under Ecological-Niche Factor Analysis (ENFA)44 analysis first, and then estimating marginality and specialization for the rare species via phylogenetic imputation. Although this procedure provides the essential niche information, at least five occurrences are necessary to convert the phylogenetically-imputed niche marginality and specialization axes into Mahalanobis distances from the available climates and then into habitat suitability values27. Here, we expanded ENphylo to work with less than five occurrences. To this aim, we started designing as pseudo-presences the cells adjacent to true presence (‘reference’) cells, according to the knight move criterion49. Then, we reduced the number of pseudo-presences by selecting among them those closest to the reference cells in terms of climate and LULC values. We first quantified the angle (i.e. the correlation coefficient) between the climatic/LULC vectors associated to reference cells and those of the pseudo-presence cells by computing the angle between their respective climatic/LULC vectors using the R package “RRphylo”50. Then, we selected n pseudo-presence cells, where n is equal to 5 minus the number of reference cells, according to their climatic/LULC similarity to the reference cells. This procedure is meant to select a minimum number of pseudo-presences as to attain to 5 potential presence cells (including the references) that could be used to convert the phylogenetically-imputed niche marginality and specialization into habitat suitability values27. To perform phylogenetic imputation of the niche marginality and specialization axes, we assembled a composite, informal supertree using the chameleon phylogeny in Tonini et al. 51 and Giles et al. 34 using the function tree.merger52 in “RRphylo”. We excluded species for which (i) geographical information was not available, (ii) occupy a single geographic cell, or (iii) phylogenetic information was not available or conflicting between the source trees. The resulting phylogeny includes 134 species (56 of them currently living in Madagascar, Supplementary Data 1). After removing duplicate occurrences per cell, the total occurrence number amounts to 6915 (Fig. 3, Supplementary Data 2).
(Top) The colours and tree refer to chameleons living in Madagascar. (Bottom) The image of Furcifer verrucosus is distributed under CC BY-SA 2.0 https://commons.wikimedia.org/wiki/File:Warty_Chameleon_(Furcifer_verrucosus)_(9628372559).jpg. The image of Brookesia superciliaris is distributed under CC0. https://commons.wikimedia.org/wiki/File:Brookesia_superciliaris_185939354.jpg. The image of Palleon nasus is distributed under CC BY-SA 4.0 https://commons.wikimedia.org/wiki/File:Palleon_nasus.JPG. The image of Calumma parsonii is distributed under CC BY-SA 4.0. https://upload.wikimedia.org/wikipedia/commons/4/42/Calumma_Parsonii_Ste_Marie_Madagascar.jpg.
ENphylo models were assessed through a random bootstrap cross-validation procedure with replacement method by splitting the data into 80–20% training/testing folds. The bootstrap was repeated 10 times. Model predictive accuracy was assessed by calculating the area under the receiver operating characteristic curve (AUC), True Skill Statistic (TSS53), and Boyce index54 and removing the models with an AUC value < 0.7. To account for phylogenetic uncertainty, the entire procedure was repeated by testing 50 alternative phylogenies, produced randomly modifying the species topology and branch lengths with the function swapOne in the R package “RRphylo”50. Then, we selected the model with the best AUC value over the 50 replicates. Since we were interested in species diversity changes in Madagascar, we retrieved only the best-fit models associated with Chamaeleonidae species which currently live in Madagascar and projected them on current Madagascar climate/LULC as well as on future scenarios. Specifically, we considered the following climate and LULC change scenarios: (i) dynamic climate while LULC was held constant (hereafter “dynamic climate”), (ii) dynamic LULC while climate was held constant (hereafter “dynamic land use”), and (iii) dynamic LULC and dynamic climate (hereafter dynamic “LULC-climate”), following the same approach described in ref. 55. Both current and future model predictions were binarized to obtain presence/absence maps by using three thresholding schemes, as to account for the effect of adopting different binarization approaches56. Specifically, we selected the ‘equalize sensitivity and specificity’ (SensSpec), ‘maximize TSS’ (MaxSens+Spec), and ‘minimum training presence’ (TenPerc) using the “PresenceAbsence” R package57.
