Abstract
The two main Afrotropical malaria vectors - Anopheles coluzzii and An. gambiae – are genetically distinct and reproductively isolated across West Africa. However, populations at the western extreme of their range are assigned as “intermediate” between the two species by whole genome sequence (WGS) data, and as hybrid forms by conventional molecular diagnostics. By exploiting WGS data from 1190 specimens collected across west Africa via the Anopheles gambiae 1000 Genomes network, we identified a putative taxon in the far-west (provisionally named Bissau molecular form), which did not arise by admixture but rather may have originated at the same time as the split between An. coluzzii and An. gambiae. Intriguingly, this taxon lacks insecticide resistance mechanisms commonly observed in the two main species. These findings lead to a change of perspective on malaria vector species in the far-west region with potential for epidemiological implications, and a new challenge for genetic-based mosquito control approaches.
Similar content being viewed by others
Introduction
Members of the Anopheles gambiae complex include the most important Afrotropical malaria vectors. Because of their huge public health relevance, they have been subject of cytogenetic, genetic and genomic studies for more than 50 years. The large amount of knowledge and data available on these species puts them at the forefront of research on genomic patterns associated with species formation. Anopheles gambiae complex species have invaluable features as model organisms in the speciation genomics field: (i) they are morphologically indistinguishable, but genetically and ecologically differentiated, showing adaptation to different breeding habitats, different geographic distribution at the continental and local scales and different seasonality1,2,3; (ii) they are characterised by a short generation time (~1–2 generations/month) enhancing opportunities to monitor genomic changes over time; (iii) they show very large effective population sizes (Ne ~ 5–6 million) and very high nucleotide diversity4,5, enhancing capacity for adaptive evolution; (iv) they occur over a broad distribution range, encompassing contrasting environments, as well as potential geographical and anthropogenic barriers6,7; (v) they have different degrees of reproductive isolation by post-mating isolation and/or pre-mating mechanisms8; (vi) they are subject to strong and contemporary selection pressures due to extensive use of insecticides during the last 70 years, which has driven adaptive introgression9,10 and allows observation of genomic evolution in short ecological timescales. However, despite long-standing ecological and genomic studies triggered by the huge public health relevance of the complex, the speciation process and its drivers are not yet completely deciphered.
Nine sibling species are presently known in the complex, three of which were only described in the last decade. Anopheles coluzzii and An. gambiae have been raised to the species status in 201311 following evidence of genetic discontinuities within An. gambiae s.s. These were initially based on different frequencies of inversion arrangements on the right arm of chromosome-2, which led to the description of five sympatric non-panmictic ‘chromosomal forms’, informally named MOPTI, SAVANNA, BAMAKO, FOREST and BISSAU1,12. Based on this evidence, a ‘chromosomal theory of ecotypic speciation’ was proposed13 (and later confirmed by genomic evidence on the BAMAKO chromosomal form14,15). In this model, chromosomal inversions play an instrumental role in speciation by protecting part of the genome from recombination at the heterozygous state, whereas inverted homozygotes found in more marginal environments allow further ecological expansions. Transitory population expansions and crashes, and the attendant genetic drift and/or strong directional selection pressures would favour genetic mechanisms like inversions that can stabilise novel, adaptive gene associations. Decades after the chromosomal forms’ first description, two assortative mating units (the M and S-molecular forms, partially overlapping with the SAVANNA and MOPTI chromosomal forms) were described based on form-specific SNPs in the rDNA IGS region16. The availability of easy genotyping of these SNPs17,18 (or of the An. coluzzii-specific insertion of a Short INterspersed Element19) allowed accumulation of evidence on the genomic and ecological differentiation of the molecular forms eventually leading to the description of the two formally named species11. The BISSAU chromosomal forms, reported from The Gambia and Guinea Bissau1,20, remains less clearly defined by contemporary population genomic approaches.
Anopheles coluzzii and An. gambiae are sympatric in sub-Saharan Africa west of the Rift Valley where they represent the major vectors of human malaria. The species’ ability to benefit from human-made habitats has linked their speciation process to human-made environmental changes21. Anopheles coluzzii has broken free from the need to rely on rain-dependent breeding sites—typical of fresh-water species of the complex, including An. gambiae - by adapting to more permanent breeding sites produced by irrigation, deforestation and urbanisation. This main feature led to a differentiation of the seasonal and the meso-/micro-geographic distribution of the two species. Furthermore, differences in survival strategies across the dry season have been described22,23. From the genomic perspective, while in An. gambiae intraspecific differentiation is extremely weak across the whole range, An. coluzzii shows a ‘bimodal’ distribution pattern with populations occupying the xeric savannah belt in the northern hemisphere differentiated from populations occupying the ribbon of coastline along the Gulf of Guinea24,25.
Anopheles coluzzii and An. gambiae are known to mate freely and produce viable progeny under laboratory conditions. However, putative hybrids (i.e. individuals showing a heterozygous pattern of diagnostic molecular markers) are rarely found at frequencies >1% in the field, due to pre-mating mechanisms (i.e. swarm segregation and within swarm assortative mating) and incompletely understood post-mating mechanisms of selection against hybrids8. However, in coastal areas at the western extreme of the two species range (from The Gambia to Guinea Bissau, hereafter, ‘far-west’) frequencies of putative hybrids >20% are stably observed26,27. Genetic studies have revealed that these ‘far-west’ populations are highly differentiated from neighbouring populations in inland areas28,29,30, leading to the hypothesis that they may have originated from extensive introgressive hybridisation between the two species30. Based on Ag1000G Phase-1 and Phase-2 whole genome sequence (WGS) data, these coastal populations were later defined as ‘intermediate’ between An. coluzzii and An. gambiae5,25. Interestingly, the previously-mentioned BISSAU chromosomal form - characterised by high frequencies of 2 Rd and low frequencies of 2La inversions, was described in the same geographical region12,20.
The aim of this work was to understand the nature of the ‘intermediate far-west’ populations focusing on their relationships with West-African An. coluzzii and An. gambiae, and to confirm or confute the hypothesis of their hybrid origin30. To this aim, we exploited Ag1000G Phase-3 data31, which include additional samples from The Gambia and Guinea Bissau compared to those available in Phase-1 and -2. Results obtained by population genomic, divergence and demographic analyses illustrate the need for a change of perspective in the current view of ongoing evolutionary processes in major malaria vector species at the western extremes of Sub-Saharan Africa, suggesting the existence of a putative taxon not originating from admixture between An. coluzzii and An. gambiae. Available data on genetic traits associated with resistance to insecticides in this taxon suggest lack of current gene-flow with both species.
