Abstract
Metabolic memory (MM) is a major factor in the delayed wound healing observed in diabetic patients. While “Nocardia rubrum cell wall skeleton” (Nr-CWS) is utilized to enhance macrophage proliferation in immune diseases, its impact on MM wounds in diabetes is unclear. This study demonstrates that transient hyperglycemia leads to prolonged damage in vascular endothelial cells by decreasing METTL3 expression, leading to decreased RNA methylation and impaired cellular metabolism. Remarkably, Nr-CWS application increases METTL3 levels in these cells, facilitating the recovery of cell function. Further in vivo and in vitro analyses demonstrate that transient hyperglycemia-induced reduction in METTL3 hinders RNA methylation of the downstream gene Cds2, impacting mitochondrial function and energy metabolism and consequently reducing angiogenic capacity in endothelial cells. This impairment significantly influences diabetic wound healing. Our findings highlight the profound impact of transient hyperglycemia on wound healing, establishing METTL3 as a significant role in vascular complications of diabetes. This study not only elucidates the pathophysiological mechanisms behind MM in diabetic wounds but also suggests Nr-CWS as a potential therapeutic agent, offering a novel approach for treating diabetic wounds.
Similar content being viewed by others
Introduction
Diabetic wound complications, particularly foot ulcers, present a substantial challenge worldwide, having far-reaching economic and social consequences for patients, their families, and healthcare systems. The prevalence of these complications is alarmingly high, with a recurrence rate exceeding 50% within three years of initial diagnosis1. However, diabetic complications continue to progress unabated even after pharmacological glycemic control is achieved2. Numerous studies have shed light on a phenomenon known as metabolic memory (MM)3, which highlights the persistent cellular dysfunction triggered by prolonged exposure to high blood sugar levels, despite subsequent efforts to maintain normal blood sugar control4,5.
The concept of “metabolic memory” was originally introduced by Nathan et al. 6 This study has significant implications in the realm of diabetes management. Their controlled clinical trial crucially found that long-term intensive treatment of diabetes did not correspondingly reduce the incidence of cardiovascular diseases, suggesting complex underlying mechanisms at play. Building upon this concept, Schner and Ruiz7 further elaborated on the definition of metabolic memory, characterizing it as the long-lasting harmful impact of elevated blood glucose levels on both microvascular and macrovascular complications in individuals with diabetes. Importantly, they noted that these complications often do not reverse, even with improved glycemic control. This expanded understanding of metabolic memory highlights the critical importance of early and sustained intervention in diabetes management, emphasizing the need to mitigate the long-term vascular complications attached to the disease.
Diabetic foot ulcers often arise from minor or repetitive injuries in patients suffering from nerve damage and peripheral arterial disease, highlight the vital importance of enhancing blood flow for effective wound healing8,9. In this context, improving blood circulation to the wound site is pivotal in diabetic wound management. Nocardia rubra cell wall skeleton (Nr-CWS) is derived from Nocardia Rubra and is composed of arabogalactan, nocardomycolic acid, and mucopeptide10. Our research team’s previous experiments showed that the secretion of mesenchymal stem cells pretreated with Nr-CWS could treat diabetic wounds by promoting angiogenesis11. However, despite these encouraging results, the exact molecular mechanisms through which Nr-CWS enhances the activity of vascular endothelial cells, particularly in the context of MM wounds, remain largely unexplored.
N6-methyladenosine (m6A) is recognized as the most prevalent internal modification in eukaryotic mRNA, significantly influencing multiple processes in RNA metabolism12,13. Its diverse functions extend to the modulation of messenger RNA (mRNA) through several critical pathways, including stability, splicing, transport, localization, translation, and interactions with RNA-binding proteins14. A deficiency in m6A RNA methylation has been linked to various diseases, such as diabetes and cancer15. A previous study demonstrated a correlation between m6A modification and type 2 diabetes mellitus (T2DM)16,17. The m6A levels in the RNAs of T2DM patients and diabetic mouse were considerably lower than those observed in the control groups18. Consequently, reduced m6A levels could serve as a new potential biomarker for T2DM. While the precise biological significance and regulatory roles of m6A methylation in the specific context of metabolic memory wounds remain incompletely understood.
In this study, we delved into the relationship between mitochondrial metabolism and METTL3 in human umbilical vein endothelial cells (HUVECs), particularly within the framework of metabolic memory. Our findings reveal that the application of Nr-CWS notably upregulates METTL3 expression and enhances m6A modification of Cds2 mRNA. This molecular modification results in enhanced protein synthesis, ultimately leading to the restoration of mitochondrial function in HUVECs. These results suggest that METTL3, influenced by Nr-CWS, could serve as a promising predictive marker and treatment target for the management and progression of diabetic wounds, opening new avenues for treatment strategies.