To model species climatic niche, we adopted the ESM approach for species reporting a number of occurrences between 15 and 30. ESMs were trained by considering all possible combinations of the environmental variables taken two at a time. Lastly, for species with >30 occurrences were modelled by applying a traditional SDM approach (i.e. including all environmental variables at the same time). For both ESMs and SDMs, we adopted an ensemble forecasting approach by testing three widely used modelling techniques: Maximum Entropy (MaxEnt58), Random Forest (RF59), and Generalized Linear Models (GLM). Models were trained by relying on the functionalities provided in the ‘biomod2’ R package60. Specifically, we set “quadratic” and interaction level = 1 for defining the GLM parameters. For the other algorithms, we maintained the default parameter settings adopted in biomod2.
To evaluate model predictive accuracy, we performed a random bootstrap cross-validation with replacement scheme splitting the data into 80–20% training/testing samples and repeating this procedure 10 times. Model accuracy was evaluated by measuring the AUC, TSS, and Boyce index. Poorly calibrated models were avoided removing those with an AUC value < 0.7. Model averaging was performed by weighting the individual model projections by their AUC values and averaging the results61. Models were projected on the current Madagascar climate/LULC and on the future scenarios described above. Model projections were subsequently binarized by using the same thresholds described before.
Since the Chamaeleonidae species are characterized by limited dispersal abilities and live in highly-fragmented habitats62, we decided to incorporate a dispersal constraint to the future projections of their distribution. We established a distance equal to 1 km as maximum annual dispersal rate, maintaining constant this value over time for all species. According to this strategy, we first calculated the minimum convex polygon enclosing all the localities where a species occurs, then created a buffer around this polygon with a radius equal to the maximum reachable distance from nowadays to the future (i.e., 60 km over a time span from 2010 to 2070). Subsequently, the current and future binary maps were cropped accordingly.
Binary maps obtained from the three model approaches were stacked among the species for current and future scenarios separately. After that, we calculated three indices: species richness (SR), species loss (L), and species gain (G), as to obtain a single vector of three values at each grid cell following the approach described in ref. 63. All indices were calculated by using the “biomod2”.
To quantify the individual and synergic effects of the climatic and LULC changes in predicting Chamaeleonidae distribution, we calculated the delta in species richness (i.e., future − current) and summed the species losses and gains per cell by applying a paired comparison between the current and all the future scenarios. Moreover, the loss and gain values obtained for the dynamic land use (a), dynamic climate (b) and dynamic land-climate future scenarios (c) were adopted to define five types of interactions among the scenarios, as follows: (i) “Synergistic”, when c > a + b, (ii) “Additive”, when c = a + b, and (iii) “Antagonistic”, when c < min(a, b) or c < max(a, b), or max(a, b) ≤ c < a + b45, (iv) “only climate” when a = 0 and b > 0, and “only LULC” when a > 0 and b = 0. All the procedure was repeated for loss and gain separately after removing the cells where no interaction occurred (i.e., all a, b, and c = 0).
Lastly, we quantified the types of interactions separately for mild and severe SPP under different future scenarios calculating the percentage of grid cells where an antagonistic, synergistic, and additive effect occurred.
The software developed to produce the rare species modelling, including ENphylo, is embedded in the R package RRdtn64.
Statistics and reproducibility
R scripts associated with this study are available as Supplementary Data 5.zip file. Files contains three annotated R scripts which describe the entire procedure used to model the Chamaeleonidae species by using the three different modelling approaches (ENphylo, ESM, and SDM). Codes are annotated to allow reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw data and the chameleon phylogenetic tree are available in the Supplementary Data files.
Code availability
R scripts associated with this study are available as Supplementary Data 5.zip file. The R package RRdtn embedding the ENphylo functions is available via Zenodo at https://doi.org/10.5281/zenodo.12734585.