Results
Genomic structure reveals clusters in FW-Africa distinct from western An. coluzzii and An. gambiae
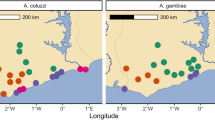
Results of PCAs based on SNPs either on single chromosomes (Fig. 1B, Supplementary Fig. 1) or on the whole genome (Supplementary Fig. 2A) consistently separate gcx1-specimens from the two clusters grouping An. coluzzii (hereafter, CO) and An. gambiae (GA, including individuals from Leibala in Guinea Bissau) specimens, respectively. All gcx2 individuals cluster together either between gcx1 and CO (based on chromosomes -X and -3 SNPs) or close to CO (based on chromosome-2 and whole genome SNPs; Supplementary Fig. 2A). When focusing the analysis on the centromeric region of chromosome-X (known as the CO and GA speciation island32) gcx1 individuals consistently cluster separately from CO+gcx2 and GA (Supplementary Fig. 2B). When focusing the analysis on the genomic regions included within chromosomal inversion 2 Rd and 2La (whose frequencies characterise the so called BISSAU chromosomal form12) PC1 separates individuals based on their 2La karyotype, while PC2, which explains similarly high variance, separates gcx1 from CO+gcx2 and GA (Supplementary Fig. 2C).
A Distribution and numbers of 1190 Ag1000G Ag3.0 individuals included in the analysis (right) with focus on the Far-West region (left), according to the Ag1000G Consortium5,25. B Principal component analysis based on SNPs on chromosome-X, -3 and -2. Circles = Anopheles coluzzii and Anopheles gambiae; crosses = Far-West individuals. C ADMIXTURE Bayesian ancestry most parsimonious models for chromosome-X (K = 3) and chromosome-3 (K = 3). Anopheles coluzzii: BFcol (Burkina Faso), CIcol (Côte d’Ivoire), GHcol (Ghana), GNcol (Guinea), MAcol (Mali); An. gambiae: BFgam (Burkina Faso), GHgam (Ghana), GNgam (Guinea), GWgam (Guinea-Bissau), MAgam (Mali).
Results of individual-based ancestry analysis (ADMIXTURE; K = 3) are largely consistent with those from PCA, suggesting a third ancestral cluster (purple) in addition to CO (red) and GA (blue) clusters (Fig. 1C; Supplementary Figs. 3 and 4). All gcx1 individuals show mostly ‘purple’ ancestry, with a limited proportion of GA-ancestry on chromosome-3 in gcx1 from The Gambia (hereafter, gcx1-GM) and some proportions of GA and CO on both chromosomes in gcx1 from Guinea Bissau (gcx1-GW). On the other hand, gcx2 individuals share gcx1 and CO ancestries. At K = 2, all FWpops and CO share the same ancestry, with a limited proportion of GA-ancestry in gcx1 (Supplementary Figs. 3 and 4).
High genomic divergence between FWpops, An. coluzzii and/or An. gambiae
In this and in the following paragraphs results are shown from analyses based on chromosome-3 euchromatic region to assess genomic divergence and gene-flow among FWpops (gcx1-GM, gcx1-GW and gcx2), CO and GA.
To assess whether FWpops form a genomic group differentiated from CO and GA, we measured the extent of divergence among populations relative to the net genetic diversity using Hudson’s FST (Fig. 2A). Within FWpops, the highest level of divergence is observed between gcx1-GM and gcx2 (0.020), while gcx1-GW exhibits similar and lower values with both gcx1-GM and gcx2 (0.009). All FWpops show levels of differentiation with GA (gcx1-GM = 0.031–0.032; gcx1-GW = 0.026–0.028; gcx2 = 0.034–0.035) in the ranges of that between CO and GA (0.029–0.040). Moreover, gcx1-GM and gcx1-GW show high (0.028–0.034) and intermediate (0.018–0.028) levels of differentiation with CO, respectively.
Genetic diversity statistics show (Fig. 2B): (i) no significant variation in nucleotide diversity among the 3 FWpops, CO and GA, suggesting similar effective population size in all groups; (ii) negative values of Tajima’s D for all populations, consistent with population expansion; (iii) Tajima’s D values for gcx1-GM comparable to those of the two CO populations from coastal areas, suggesting less extreme population growth in this region; (iv) lower Tajima’s D values for gcx1-GW and gcx2 compared to gcx1-GM; (v) non-significant differences in absolute genetic divergence (Dxy) between FWpops and either CO or GA (Supplementary Fig. 5).
Patterns of genomic divergence and admixture in FW-Africa
We applied different methods (F3 statistics, Patterson’s D-statistics and Tree-Mix) to test for the possible origin of FWpops from admixture between CO and GA and assess their reciprocal genomic relationships.
Results of admixture-F3 tests reject the hypothesis of FWpops’s origin by hybridisation between CO and GA (Z-values » 5 in all 75 tests; Supplementary Table 1). On the other hand, results of the second F3 analysis testing the hypothesis of hybridisation between CO and FWpops suggest that gcx1-GW might have originated from admixture between gcx1-GM and either gcx2 or CO (Supplementary Fig. 6). Outgroup-F3 performed to calculate shared genetic drift of FWpops and either CO or GA, as reference populations, showed a closer relationship of FW with CO (F3 = 0.018–0.030).
Results of Patterson’s D statistics - performed to test for excesses of shared derived polymorphisms - showed that regardless of topology, all FWpops show statistically significant levels of gene-flow with CO (D < 0; Z score > −5), but not with GA (Supplementary Table 2).
Genomic relationships between FWpops, CO and GA were assessed by building a population tree by using TreeMix, based on genome-wide allele frequency data derived from allele counts. For this purpose, CO and GA individuals collected in different sites from the same country and in different years were grouped together. Anopheles christyi was used as outgroup. For every migration (m) value modelled (from 0 to 3), TreeMix placed FWpops as sister to CO relative to GA and An. christyi, with a FW-branch splitting from CO not long after the split from the GA lineage. In the optimal model (1 migration edge, 99% of variance explained; Fig. 3; (Supplementary Fig. 7, Supplementary Fig. 8), all FWpops form a clade sister to CO, with migration from GA to gcx1, but not to gcx2. This topology suggests there was a single FW ancestral population similar to CO, with subsequent divergence between gcx1 and gcx2 driven in part by GA introgression into gcx1, leaving gcx2 retaining more affinity to CO. Models with larger migration values have higher likelihoods, but not sufficiently so to justify the more complex topologies and are largely congruent with the optimal model. For example, the m = 2 model suggest that gcx2 has ancestry from both a CO-like branch and a gcx1-like branch, but it remains true that all FWpops share a common ancestry and that gcx2 is closer to CO than gcx1 is. This interpretation is largely consistent with scenarios supported by results from PCA and ADMIXTURE.
Demographic history in West Africa
Folded allele frequency spectra results show that FWpops have an excess of rare variants, suggesting a population expansion equivalent to that observed in CO and GA (Supplementary Fig. 9).
We here focused on inferring the demography of gcx1-GM cluster, which previous results suggest as the FWpop most isolated from CO and GA. To this aim, ∂a∂i analysis was performed with the goal to discriminate among three alternative hypotheses, i.e. whether gcx1-GM: (1) originated from admixture between CO and GA; (2) split simultaneously from CO and GA; or (3) experienced divergence from either CO or GA (Fig. 4). Lowest AIC values support the second hypothesis (Supplementary Table 3, θ = 881.21; ΔAIC ≥ 82,840 for all other models) suggesting that the divergence of the gcx1-GM population is contemporary to the split between CO and GA.