Results
Nr-CWS has therapeutic effects on delayed wound healing caused by transient hyperglycemia in vivo and in vitro
To replicate transient hyperglycemic conditions in vitro, HUVECs were initially cultured in high-glucose (HG) medium for 24 h, followed by incubation in normal glucose (NG) medium for different periods: 1 day (MM1), 3 days (MM2), and 5 days (MM3). As controls, a set of HUVECs was continuously cultured in NG to represent baseline conditions. Additionally, cells maintained exclusively in HG were used to model the scenario of diabetes without stringent glucose control. (Supplementary Fig. 1A).
Following HG treatment, we observed a significant reduction in the ability of endothelial cells to form capillary-like structures, along with a decrease in their proliferation capacity (Supplementary Fig. 1B–E). This reduction persisted for 6 days, even after the cells were transitioned back to an NG environment. This prolonged impact underscores the lasting influence of transient hyperglycemic conditions on endothelial cell functionality. Additionally, we conducted a preliminary assessment of energy metabolism in vascular endothelial cells. Following treatment with HG, we observed a notable decrease in mitochondrial potential energy, suggesting an impairment in mitochondrial function. Simultaneously, a significant rise in reactive oxygen species (ROS) levels was observed in these cells following HG treatment. Intriguingly, these elevated ROS levels did not return to baseline but remained high even after the cells were exposed to NG. (Supplementary Fig. 1F–I). This persistent elevation of ROS levels implies a lasting impact of hyperglycemic conditions on cellular oxidative stress.
To evaluate the therapeutic efficacy of Nr-CWS on metabolic memory, we focused on the MM3 group from our previous experiments (Fig. 1A). This choice was based on the prolonged exposure to NG conditions in this group, which closely mimics the clinical scenario of metabolic memory in diabetic patients. In our in vitro assessments, the application of Nr-CWS to the MM3 group led to significant improvements in endothelial cell function. Notably, a significant boost in the cells’ capability to form capillary-like structures was observed, along with a pronounced elevation in their proliferation rate (Fig. 2B–D). Upon re-evaluating the indicators of energy metabolism, we noted a considerable reduction in ROS levels following Nr-CWS treatment (Fig. 2E). These findings suggest that Nr-CWS may effectively counteract the detrimental effects of prolonged hyperglycemia on vascular endothelial cells.
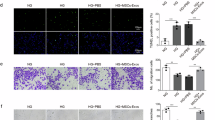
A Schematic representation of the experimental model in vitro. (Image produced by Figdraw). B, C The tube formation assay was used to assess the ability of endothelial cells to form three-dimensional capillary-like structures in vitro (B, scale bars = 100 µm) and to quantify the tube formation and cell growth (C) (n = 3 per group). D EDU staining of HUVECs from different treatment groups to detect cell proliferation function. Positive cells were imaged (scale bars = 100 µm, left panel) and quantified (right panel) (n = 3 per group). E Cellular ROS levels in Control, DM, MM and Nr-CWS of HUVECs. The ROS was quantified (right panel) (n = 3 per group). F Schematic representation of the experimental model in vivo. G Representative wound images at day 0 and 14 during the healing process, And KRT14 immunohistochemical staining of 14-day wound tissue. The wound-healing rate was measured (right panel) every d and is shown as the percentage of the initial wound area (n = 8 per group). H, I Histological changes and collagen deposition in wounds were evaluated by H H&E staining and I Masson staining. bars = 100 µm. J CD31 and CDH5 (red for CD31; green for CDH5; blue for nuclei) in skin sections from different experimental groups. bars = 100 µm. K, L A laser Doppler imager show the blood flow perfusion in the wounds that were subjected to different treatments (K) and quantification of wound perfusion in the wounds (L) (n = 8 per group). M Epithelial thickness measured by H&E staining (n = 3 per group). The data are represented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 are considered significant. HUVECs human umbilical vein endothelial cells, Nr-CWS Nocadia rubra cell wall skeleton preparation, DM diabetic memory, MM metabolic memory.