References
Ellis, E. C. The Anthropocene condition: evolving through social–ecological transformations. Philosophical Transactions of the Royal Society B: Biological Sciences 379, (2024).
Ceballos, G. & Ehrlich, P. R. Mutilation of the tree of life via mass extinction of animal genera. Proc. Natl Acad. Sci. 120, e2306987120 (2023).
Dirzo, R. et al. Defaunation in the Anthropocene. Science 345, 401–406 (2014).
Barnosky, A. D. et al. Has the Earth’s sixth mass extinction already arrived? Nature 471, 51–57 (2011).
Ceballos, G., Ehrlich, P. R. & Raven, P. H. Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proc. Natl Acad. Sci. USA 117, 13596–13602 (2020).
Plotnick, R. E., Smith, F. A. & Lyons, S. K. The fossil record of the sixth extinction. Ecol. Lett. 19, 546–553 (2016).
Ripple, W. J. et al. Extinction risk is most acute for the world’s largest and smallest vertebrates. Proc. Natl Acad. Sci. USA 114, 10678–10683 (2017).
Fritz, S. A., Bininda-Emonds, O. R. P. & Purvis, A. Geographical variation in predictors of mammalian extinction risk: Big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 (2009).
Ralimanana, H. et al. Madagascar’s extraordinary biodiversity: Threats and opportunities. Science (1979) 378, (2022).
Godfrey, L. R. et al. Teasing apart impacts of human activity and regional drought on Madagascar’s large vertebrate fauna: insights from new excavations at tsimanampesotse and antsirafaly. Front Ecol. Evol. 9, 605 (2021).
Cox, N. et al. A global reptile assessment highlights shared conservation needs of tetrapods. Nature 2022 1–6 https://doi.org/10.1038/s41586-022-04664-7 (2022).
Antonelli, A. et al. Madagascar’s extraordinary biodiversity: evolution, distribution, and use. Science (1979) 378, (2022).
Coldrey, K. M. & Turpie, J. K. The future representativeness of Madagascar’s protected area network in the face of climate change. Afr. J. Ecol. 59, 253–263 (2021).
Hannah, L. et al. 30% land conservation and climate action reduces tropical extinction risk by more than 50. Ecography 43, 943–953 (2020).
Hannah, L. et al. Climate change adaptation for conservation in Madagascar. Biol. Lett. 4, 590–594 (2008).
Brown, K. A., Parks, K. E., Bethell, C. A., Johnson, S. E. & Mulligan, M. Predicting plant diversity patterns in Madagascar: understanding the effects of climate and land cover change in a biodiversity hotspot. PLoS One 10, e0122721 (2015).
Jenkins, R. K. B. et al. Extinction risks and the conservation of Madagascar’s reptiles. PLoS One 9, e100173 (2014).
Wiens, J. J. & Zelinka, J. How many species will Earth lose to climate change? Glob. Chang Biol. 30, e17125 (2024).
Breiner, F. T., Nobis, M. P., Bergamini, A. & Guisan, A. Optimizing ensembles of small models for predicting the distribution of species with few occurrences. Methods Ecol. Evol. 9, 802–808 (2018).
Breiner, F. T., Guisan, A., Bergamini, A. & Nobis, M. P. Overcoming limitations of modelling rare species by using ensembles of small models. Methods Ecol. Evol. 6, 1210–1218 (2015).
Roll, U. et al. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat. Ecol. Evol. 1, 1677–1682 (2017).
Biber, M. F., Voskamp, A. & Hof, C. Potential effects of future climate change on global reptile distributions and diversity. Global Ecology and Biogeography https://doi.org/10.1111/geb.13646 (2023).
Raxworthy, C. J. et al. Predicting distributions of known and unknown reptile species in Madagascar. Nature 426, 837–841 (2003).
Stewart, P. S. et al. Global impacts of climate change on avian functional diversity. Ecol. Lett. 25, 673–685 (2022).