Identification of gcx1-GM related SNPs
Here we focused on gcx1-GM as representative of the most divergent FWpop in order to identify its most distinctive genomic characters and create the basis for the future development of a molecular diagnostic method.
We identified 44 SNPs showing Fst values > 80% between gcx1-GM and both CO and GA (Supplementary Table 4; Supplementary Fig. 10). These are 29 in coding regions aligned to the PEST genome (17 on chromosome-2R, of which 15 cluster in a 60 Kbp region of AGAP004008 gene encoding for a myosin and a protein kinase domain-containing protein; 3 on chromosome-2L, 2 on chromosome-3R; and 7 on chromosome-3L, of which 6 cluster in a 46 Kbp region of AGAP029482 gene encoding for a TPR region domain-containing protein) and 15 in positions out of PEST genome coding regions (6 on chromosome-2R; 1 on chromosome-3R; 7 on chromosome-3L, 6 of which cluster in a 70 Kbp region; and 1 on chromosome-X). The latter could represent the most gcx1-GM distinctive SNPs with respect to both CO and GA.
Some of the above SNPs (in bold in Supplementary Table 5) could be used to design a molecular diagnostic assay, although no fixed SNPs are identified and allele frequency differences (DAF) between gcx1-GM and both CO and GA are rarely >80%. However, a more specific two-step approach could be envisaged to identify gcx1-GM. The first step would be to separate gcx1-GM from CO and gcx2 based on Ancestry Informative Markers on chromosome-X (sensu Ag1000G5,33). The second step would be to focus on allele frequency differences (DAF) between gcx1-GM and GA (Supplementary Table 5). To this aim, we identified three SNPs on chromosome-2 fixed in gcx1-GM and showing frequencies <12% in GA and further three SNPs showing a DAF > 98% (all on chromosomal arm 2 R). Arbitrarily lowering the threshold to >85% leads to the identification of 201 gcx1-GM-related SNPs, 85% of which are on chromosome-2 and 1.5% on chromosome-X. Almost all of the identified chromosome-2 SNPs are clustered in two regions on chromosome arm 2 R (15% within 28.01–28.53 Mb; 45% within 47.79–47.85 in Mb) and in two regions on chromosome arm 2L (16% within 1.95–2.53 Mb; 17% within 46.60–46.74 Mb). Supplementary Fig. 11 shows the location of chromosome-2 gcx1-GM-related SNPs on a graph showing chromosome-2 FST per 1 bp between gcx1-GM and An. gambiae. Supplementary Table 5 also reports the genes included in these four chromosomal regions.
Computation of individual allelic percentages (Fig. 5) shows that 74 out of 77 gcx1-GM specimens carry >80% of the 201 gcx1-GM-related alleles, while all 449 GA specimens carry <20% of gcx1-GM-related alleles, suggesting these values as possible cut-offs for the individual allelic percentage to discriminate gcx1-GM from GA with >99.4% accuracy.
Rows represent individual mosquitoes (grouped by population) and columns represent SNPs (grouped by chromosome arm). Approximate chromosomal position is given below the graph. Colours represent gcx1-GM related (Purple), An. gambiae (GA; Green), gcx1-GM/GA (cyan) heterozygous genotypes; Missing genotypes in black. The column at the far right shows the individual allelic percentage (A.P.).
Ranges of individual allelic percentages of the above gcx1-GM related alleles for the other populations are: (i) 6–60% for all CO; (ii) 31–90% for all gcx2 and (iii) 61–80% for 16 out of 93 gcx1-GW specimens, with the remaining 77 specimens showing values >80%. Thus, no CO would be falsely identified as gcx1-GM exceeding an 80% cut-off, and no gcx2 or gcx1-GW would be falsely identified as GA under a 20% cut-off, though some would be identified as gcx1-GM (Supplementary Fig. 12, also showing snapshots of the two chromosomal regions with low CO vs GA divergence including clusters of SNPs with highest Fst values between gcx1-GM and CO + GA).
Insecticide resistance in FW-pops
Amino acid changes at target sites known to be associated with insecticide resistance (i.e. Vgsc, Rdl and Ace-1) are virtually absent in FWpops, although they are commonly observed—sometimes at very high frequencies - in all CO and GA populations analysed (Supplementary Fig. 13).
Analogously, CNVs associated with metabolic insecticide resistance are common in CO and GA populations, but are either absent (detoxification gene GST) or present at much lower frequencies (CYP6AA1, CYP6AA2, CYP6AA/P, CYP6M2 and CYP9K1) in FWpops (Supplementary Fig. 14). Both gcx1-GM and gcx1-GW lack evidence of CNVs associated with insecticide resistance. Although an amplification frequency of up to 57% was observed at CYP9K1, the presence of a duplication was not accompanied by a supporting signal of selection based on the H12 scans (see below), unlike for other species. CYP6AA1, CYP6AA2 (overexpressed in pyrethroid resistant populations) and CYP9K1 was observed at low frequencies (1–7%) in gcx2.
In contrast to what was observed in CO and GA, results from the H12 haplotype diversity selection scans show negligible evidence of positive selection in the vicinity of IR amino acid substitutions across the genome in FWpops. In gcx1-GM the only selective peak is observed at CYP6AA/P and is not associated with any amino acid change at the locus (see above). In gcx2, two selective peaks are observed at Rdl and around the CYP9K1 gene (Supplementary Fig. 15).
Discussion
Analysis of Phase-3 Ag1000G populations from West Africa suggests the existence of a demographically stable cryptic taxon of the An. gambiae complex in the coastal region of far-west Africa. This taxon is best represented by gcx1, and it is hereafter provisionally named Bissau molecular form, due to the possible partial overlap with the BISSAU chromosomal form (see below). This conclusion represents a change of perspective in the interpretation of ‘intermediate’ FW-populations.
Several lines of evidence support the hypothesis that this taxon is distinct from any described species sensu stricto.
In The Gambia, the Bissau molecular form (corresponding to gcx1-GM) is characterised by: (i) a unique ancestral gene-pool (purple in ADMIXTURE analysis; Fig. 1) and genetic isolation from West-African An. coluzzii and An. gambiae (as revealed by PCA results); (ii) levels of divergence from West-African An. coluzzii (FST = 0.028–0.034) and An. gambiae (FST = 0.026–0.033) in the range of that observed between the two species (FST = 0.029–0.041); (iii) differentiation from An. gambiae by 201 SNPs mainly on chromosome-2 close to fixation (i.e. showing differences in allele frequencies >85%). Notably, results of PCA analyses of novel samples collected along the Gambia river in 2019 and 202134,35 strongly support the Bissau molecular form as a cryptic group spread from the coast of The Gambia to the eastern border with Senegal and stable in the country for at least 15 years (Supplementary Fig. 16).