A mRNA isolated from patients with diabetes wounds and paired normal skin were used in Dot blot analysis with an anti-m6A antibody, and MB (methylene blue) staining as load control (Representative image in the left panel). Calculate the relative content of messenger ribonucleic acid m6A in diabetes wound tissue and paired normal skin tissue (Right panel) (n = 5 per group). B The mRNA isolated from HUVECs of different treatment groups was subjected to dot blot analysis using the m6A antibody (upper panel). MB staining serves as a loading control and the results were quantified and processed (right panel) (n = 3 per group). C The heatmap of cluster analysis based on transcriptome RNA-Seq of MM and Nr-T. D Venn diagram of differential genes and m6A-related genes in vascular endothelial cells of different treatment groups compared to control cells. E METTL3 protein levels were measured in HUVECs of different treatment groups by western blotting and the results were quantified and processed (top panel). and Nr-CWS treatment group by western blotting and the results were quantified and processed (down panel) (n = 3 per group). F The protein levels of METTL3 in HUVECs with METTL3 overexpression were measured by western blotting (n = 3 per group). G Tube formation assay and cell growth in HUVECs with METTL3 overexpression or from the corresponding control cells. Tubes were imaged (scale bars = 100 µm, left panel), and tube formation and cell growth were quantified (right panel) (n = 3 per group). H EDU staining of HUVECs with Mettl3 overexpression or from the corresponding control cells. Positive cells were imaged (scale bars = 100 µm, left panel) and quantified (right panel) (n = 3 per group). I The protein levels of METTL3 in HUVECs with wild-type or catalytic mutant METTL3 overexpression were measured by western blotting (left panel). The mRNAs isolated from wild-type or catalytic mutant Mettl3-overexpressing HUVECs were used in dot blot analyses with m6A antibody (right panel). MB staining served as a loading control (n = 3 per group). J The tube formation assay in HUVECs with wild-type or catalytic mutant Mettl3 overexpression was determined (left panel). The tubes were imaged (scale bar = 100 µm, left panel) and tube formation was quantified (right panel) (n = 3 per group). K Mettl3 expression was knocked down by siRNA in HUVECs and measured by western blotting (upper panel). The mRNAs isolated from si-Mettl3 or si-Control HUVECs were used in dot blot analyses with m6A antibody (bottom panel). MB staining served as a loading control. L The tube formation assay of Mettl3 knocked down by siRNA or si-Control in HUVECs. Tubes were imaged (scale bars = 100 µm, left panel), and tube formation was quantified (right panel) (n = 3 per group). M EDU staining of HUVECs with Mettl3 knocked down by siRNA or si-Control. Positive cells were imaged (scale bars = 100 µm, left panel) and quantified (right panel) (n = 3 per group). N Total cellular ATP levels were measured in HUVECs with Mettl3 knocked down by siRNA or si-Control starved for the indicated times (n = 3 per group). O Cellular ROS levels in si-control and Mettl3 knockdown HUVECs (left panel). The ROS was quantified (right panel) (n = 3 per group). P Representative wound images at d 0 and 14 during the healing process in animals treated with sense or AS-ODN. And KRT14 immunohistochemical staining of 14-day wound tissue. Wound diameters were measured on indicated days post-wounding from days 0–14 (the number of injections is 2) (n = 4 per group). Q METTL3 protein in these wounds was analyzed by western blotting, and relative METTL3 expression was normalized to β-actin using ImageJ densitometry (n = 4 per group). R Wound sections (day 14) were stained for CD31 (red) and α-SMA (green) to visualize stratified epithelial cells and counterstained with DAPI (scale bars = 50 µm). S A laser Doppler imager show the blood flow perfusion in the wounds that were subjected to different treatments (upper panel) and Quantification of wound perfusion in the wounds (down panel) (n = 4 per group). The data are represented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 are considered significant. α-SMA α-smooth muscle actin, HUVECs human umbilical vein endothelial cell.
In our in vivo studies using a diabetic mouse model (Fig. 1F), we observed that wound healing was impeded in both the diabetes mellitus (DM) and metabolic memory groups when compared to the control group. However, upon treatment with Nr-CWS, a significant improvement in healing was noted in the MM group. On the 7th day after treatment, the wound healing rate was 50% in the control group and 30% in the Nr-CWS-treated (Nr-T) group. In contrast, the groups of DM and MM exhibited less than 15% wound closure. Notably, by day 14, the control group showed complete wound closure, whereas the Nr-T group exhibited 80% closure. In comparison, the DM and MM groups had 49% and 42% of their wound areas still unhealed. At the same time, the expression of Keratin 14 (KRT14) in the epithelium of mice from the groups of DM and MM was significantly lower than in the control group but improved following Nr-CWS treatment (Fig. 1G).