Newbold, T. Future effects of climate and land-use change on terrestrial vertebrate community diversity under different scenarios. Proc Biol Sci 285, (2018).
Lomba, A. et al. Overcoming the rare species modelling paradox: A novel hierarchical framework applied to an Iberian endemic plant. Biol. Conserv 143, 2647–2657 (2010).
Mondanaro, A. et al. ENphylo: a new method to model the distribution of extremely rare species. Methods Ecol. Evol. 14, 911–922 (2023).
Rolland, J. et al. The impact of endothermy on the climatic niche evolution and the distribution of vertebrate diversity. Nat. Ecol. Evol. 2, 459–464 (2018).
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B. & Kent, J. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000).
Raxworthy, C. J., Forstner, M. R. J. & Nussbaum, R. A. Chameleon radiation by oceanic dispersal. Nature 415, 784–787 (2002).
Tiandraza, F. et al. Response of ecosystem services to land use change in Madagascar island, Africa: a multi-scale perspective. Int J. Environ. Res Public Health 20, 3060 (2023).
Harper, G. J., Steininger, M. K., Tucker, C. J., Juhn, D. & Hawkins, F. Fifty years of deforestation and forest fragmentation in Madagascar. Environ. Conserv 34, 325–333 (2007).
Vieilledent, G. et al. Combining global tree cover loss data with historical national forest cover maps to look at six decades of deforestation and forest fragmentation in Madagascar. Biol. Conserv 222, 189–197 (2018).
Giles, S. A. W. & Arbuckle, K. Diversification dynamics of chameleons (Chamaeleonidae). J. Zool. 318, 241–252 (2022).
Jiménez-Valverde, A. Sample size for the evaluation of presence-absence models. Ecol. Indic. 114, 106289 (2020).
Razafindratsima, O. H., Mehtani, S. & Dunham, A. E. Extinctions, traits and phylogenetic community structure: insights from primate assemblages in Madagascar. Ecography 36, 47–56 (2013).
Burney, D. A chronology for late prehistoric Madagascar. J. Hum. Evol. 47, 25–63 (2004).
Andermann, T., Faurby, S., Turvey, S. T., Antonelli, A. & Silvestro, D. The past and future human impact on mammalian diversity. Sci. Adv. 6, eabb2313 (2020).
Hending, D., Holderied, M., McCabe, G. & Cotton, S. Effects of future climate change on the forests of Madagascar. Ecosphere 13, (2022).
Shirk, P. Sex-Specific Habitat Use and Responses to Fragmentation in an Sex-Specific Habitat Use and Responses to Fragmentation in an Endemic Chameleon Fauna Endemic Chameleon Fauna Downloaded from Downloaded From. https://scholarscompass.vcu.edu/etd/390 (2012).
Erickson, K. D. & Smith, A. B. Modeling the rarest of the rare: a comparison between multi‐species distribution models, ensembles of small models, and single‐species models at extremely low sample sizes. Ecography 2023, e06500 (2023).
van Proosdij, A. S. J., Sosef, M. S. M., Wieringa, J. J. & Raes, N. Minimum required number of specimen records to develop accurate species distribution models. Ecography 39, 542–552 (2016).
Clark, J. D., Dunn, J. E. & Smith, K. G. A multivariate model of female black bear habitat use for a geographic information system. J. Wildl. Manag. 57, 519 (1993).
Hirzel, A. H., Hausser, J., Chessel, D. & Perrin, N. Ecological-niche factor analysis: how to compute habitat-suitability maps without absence data? Ecol. 83, 2027 (2002).
Karger, D. N. et al. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 4, 1–20 (2017).
Chen, M. et al. Global land use for 2015–2100 at 0.05° resolution under diverse socioeconomic and climate scenarios. Sci. Data 7, 320 (2020).
Naimi, B., Hamm, N. A. S., Groen, T. A., Skidmore, A. K. & Toxopeus, A. G. Where is positional uncertainty a problem for species distribution modelling? Ecography 37, 191–203 (2014).