Results from demographic analyses suggest a complex history, with distinct methods revealing different population aspects. It is clear that the Bissau molecular form did not originate from simple admixture between An. coluzzii and An. gambiae (as previously hypothesised by Vicente et al. 30), as this model is not supported by any of the methods that could detect it, including ADMIXTURE, F3, TreeMix and ∂a∂i. Instead, the Bissau molecular form appears to have originated as a distinct lineage from the same ancestral population that produced An. coluzzii and An. gambiae. The specific nature of divergence and subsequent gene flow leading to the distinctive Bissau taxon remains somewhat equivocal. While ∂a∂i predicts a perfect polytomy, TreeMix assumes a bifurcating topology and thus forces the Bissau molecular form closer to one species (An. coluzzii) than to the other (An. gambiae), though with no formal support for internal branches. We thus infer that divergence among the three lineages happened very quickly, though it is difficult to conclude whether it was truly simultaneous, or whether the original divergence has been obscured by later gene flow. Similarly, ∂a∂i does not detect migration, though there is strong evidence of gene flow via both TreeMix and Patterson’s statistics. Patterson’s statistics will be significant if there is inter-branch migration or if the topology is incorrect; however, no topology is consistent with complete reproductive isolation. Gene-flow from An. gambiae seems to be relatively minor, as supported by ADMIXTURE and TreeMix, while gene-flow from An. coluzzii may have been substantial although difficult to distinguish from a more recent split from An. coluzzii than from An. gambiae.
In coastal Guinea Bissau, the Bissau molecular form seems to have undergone admixture, leading to a demographically stable population (corresponding to gcx1-GW), which: (i) clusters with gcx1-GM in PCA; (ii) is characterised by admixed gcx1-GM and gcx2-ancestries (as revealed by ADMIXURE with K > 3); and (iii) shows a high level of divergence with An. gambiae (FST = 0.026–0.028) and intermediate ones with An. coluzzii (FST = 0.018–0.024). F3 analysis suggests an origin of this population from an admixture between Bissau molecular form and coastal An. coluzzii populations (including gcx2). Closeness to gcx1-GM and gene-flow with An. coluzzii is also confirmed by Patterson’s statistics (Supplementary Table 2).
In The Gambia, the Bissau molecular form is found in sympatry with gcx2 (hereafter FW-An. coluzzii), which appears as a peculiar local An. coluzzii-like population (as revealed by PCA, ADMIXTURE, F3). Interestingly, further evidence of the presence of FW-An. coluzzii at the western extreme of the country come from Ag3.5 dataset of the MalariaGEN Vector Observatory, i.e. the project succeeding Ag1000G35.
It is relevant to note that while most analyses were carried out on the euchromatic region of chromosome-3 - which is well known to cluster individuals by geographical region, rather than to be associated with interspecific divergence between An. coluzzii and An. gambiae5- genomic structure analyses (PCA and ADMIXTURE) were carried out on the whole genome, on each of the three chromosomes and on specific chromosomal regions separately, providing consistent results. The first specific region is the pericentromeric region of chromosome-X (Supplementary Fig. 2B), a region of reduced recombination known to be associated with reproductive isolation between An. coluzzii and An. gambiae36. The second includes the euchromatic regions within 2Rd and 2La paracentric inversions of chromosome-2 (Supplementary Fig. 2C), which have been included in the analysis, as differences in the frequencies of the two inversion polymorphisms were associated with BISSAU chromosomal form in the same geographic regions12,20. PCA results on SNPs within the two inversions suggest a major role of these genomic regions in separating the Bissau molecular form from An. coluzzii, FW-An. coluzzii and An. gambiae. This latter result suggests a possible partial overlap between the BISSAU chromosomal form and the Bissau molecular form, analogous to the relationship between the SAVANNA and MOPTI chromosomal forms and the S- and M-molecular forms, known today as An. gambiae and An. coluzzii16,37. In fact, although cytogenetic evidence was instrumental in revealing lack of panmixia within An. gambiae s.s., only genomic data allow the full characterisation of the different units, as chromosomal forms shared inversion polymorphisms (2Rd and 2La in the case of the BISSAU and FOREST/SAVANNA chromosomal forms) which impede the assignment of each specimen to a given form.
The actual existence of the Bissau molecular form as a cryptic taxon within the An. gambiae complex is further supported by lack of genomic evidence of insecticide resistance mechanisms commonly observed in An. gambiae and An. coluzzii across their entire range, sometimes also at very high frequencies. First, amino acid changes associated with target site resistance mechanisms (i.e. Vgsc, Rdl and Ace1) are virtually absent in all FW-pops (Supplementary Fig. 13). Analogously, CNVs associated with metabolic insecticide resistance are common in An. coluzzii and An. gambiae populations, but are either absent (detoxification gene GST) or present at much lower frequencies (CYP6-related) in FW-pops (Supplementary Fig. 14). Finally, H12 haplotype diversity selection scans show negligible evidence of positive selection in the vicinity of IR amino acid substitutions. This was already reported by Clarkson et al. 38 for fewer FWpops (Ag1000G Phase-2), despite lack of evidence of lower insecticide pressure in coastal FW-region compared to inland western regions. These genomic results not only support the ancestral divergence of the Bissau molecular form from the two species, but also suggest that the novel taxon may either be more susceptible to insecticides or have different insecticide resistance mechanisms. As the analysed specimens were collected as adults indoors, the Bissau molecular form is likely to be exposed to malaria vector control interventions by either LLIN or IRS, commonly implemented in the FW-region as well as across sub-Saharan Africa. Bioassay surveys, to determine insecticide resistance phenotype, will be necessary to establish if the Bissau molecular form is susceptible to insecticides or carries new resistance conferring mutations.
Interestingly, recent studies on populations collected in The Gambia >4 years after those analysed in the present work report presence of the 995F kdr resistant allele in An. coluzzii, in An. gambiae and in putative hybrids (likely belonging to the Bissau molecular form), both in the coastal area (frequencies up to 17%) and inland eastern sites (frequencies up to 28%)39. This may suggest adaptive introgression in the Bissau molecular form (as extensively shown between An. coluzzii and An. gambiae9,10,40), but lack of genomic identification of the sampled individuals precludes confirming this speculation. In fact, conventional species molecular identification provides indications of ‘unusual’ situations when heterozygous patterns are observed (as in the Far-West region), but does not allow discrimination between An. gambiae and Bissau molecular forms individuals. The SNPs identified as distinctive of the Bissau form will be instrumental for the development of a diagnostic method, possibly exploiting a machine-learning assay (e.g. decision tree) to quantify the minimum number of SNPs required. This would allow studying the Bissau molecular form’s ecology, behaviour and epidemiological significance.