Re-epithelialization plays a vital role in wound healing. In H&E staining, we evaluated the epithelial tongue length and found that Nr-T group showed a greater capacity to enhance re-epithelialization than the MM group (Fig. 1H, M). These pathological changes, indicative of chronic inflammation and impaired healing typical in diabetic wounds, showed marked improvement following Nr-CWS treatment. Additionally, we observed increased collagen deposition in the DM and MM groups, but the collagen fibers were found to be disorganized (Fig. 1I). This disorganization is a hallmark of dysfunctional wound healing. In contrast, the Nr-T group demonstrated more organized collagen deposition, suggesting a more effective healing process.
The proliferating capillary index, represented by the proportion of CD31-positive microvessels, was used to quantify angiogenesis. Additionally, intercellular adhesion in the wound area was assessed through CDH5 expression (Fig. 1J). This index was significantly decreased in both the DM and MM groups when compared to the control group, indicating a compromised angiogenesis response and diminished intercellular adhesion under these conditions. Vascular imaging revealed a marked increase in the proliferating capillary index in the Nr-T group, indicating enhanced microvascular proliferation, especially when compared to the MM and HG groups (Fig. 1K, L). These findings highlight the potential of Nr-CWS in promoting angiogenesis in diabetic wound healing scenarios.
Decreased METTL3 expression is linked to poor prognosis in patients with diabetic wounds
To explore the functional role of m6A modification in metabolic memory, we initially measured m6A RNA levels in wound tissues from five MM subjects and compared them with those in their corresponding normal skin tissues. Using a dot blot assay, we detected a significant decrease in m6A RNA levels in the MM wound tissues (Fig. 2A). For further verification, we isolated and analyzed RNA from vascular endothelial cells, reaffirming this decrease in m6A levels (Fig. 2B left panel). Interestingly, upon treating the MM group with Nr-CWS, we noted an elevation in m6A levels (Fig. 2B right panel). These findings suggest that m6A modification may have a regulatory role in diabetic wound healing and metabolic memory, especially in response to Nr-CWS treatment.
To investigate the underlying factors responsible for the changes in m6A levels within the Nr-T and MM groups, we performed RNA sequencing on vascular endothelial cells derived from these groups (Fig. 2C, D). The comprehensive RNA-seq analysis revealed that among the various factors assessed, METTL3 emerged as the primary influence on the dynamic alterations in m6A levels observed in both the Nr-T and MM groups.
To validate the molecular changes indicated by our earlier findings, we conducted Western blot analysis to assess METTL3 protein levels. The results revealed a notable decrease in METTL3 levels in cells subjected to HG conditions and in the MM group. Notably, METTL3 levels were elevated after Nr-CWS treatment, suggesting a potential restorative effect (Fig. 2E).
To further explore the role of METTL3 in HUVECs, we successfully generated stable cell lines with METTL3 overexpression (Fig. 2F). The overexpression of METTL3 significantly boosted both angiogenesis and the proliferation capacity of these cells, as evidenced by the results shown in Fig. 2G, H. Additionally, in an environment characterized by metabolic memory impairment, METTL3 overexpression was able to restore the angiogenic capability of HUVECs (Supplementary Fig. 2A). Further exploring the molecular mechanisms, we examined whether the promotion of HUVEC proliferation by METTL3 was dependent on METTL3 m6A catalytic activity. For this purpose, Plasmids encoding wild-type METTL3 and its mutant form (aa395-398, DPPW-APPA) were constructed according to the protocols described in a previous study19. Notably, HUVECs expressing the METTL3 mutant showed a significant decrease in m6A levels compared to those with wild-type METTL3 (Fig. 2I). This finding underscores the critical role of METTL3’s m6A catalytic activity in enhancing angiogenesis in HUVECs, a conclusion further supported by the results in Fig. 2J.
To explore the functional role of METTL3 in HUVECs, we specifically targeted the gene using two different siRNAs to achieve efficient gene silencing. This intervention resulted in a substantial decrease in both the protein and m6A levels of METTL3 (Fig. 2K). The knockout of Mettl3 notably impeded angiogenesis, cell growth, and proliferation capacity in HUVECs (Fig. 2L, M). Given the reliance of cellular activities on energy metabolism, we subsequently measured ATP and ROS levels in Mettl3-depleted cells. Remarkably, we observed a significant decline in ATP levels following 2 and 5 h of cell starvation post-Mettl3 knockout. Concurrently, ROS levels exhibited a marked increase (Fig. 2N, O). These findings underscore the essential role of METTL3 in maintaining cellular energy homeostasis and its critical influence on angiogenic and proliferative processes in endothelial cells.