Olson, D. et al. Terrestrial ecoregions of the world: a new map of life on Earth. Bioscience 51, 933–938 (2001).
Hijmans, R. Terra: spatial data analysis. R package version 1.7-74. (2024).
Castiglione, S. et al. A new method for testing evolutionary rate variation and shifts in phenotypic evolution. Methods Ecol. Evol. 9, 974–983 (2018).
Tonini, J. F. R., Beard, K. H., Ferreira, R. B., Jetz, W. & Pyron, R. A. Fully-sampled phylogenies of squamates reveal evolutionary patterns in threat status. Biol. Conserv 204, 23–31 (2016).
Castiglione, S., Serio, C., Mondanaro, A., Melchionna, M. & Raia, P. Fast production of large, time‐calibrated, informal supertrees with tree.merger. Palaeontology 65, e12588 (2022).
Allouche, O., Tsoar, A. & Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232 (2006).
Hirzel, A. H., Le Lay, G., Helfer, V., Randin, C. & Guisan, A. Evaluating the ability of habitat suitability models to predict species presences. Ecol. Model. 199, 142–152 (2006).
Zhao, X. et al. Additive, antagonistic and synergistic interactions of future climate and land use change on Theaceae species assemblages in China. Glob. Ecol. Conserv 48, e02750 (2023).
Jamwal, P. S., Di Febbraro, M., Carranza, M. L., Savage, M. & Loy, A. Global change on the roof of the world: Vulnerability of Himalayan otter species to land use and climate alterations. Divers Distrib. 28, 1635–1649 (2022).
Moisen, G. & Freeman, E. Presence Absence: an R package for presence absence analysis. J Stat Softw 23, (2008).
Elith, J. et al. A statistical explanation of MaxEnt for ecologists. Divers Distrib. 17, 43–57 (2011).
Breiman, L. Random forests. Mach. Learn 45, 5–32 (2001).
Thuiller, W., Lafourcade, B., Engler, R. & Araújo, M. B. BIOMOD - A platform for ensemble forecasting of species distributions. Ecography 32, 369–373 (2009).
Marmion, M., Parviainen, M., Luoto, M., Heikkinen, R. K. & Thuiller, W. Evaluation of consensus methods in predictive species distribution modelling. Divers Distrib. 15, 59–69 (2009).
da Silva, J. M. & Tolley, K. A. Diversification through ecological opportunity in dwarf chameleons. J. Biogeogr. 44, 834–847 (2017).
Thuiller, W., Lavorel, S., Araújo, M. B., Sykes, M. T. & Prentice, I. C. Climate change threats to plant diversity in Europe. Proc. Natl Acad. Sci. 102, 8245–8250 (2005).
RRdtn - Species Distribution Modelling for Rare Species. R package https://doi.org/10.5281/zenodo.12734585.
Acknowledgements
We are grateful to Francesco Carotenuto for his precious advice given in commenting some of the statistical approaches developed here.
Author information
Authors and Affiliations
Contributions
AM, MDF and PR conceived the study. AMB, GG, MM, CS and AE collected the data and helped with the preparation of the manuscript. AM conducted the main analyses with inputs by SC and MDF. PR and AM wrote the first draft with significant inputs by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Communications Biology thanks Luciano Bosso and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Luke Grinham, Christina Karlsson Rosenthal, and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mondanaro, A., Di Febbraro, M., Castiglione, S. et al. Modelling reveals the effect of climate and land use change on Madagascar’s chameleons fauna. Commun Biol 7, 889 (2024). https://doi.org/10.1038/s42003-024-06597-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06597-5
This article is cited by
-
Habitat use, selection, and population density of chameleon communities (Calumma spp.) in the fragmented central highlands forests of Madagascar
BMC Ecology and Evolution (2025)
-
Impact of global change on water availability in Madagascar: evaluation of ISIMIP3b GCM performance and comparison with Soil and Water Assessment Tool modeling with Inter- and Intra-basin approaches
Climatic Change (2025)