It is not the first time that a novel putative cryptic taxon has been found in the An. gambiae complex following observations of unusually high frequencies of individuals characterised by polymorphic An. coluzzii/ An. gambiae molecular diagnostic markers. In Burkina Faso, a subgroup genetically distinct from sympatric An. coluzzii was observed and named GOUNDRY41,42,43. Subsequently a second group genetically divergent from An. coluzzii and all other An. gambiae complex species (named Anopheles TENGRELA) was observed in the same geographic region and GOUNDRY was shown to be a recently diverged population from the TENGRELA lineage via introgression from An. coluzzii44. TENGRELA is characterised by taxon-specific variants44 which are absent in Bissau molecular form. In addition to being distinct from TENGRELA, Bissau molecular form has unique features. First, it is the first time that adult field females are suggested to belong to a new taxon close to An. coluzzii and An. gambiae, as both TENGRELA and GOUNDRY were collected exclusively as larvae and no adults have ever been found despite extensive and long-term efforts. Second, present results (and additional reports from 2019 and 2021; Supplementary Fig. 16) show that the size of the geographic range of the Bissau molecular form is much larger than that of TENGRELA and GOUNDRY. Third, several recurrent observations support the presence of stable populations with unique gene-pools in the field (likely corresponding to the Bissau molecular form): (i) admixed An. gambiae populations observed in coastal The Gambia since 200527,45; (ii) frequencies >20% of individuals characterised by An. coluzzii/An. gambiae heterozygous genotypes observed in coastal Guinea Bissau since 199510,26,45; (iii) data on chromosome -X and -3 microsatellites and on chromosome-2R paracentric inversion polymorphisms showing that these admixed coastal An. gambiae populations were highly differentiated from An. gambiae populations from inland areas both in The Gambia28 and in Guinea Bissau30. Even more importantly, results of PCA analyses of novel samples repeatedly collected along the Gambia River in 2019 and 202134, strongly support the Bissau molecular form as a cryptic group spread from the coast of The Gambia to the eastern border with Senegal and stable in the region since at least 15 years (Supplementary Fig. 16).
The results presented lead to a change of perspective on the major malaria vector species at the extreme west of their range, supplanting previous hypotheses of either species radiation promoted by massive genomic introgression between An. coluzzii and An. gambiae (based on the genotyping of few species-specific markers29,30), or of an ‘intermediate’ far-west taxon (based on WGS data from Phase-1 Ag1000G dataset5). At this stage, it is not possible to assess the implications of this change of perspective on malaria transmission and conventional vector control in the region. However, it is already evident that the existence of a possibly widespread putative taxon in addition to those representing the actual target of future gene-drive based control interventions46,47, will represent a new challenge for these innovative mosquito control approaches.
In conclusion, there is still much to understand on Bissau molecular form taxonomic status, demographic history and degree of isolation from An. coluzzii and An. gambiae and other sympatric members of the An. gambiae complex, such as An. arabiensis and An. melas. Deeper studies will be soon possible thanks to new releases of WGS data by the MalariaGEN Vector Observatory, including those from additional Bissau molecular form populations in The Gambia mentioned in the Discussion and shown in Supplementary Fig. 16. There is more to learn about the Bissau form geographical range, its larval and adult bionomics and, most importantly, its epidemiologically relevant behavioural traits. These studies are dependent upon the development of a high-throughput and cheap molecular method to identify Bissau from individuals in the field in the simplest way possible. The preliminary identification of Bissau molecular form-associated SNPs (as well as the identification of new specific ones by analysis of unfiltered genome data) opens the possibility for the development of a multi-locus genotyping approach to identify Bissau form individuals at all stages of its life cycle.
Methods
West-African Ag1000G Phase-3 data
The sample data set utilised in the present study includes the genome sequences and associated metadata of 1190 mosquito females collected between 2004 and 2014 at 36 sites from 7 different countries in West Africa provided by MalariaGEN Anopheles gambiae 1000 Genomes (Ag1000G) Project Phase-3 data resource (Ag3.0) (Fig. 1A; Table 1; Supplementary Data 1). Details of the samples and associated references are provided in the Ag1000G partner studies page (https://malariagen.github.io/vector-data/studies-ag1000g.html). Analysed samples include: 374 An. coluzzii individuals from Burkina Faso, Côte d’Ivoire, Ghana, Guinea, Mali; 449 An. gambiae individuals from Burkina Faso, Ghana, Guinea, Mali and inland Guinea-Bissau; 274 individuals collected indoors in The Gambia and 93 in coastal Guinea-Bissau (referred to as Far-West, FW). All FW-populations (FWpops) were classified as ‘intermediate’ according to the Ag1000G Consortium5,25, and named gcx1-GM or gcx2 (in the case of populations from The Gambia) and gcx1-GW (in the case of populations from Guinea Bissau)5,24
Sequence data utilised to explore the genetic variation and structure are available from the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home). ENA accession numbers for the specific samples and sequencing runs used in this study are provided as Supplementary Table 1. Further information on availability of these data is available from the MalariaGEN website at https://malariagen.github.io/vector-data/ag3/ag3.0.html.’
SNPs on the three chromosomes were first filtered by the ‘gamb_colu’ site filters to reduce sequencing and alignment error as defined by Ag1000G Consortium. This filtering procedure is based on the accuracy of genotyping at each position and is not expected to remove fixed alleles. A variant set/chromosome was then generated by selecting biallelic variants with a minor allele frequency ≥1% and randomly down-sampling to 100,000 SNPs per chromosomal arm. Subsequently we pruned for linkage disequilibrium by excluding variants SNPs above an r2 threshold of 0.01 in moving windows of 500 SNPs with a step size of 250 SNPs using the locate_unlinked function in scikit-allel Python package48.
To carry out genomic window-level analyses, a second biallelic SNP set was generated for the euchromatic region chromosome-3 (i.e. 34,707,855 SNPs in 3R: 1–24 Mbp; 3L: 15–41 Mbp). This region is recognised to provide the most coherent view of population structure due to the absence of polymorphic inversions49 and to cluster populations by geographical region rather than by An. coluzzii and An. gambiae species5.
Finally, for the analysis of genomic diversity and divergence across the whole genome, all 191,422,813 SNPs under ‘gamb_colu’ site filters were assessed.
Supplementary Table 6 summarises the details of the above datasets and the analyses carried out on each of them, which are detailed in the following paragraphs.
Genetic variation among West African populations
Patterns of genetic variation were explored by PCA50, implemented by scikit-allel with the function allel.pca (gn, n_components = 10, copy = True, scaler = ‘patterson’, ploidy = 2).
Estimates of individual ancestries and detection of population structure and admixture were assessed by the maximum likelihood approach implemented in ADMIXTURE 1.3.051. A 5-fold cross-validation for K values from 1 to 8 was run.
All following analyses were run on PCA inferred clusters.
Nucleotide diversity and divergence in FW
The average pairwise nucleotide diversity (i.e., π52), Tajima’s D53, Watterson’s θ54 and Hudson’s FST on 100,000 accessible bases55 were computed following Bhatia et al. 56. Each population was harmonised with a random sample size of 30 individuals: a sample size which has provided good resolution for a range of population genomic analyses in other species5,57,58. Absolute genetic divergence (i.e., Dxy) on 100,000 accessible bases was computed using scikit-allel48.