In light of mitochondria being the central hub of energy metabolism in human cells, we utilized MitoTracker staining to visualize the mitochondrial morphology in HUVECs, particularly focusing on Mettl3 knockout cells. Our observations, as depicted in Supplementary Fig. 2B, revealed a striking morphological change in the mitochondria of METTL3-depleted HUVECs. We noted a transition from their typical elongated rod-like structures to fragmented, dotted formations. To clarify the morphological changes in mitochondria more clearly, we used transmission electron microscopy to observe the cells with si-Mettl3. We found that after Mettl3 was silenced, the inner cristae of the mitochondria disappeared, and the morphology became fragmented (Supplementary Fig. 2C), consistent with the results observed by confocal microscopy. Subsequent Western blot experiments demonstrated that silencing Mettl3 resulted in a notable reduction in the mitochondrial inner membrane protein OPA1, which is essential for mitochondrial fusion, the formation of mitochondrial cristae, and overall mitochondrial quality control. In contrast, the mitochondrial division protein DRP1 did not exhibit any significant changes (Supplementary Fig. 2D). Therefore, we believe that the reduction in OPA1 is a key factor contributing to the observed mitochondrial abnormalities. This change in mitochondrial morphology is indicative of mitochondrial dysfunction, which is often associated with impaired cellular energy metabolism and can have significant implications for cellular health and function, especially in the context of endothelial cells, where mitochondrial integrity is crucial for angiogenic processes.
To assess the function of METTL3 in the context of wound healing in vivo, we employed antisense oligonucleotides (AS-ODNs) to selectively reduce METTL3 expression in excisional skin wounds (Fig. 2Q). Starting from day 2 post-wounding, the wounds were treated daily with Mettl3 AS-ODN. After 14 days, The ASO-Mettl3-treated group showed a significant delay in the wound healing process compared to the control group, accompanied by markedly lower KRT14 expression in the epithelium of the ASO-Mettl3-treated group (Fig. 2P). Furthermore, we assessed the angiogenic process in these wounds and found a significant repression of angiogenesis in the ASO- Mettl3 group (Fig. 2R, S). These findings suggest that METTL3 is essential for wound healing and angiogenesis, and its inhibition by AS-ODN results in impaired wound closure and diminished angiogenic response.
Nr-CWS downregulates KLF9 and activates METTL3 transcription in HUVECs by H3K27ac
To unravel the mechanism behind the diminished expression of METTL3 in MM, we initiated our study by examining the Mettl3 promoter region. Utilizing the UCSC Genome Bioinformatics site for this analysis, we identified a high density of H3K27 acetylation signals (H3K27ac) within this region (Fig. 3A). To corroborate these findings, we conducted chromatin immunoprecipitation (ChIP) assays. These assays confirmed the presence of H3K27ac signals in the Mettl3 promoter region (Fig. 3B). Following this, we assessed the protein expression levels of H3K27ac under various conditions: NC, HG, MM, and Nr-T. This evaluation revealed a notable reduction in H3K27ac expression in both the HG and MM groups (Fig. 3C), underscoring the potential regulatory role of H3K27ac in METTL3 expression under these conditions.
A Data from the UCSC genome bioinformatics website (http://genome.ucsc.edu/) showed high enrichment of H3K27ac in the promoter of Mettl3. B ChIP-Qpcr analysis identified enrichment of H3K27ac in the promoter of Mettl3 in HUVEC cells. C Examination of the expression levels of the H3K27ac protein in HUVECs across different environments (n = 3 per group). D Venn diagram displays Mettl3-regulated genes from the tftarget and gencards database. And QPCR validation in different environments. E Protein levels of KLF9, H3K27ac, and METTL3 in different environments (n = 3 per group). F KLF9, H3K27ac, and METTL3 protein levels under different Nr-CWS treatment times (n = 3 per group). G After silencing KLF9 using small interfering RNA in the MM environment, the protein content of KLF9, H3K27ac, and METTL3 changed (n = 3 per group). H After overexpression of KLF9, the protein content of KLF9, H3K27ac, and METTL3 changed (n = 3 per group). I Schematic representation of KLF9 binding sites and Mettl3 promoter region with transcription start site (TSS). J Sequence logo representing the consensus Klf9 binding motif. K ChIP-qPCR validation of Klf9 binding at two sites within the METTL3 promoter, with siRNA-mediated Klf9 knockdown. L Luciferase reporter assay results showing relative promoter activity of wild-type and mutant Mettl3 promoters. M Western blot analysis demonstrating the effect of Klf9 knockdown on H3K27ac and METTL3 expression (n = 3 per group). N Graphical summary illustrating the proposed mechanism where KLF9 regulates H3K27ac and METTL3 in high-glucose and Nr-CWS conditions. The data are represented as mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 are considered significant. H3K27ac H3K27 acetylation; ChIP chromatin immunoprecipitation.