Inference of gene flow and introgression in FW clusters
PCA and ADMIXTURE analyses are powerful tools for detecting population substructure at the individual level, but they cannot discriminate among multiple possible population histories. In order to shed light on the latter, we applied methods (F3 statistics, Patterson’s D-statistics and Tree-Mix) that formally test for a history of population admixture and allow researchers to build models of population relationships that fit genetic data. All of these admixture tests assume bifurcating branch splits, even if later re-joined by gene flow, but they do not allow for polytomies caused by simultaneous divergence of multiple branches.
The F3 statistic59 is a formal test of admixture that estimates whether allele frequency differs between the target population X and populations A and B and indicates incomplete lineage sorting expected from a species tree or results from admixture, even if the gene flow events occurred hundreds of generations ago. First, we tested the hypothesis that FW clusters are the result of admixture between West-African CO and GA. For a simple bifurcating tree, the product of the frequency differences (F3 (FWpops; COpops, GApops) between COpops and FWpops and GApops and FWpops is expected to be positive. The product can be negative only if each FWpop has equal ancestry related to both CO and GA. Thus, a significantly negative F3 value (Z < 5) would provide evidence of admixture in FWpop history. Second, we tested the hypothesis that a FWpop is the result of admixture between CO or GA and another FWpop.
After the admixture test, we conducted outgroup-F3-statistics in the form of F3 (FWpops, reference population; outgroup) to test shared genetic drift between FWpops and a reference population since their divergence from the outgroup.
Patterson’s D-statistics60 provide evidence for admixture as well as some insights about the directionality of the gene flow. These statistics test whether an unrooted phylogenetic tree applies to the data, assuming that the analysed SNPs are ascertained as polymorphic in a population that is an outgroup (in our case a Anopheles christyi, a non-malaria vector closely related to the An. gambiae complex61 used as an outgroup following ref. 62) to both populations (in our case CO and GA) that make up one of the clades (in our case a single FWpop). The method (also known as ABBA BABA) compares the number of ABBA and BABA sites, parsimony-informative sites that support a different phylogeny than the species tree and determine whether they are statistically equal in number. The two genealogies discordant with the species tree, ABBA and BABA are equally likely to be produced by incomplete lineage sorting (ILS); therefore, they should not differ in number if only ILS, but not gene flow is present. A significant difference between ABBA and BABA sites indicates that two non-sister species are more similar to each other than expected, which is interpreted as a signal of gene flow. Three tree topologies were ascertained with GA [(COpops, FWpops), GApops; An. christyi], CO [(GApops, FWpops), COpops; An. christyi], or FW [(COpops, GApops), FWpops; An. christyi] as the source of gene flow.
Patterson’s D statistic with a jack-knife resampling approach was applied with a block-size of 100k SNPs for each test. All the tests were done using Python package scikit-allel48 with the specific function ‘allel.blockwise_patterson_d(aca, acb, acc, acd, blen=blen)’.
TreeMix63 allows evaluation of population splitting, drift and introgression based on frequency data from large numbers of SNPs, using a graph representation that allows both population splits and migration events, by building a maximum likelihood tree of sampled populations using a Gaussian approximation of genetic drift of allele frequencies. Subsequently, it identifies populations whose genetic covariance is underestimated by the model and migration events are added to improve fitness of the model. Ten TreeMix replicates were run for each m value (number of migration edges) up to 3 including -noss parameter and random seeds. To avoid converging on the same composite likelihood for each replicate, the number of SNPs per window (-k) was varied across runs from 500-1000 in 100 SNP increments using An. christyi as root. To evaluate the best m parameter, TreeMix output files were analysed with OptM v0.1.664, which modifies the approach of Evanno et al. 65 to choose an optimal model, using default parameters and plotted for m = 1, 2, 3 simulated models. All analyses were performed in R v3.6.3. The best tree model was plotted using the python module Toytree v.1.066.
Demographic inference of West African clusters
Demographic history can provide important insights into the underlying evolutionary processes that shape genetic differentiation, such as population bottlenecks, migration and admixture.
First, a site frequency spectrum (SFS) was computed for FWpops, CO and GA, using biallelic counts in SNPs from chromosome 3, using scikit-allel48.
To explore alternative demographic models for gcx1-GM, we used the diffusion approximation method of ∂a∂i67 to analyse joint site frequency spectra, i.e. the joint distribution of allele frequencies across biallelic variants. To guide the development of three-population models, three different demographic models were fitted by using ∂a∂i_pipeline v3.1.568 to test whether gcx1-GM (1) originated from admixture between CO and GA (admix_origin_no_mig); (2) split simultaneously to CO and GA (sim_split_no_mig); (3) experienced divergence from either CO or GA (split_nomig). Hypotheses were tested to generate a plausible demographic model, without considering gene-flow. For all models, consecutive rounds of optimisations were performed following Portik et al. 68. Across all analyses, the optimised parameter sets of each replicate were used to simulate the 3D-JSFS and the multinomial approach was used to estimate the log-likelihood of the 3D-JSFS given the model. Models were compared using the Akaike information criterion (AIC), and the replicate with the highest likelihood for each model was used to calculate AIC scores and ΔAIC scores69. To assess the robustness of our results, the best supported model was run establishing parameter bounds to ensure that the search remains within a feasible range of values. These bounds are crucial because optimisation methods often explore a wide range of parameter values, occasionally exceeding the permissible limits.
Identification of gcx1-GM related SNPs
To identify signals of highest differentiation between the most separated FWpop (gcx1-GM) and both CO and GA, genome-wide FST values were assessed at a resolution of 1 bp following the procedure by Bhatia et al. 56.
To identify a set of markers that can be used downstream for taxonomic assignment, differences in Allele Frequency (DAF) between gcx1-GM and An. gambiae were computed considering only biallelic SNPs. SNPs with overall genotyping success below 10% were excluded. The percentage of gcx1-GM-related alleles per individual (individual allelic percentage) was computed in order to define a cut-off able to discriminate gcx1-G-GM and An. gambiae specimens in the far-west African region.
Haplotype phasing
Haplotypes were phased from genotypes at biallelic SNPs with a combination of read-backed and statistical phasing as recommended by the AG1000G phase 3 project. Read-backed phasing was performed on individual samples using WhatsHap version 1.070.
Target-site insecticide resistance
Amino acid substitution frequencies were calculated for each population cohort using functions built into the malariagen_data python package. Predicted amino acid changes were identified based on a specified gene transcript and observed non-synonymous SNP frequencies. Frequencies were calculated for genes with an established association with target site resistance including the Voltage-gated sodium channel (Vgsc), the GABA-gated chloride channel subunit (Rdl) and acetylcholinesterase (Ace1), all found on chromosome two. To account for variation due to sequencing error, only substitutions present in at least one population cohort with a frequency >5% were retained.
Metabolic insecticide resistance
Genes known to be associated with metabolic resistance in Anopheles were targeted for Copy Number Variant (CNV) discovery. These included the CYP6AA/P cluster on chromosome two, CYP6M2 and glutathione S-transferase (GST) on chromosome three and CYP9K1 on the X chromosome. Copy number state was calculated across the genome for individuals as detailed in Lucas et al. 71. In summary, a Gaussian HMM model was implemented to calculate coverage over 300 bp windows, normalised to account for bias in GC content. Regions with high GC content or low mapping quality were filtered. CNVs were characterised when five or more adjacent windows had a copy number state greater than two (or greater than one for males on the X chromosome). Samples with high coverage variance or CNVs with a low HMM likelihood of CNV state were removed from analyses. The proportion of individuals within each population cohort with any CNV amplification or deletion was then calculated. This frequency is based on the presence/absence of a CNV regardless of the number of copies present.
H12 haplotype diversity statistics
We calculated the H12 measure of haplotype homozygosity across windows spanning each chromosome. The H12 statistic is described by Garud et al. 72 and is modified from the common haplotype diversity statistic H1 to combine the first and second most common haplotype frequencies. The statistic has increased sensitivity to detect both hard and soft sweeps. To account for variation in demographic history, the window size was calibrated for each population cohort. We identified the optimal window size by plotting the distribution of H12 values across a range of window sizes and identifying the value where the 95% percentile of the H12 values was at or below 0.1. Values were plotted to identify statistical peaks suggestive of a selective sweep.
Statistics and reproducibility
1190 mosquito females collected between 2004 and 2014 at 36 sites from 7 different countries in West Africa were used for this study. All bioinformatics analyses were performed in Python, additional informations are provided under ‘Code availability.’
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The sequencing and variation data utilised in this study are part of the MalariaGEN Anopheles gambiae 1000 Genomes Project Phase-3 data resource. Sequence data are available from the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home). ENA accession numbers for the specific samples and sequencing runs used in this study are provided as Supplementary Data 1.
Further information on availability of these data is available from the MalariaGEN website at https://malariagen.github.io/vector-data/ag3/ag3.0.html.
Code availability
Codes used for analysis are available on GitHub page: https://github.com/randomxsk8/caputo-et-al-2024/.
References
Coluzzi, M., Sabatini, A., della Torre, A., Di Deco, M. A. & Petrarca, V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298, 1415–1418 (2002).
Neafsey, D. E. et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1258522 (2015).
Powell, J. R., Petrarca, V., della Torre, A., Caccone, A. & Coluzzi, M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia 41, 101–113 (1999).
Leffler, E. M. et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10, e1001388 (2012).
The Anopheles gambiae 1000 Genomes Consortium. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017).
Pinto, J. et al. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evol. Appl. 6, 910–924 (2013).
Kamdem, C., Fouet, C., Gamez, S. & White, B. J. Pollutants and insecticides drive local adaptation in African malaria mosquitoes. Mol. Biol. Evol. 34, 1261–1275 (2017).
Pombi, M. et al. Dissecting functional components of reproductive isolation among closely related sympatric species of the Anopheles gambiae complex. Evol. Appl 10, 1102–1120 (2017).
Clarkson, C. S. et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat. Commun. 5, 4248 (2014).
Lee, Y. et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc. Natl Acad. Sci. USA 110, 19854–19859 (2013).
Coetzee, M. et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619, 246–274 (2013).
Coluzzi, M. et al. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Ital. J. Zool. 52, 45–63 (1985).
Coluzzi, M. & Barigozzi, C. Mechanisms of speciation. Liss 143, 153 (1982).
Manoukis, N. C. et al. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc. Natl Acad. Sci. USA 105, 2940–2945 (2008).
Love, R. R. et al. Chromosomal inversions and ecotypic differentiation in Anopheles gambiae: the perspective from whole‐genome sequencing. Mol. Ecol. 25, 5889–5906 (2016).
della Torre, A. et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol. Biol. 10, 9–18 (2001).
Fanello, C., Santolamazza, F. & della Torre, A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med. Vet. Entomol. 16, 461–464 (2002).
Santolamazza, F., della Torre, A. & Caccone, A. Short report: A new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am. J. Trop. Med. Hyg. 70, 604–606 (2004).
Santolamazza, F. et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar. J. 7, 163 (2008).
Bryan, J. H., Di Deco, M. A., Petrarca, V. & Coluzzi, M. Inversion polymorphism and incipient speciation in Anopheles gambiae s.str. in The Gambia, West Africa. Genetica 59, 167–176 (1982).
Kamdem, C. et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS One 7, e39453 (2012).
Huestis, D. L. et al. Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature 574, 404–408 (2019).
Faiman, R. et al. Isotopic evidence that aestivation allows malaria mosquitoes to persist through the dry season in the Sahel. Nat. Ecol. Evol. 6, 1687–1699 (2022).
Fossog, B. T. et al. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol. Appl 8, 326–345 (2014).
The Anopheles gambiae 1000 Genomes Consortium. Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. Genome Res. 30, 1533–1546 (2020).
Oliveira, E. et al. High Levels of Hybridization between Molecular Forms of Anopheles gambiae from Guinea Bissau. J. Med. Entomol. 45, 1057–1063 (2008).
Caputo, B. et al. Anopheles gambiae complex along the Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar. J. 7, 1–17 (2008).
Caputo, B. et al. The “far-west” of Anopheles gambiae molecular forms. PLoS One 6, e16415 (2011).
Caputo, B. et al. Prominent intraspecific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol. Ecol. 23, 4574–4589 (2014).
Vicente, J. L. et al. Massive introgression drives species radiation at the range limit of Anopheles gambiae. Sci. Rep. 7, 1–13 (2017).
The Anopheles gambiae 1000 Genomes Consortium. Ag1000G phase 3 SNP data release. MalariaGEN https://www.malariagen.net/data/ag1000g-phase3-snp (2021).
Turner, T. L., Hahn, M. W. & Nuzhdin, S. V. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3, e285 (2005).
Caputo, B. et al. Novel genotyping approaches to easily detect genomic admixture between the major Afrotropical malaria vector species, Anopheles coluzzii and An. gambiae. Mol. Ecol. Resour. 21, 1504–1516 (2021).
MalariaGEN Vector Observatory. Ag3.9: Anopheles gambiae data resource. MalariaGEN https://malariagen.github.io/vector-data/ag3/ag3.9.html (2024).
MalariaGEN Vector Observatory. Ag3.5: Anopheles gambiae data resource. MalariaGEN https://malariagen.github.io/vector-data/ag3/ag3.5.html (2023).
Tripet, F., Dolo, G. & Lanzaro, G. C. Multilevel analyses of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting mosquito projects. Genetics 169, 313–324 (2005).
della Torre, A., Tu, Z. & Petrarca, V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem. Mol. Biol. 35, 755–769 (2005).
Clarkson, C. S. et al. The genetic architecture of target‐site resistance to pyrethroid insecticides in the African malaria vectors Anopheles gambiae and Anopheles coluzzii. Mol. Ecol. 30, 5303–5317 (2021).
Opondo, K. O. et al. Status of insecticide resistance in Anopheles gambiae (s.l.) of The Gambia. Parasit. Vectors 12, 1–8 (2019).
Weill, M. et al. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol. Biol. 9, 451–455 (2000).
Riehle, M. M. et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science 331, 596–598 (2011).
Crawford, J. E. et al. Reticulate speciation and barriers to introgression in the Anopheles gambiae species complex. Genome Biol. Evol. 7, 3116–3131 (2015).
Crawford, J. E. et al. Evolution of GOUNDRY, a cryptic subgroup of Anopheles gambiae s.l., and its impact on susceptibility to Plasmodium infection. Mol. Ecol. 25, 1494–1510 (2016).
Tennessen, J. A. et al. A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex. Mol. Ecol. 30, 775–790 (2021).
Nwakanma, D. C. et al. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics 193, 1221–1231 (2013).
Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
Eckhoff, P. A., Wenger, E. A., Godfray, H. C. J. & Burt, A. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl Acad. Sci. USA 114, E255–E264 (2017).
Miles, A. et al. scikit-allel: A Python package for exploring and analysing genetic variation data. https://doi.org/10.5281/zenodo.597309 (2023).
Pombi, M. et al. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol. Biol. 8, 309 (2008).
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Nei, M. & Li, W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269–5273 (1979).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Watterson, G. A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975).
The Anopheles gambiae 1000 Genomes Consortium. Ag1000G phase 3 data resource paper. https://malariagen.github.io/ag1000g-phase3-data-paper/manuscript.pdf (2021).
Bhatia, G., Patterson, N., Sankararaman, S. & Price, A. L. Estimating and interpreting FST: the impact of rare variants. Genome Res. 23, 1514–1521 (2013).
Lack, J. B., Lange, J. D., Tang, A. D., Corbett-Detig, R. B. & Pool, J. E. A thousand fly genomes: an expanded drosophila genome nexus. Mol. Biol. Evol. 33, 3308–3313 (2016).
Martin, S. H. et al. Natural selection and genetic diversity in the butterfly Heliconius melpomene. Genetics 203, 525–541 (2016).
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012).
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710 (2010). (1979).
Newstead, R. & Carter, H. F. On some new species of African mosquitos (Culicidae). Ann. Trop. Med Parasitol. 5, 233–244 (1911).
Fontaine, M. C. et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8, e1002967 (2012).
Fitak, R. R. OptM: estimating the optimal number of migration edges on population trees using Treemix. Biol. Methods Protoc. 6, bpab017 (2021).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Eaton, D. A. R. Toytree: a minimalist tree visualization and manipulation library for Python. Methods Ecol. Evol. 11, 187–191 (2020).
Gutenkunst, R. N., Hernandez, R. D., Williamson, S. H. & Bustamante, C. D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 5, e1000695 (2009).
Portik, D. M. et al. Evaluating mechanisms of diversification in a Guineo-Congolian tropical forest frog using demographic model selection. Mol. Ecol. 26, 5245–5263 (2017).
Anderson, D. R. & Burnham, K. P. Avoiding pitfalls when using information-theoretic methods. J. Wildl. Manag. 66, 912–918 (2002).
Martin, M. et al. WhatsHap: fast and accurate read-based phasing. bioRxiv 085050. https://doi.org/10.1101/085050 (2016).
Lucas, E. R. et al. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 29, 1250–1261 (2019).
Garud, N. R., Messer, P. W., Buzbas, E. O. & Petrov, D. A. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11, 1–32 (2015).
Acknowledgements
The authors thank ELIXIR-IIB (www.elixir-italy.org), the Italian Node of the European ELIXIR infrastructure (www.elixir-europe.org) and CINECA for supporting the providing the computing and bioinformatics facilities. The Authors also thanks Jeffrey Powell and João Pinto for useful discussions on the paper results. This study was supported by the MalariaGEN Vector Observatory which is an international collaboration working to build capacity for malaria vector genomic surveillance, and involves contributions by the following institutions and teams. Wellcome Sanger Institute: Lee Hart, Anastasia Hernandez-Koutoucheva, Jon Brenas, Menelaos Ioannidis, Julia Jeans, Paballo Chauke, Victoria Simpson, Eleanor Drury, Osama Mayet, Sónia Gonçalves, Katherine Figueroa, Tom Madison, Kevin Howe, Mara Lawniczak; Liverpool School of Tropical Medicine: Eric Lucas, Sanjay Nagi, Martin Donnelly; Broad Institute of Harvard and MIT: Jessica Way, George Grant; Pan-African Mosquito Control Association: Jane Mwangi, Edward Lukyamuzi, Sonia Barasa, Ibra Lujumba, Elijah Juma. The authors would like to thank the staff of the Wellcome Sanger Genomic Surveillance unit and the Wellcome Sanger Institute Sample Logistics, Sequencing and Informatics facilities for their contributions. The MalariaGEN Vector Observatory is supported by funding awarded to Dominic Kwiatkowski and Mara Lawniczak from Wellcome (220540/Z/20/A, ‘Wellcome Sanger Institute Quinquennial Review 2021–2026’), the Bill and Melinda Gates Foundation (INV-001927). SAPIENZA research group is funded by Progetti Ateneo 2018 and 2020. The Liverpool School of Tropical Medicine’s participation was supported by the National Institute of Allergy and Infectious Diseases ([NIAID] R01-AI116811), with additional support from the Medical Research Council (MR/P02520X/1). The latter grant is a UK-funded award and is part of the EDCTP2 programme supported by the European Union. The Pan-African Mosquito Control Association’s participation was funded by the Bill and Melinda Gates Foundation (INV-031595). Recent specimen collections by Medical Research Council Unit The Gambia at LSHTM was supported by the Pan-African Malaria Genetic Epidemiology Network (PAMGEN) Human Hereditary and Health is Africa award (H3A/15/002) from African Academy of Science/Science for Africa program, Genomic Surveillance of Malaria in West Africa (GSM, NIHR134717) award from National Institute of Health Research (NIHR), and a BMGF PAMCA grant.
Author information
Authors and Affiliations
Contributions
Conceptualisation: A.d.T., A.M., B.C., C.M.D.M., D.W.; Field collections: A.A.-N., S.B.A., K.O.O.; Methodology: A.d.T., A.M., B.C., C.M.D.M., D.W.; Data analysis: B.C., C.S.C., C.M.D.M., G.B., J.A.T., K.L.B., V.P.; First draft writing: A.d.T., A.M., B.C., C.S.C., C.M.D.M., D.W., J.A.T., K.L.B., V.P.; All authors commented on all versions of the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Tovi Lehmann and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Luciano Matzkin and Christina Karlsson Rosenthal. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Caputo, B., De Marco, C.M., Pichler, V. et al. Population genomic evidence of a putative ‘far-west’ African cryptic taxon in the Anopheles gambiae complex. Commun Biol 7, 1115 (2024). https://doi.org/10.1038/s42003-024-06809-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-06809-y