In our quest to determine the factors responsible for attenuating the H3K27ac signal at the Mettl3 locus, we leveraged our sequencing data in conjunction with the TFTarget and GeneCards databases. This integrated approach facilitated the identification of four transcription factors of interest. Crucially, we discovered that the binding sites for Klf9 and Bcl6 are positioned on the H3K27ac signal region of Mettl3. To further our understanding, we proceeded to validate the mRNA expression levels of Klf9 and Bcl6 across four distinct environments. This validation revealed a notably more significant variation in the expression of Klf9 (Fig. 3D), suggesting its prominent role in modulating the H3K27ac signal and thereby influencing METTL3 expression (Supplementary Data 5).
In HG and MM conditions, there was a notable decrease in H3K27ac and METTL3 levels, accompanied by an upregulation of KLF9 expression (Fig. 3E). Furthermore, we observed a reciprocal relationship wherein prolonged application of Nr-CWS led to a reduction in KLF9 levels (Fig. 3F). This reduction in KLF9 was correlated with an elevation in the protein levels of H3K27ac and METTL3. Extending our exploration, we silenced the KLF9 gene specifically within a metabolic memory context. This intervention resulted in an upsurge in the protein levels of H3K27ac and METTL3 (Fig. 3G). Conversely, upon overexpressing KLF9, we noted a decline in both H3K27ac and METTL3 proteins, thus demonstrating an inverse trend (Fig. 3H). These findings collectively highlight the critical role of KLF9 in modulating the expression of H3K27ac and METTL3. Subsequently, we predicted the schematic representation of KLF9 binding sites and METTL3 promoter region with transcription start site (TSS) (Fig. 3I). Sequence logo representing the consensus Klf9 binding motif (Fig. 3J). ChIP analysis confirmed two Klf9 binding sites in the Mettl3 promoter region (Fig. 3K). Additionally, we constructed luciferase reporter plasmids with mutations in these binding sites within the Mettl3 promoter and obtained similar results (Fig. 3L). Subsequently, a retrospective validation experiment was conducted, revealing that within the MM, treatment with Nr-CWS resulted in a substantial decrease in KLF9 protein expression. This reduction led to an increase in H3K27ac protein content and, consequently, a subsequent elevation in METTL3 protein content (Fig. 3M). The mechanism of action of Nr-CWS on the METTL3 protein is illustrated in Fig. 3N.
METTL3-mediated m6A modification of Cds2 mRNA to maintain its stability
To elucidate how METTL3 promotes progression in HUVECs, we utilized RNA-seq and m6A-modified RNA immunoprecipitation sequencing (MeRIP-seq) data from the Gene Expression Omnibus Database (GEO) (datasets GSE157544)20,21. Our objective was to identify differentially expressed genes between control HUVECs and those with Mettl3 silencing. Given previous indications that METTL3 depletion affects HUVEC energy metabolism, we cross-referenced these datasets with an energy metabolism gene dataset (Supplementary Data 3) from GeneCards (Fig. 4A). This integrative analysis resulted in the identification of 141 differentially expressed genes (DEGs). From this list, we selected the top 10 genes for validation using quantitative real-time PCR (qRT‒PCR), with Cds2 identified as the most significantly altered gene (Fig. 4B). Subsequent Western blot analysis confirmed that CDS2 protein levels are positively regulated by METTL3 in HUVEC lines (Fig. 4C). Interestingly, overexpression of the catalytic mutant METTL3 failed to replicate this regulatory effect on CDS2 at the protein level (Fig. 4D). Furthermore, MeRIP-qPCR assays corroborated these findings, demonstrating a decreased abundance of m6A modification on Cds2 mRNA following METTL3 deletion (Fig. 4E).
A RNA-seq and MeRIP-seq identified differentially expressed genes in Mettl3 stable knockout cells when compared with their corresponding controls, and analyzed genes related to energy metabolism. B The mRNA levels of Top 10 in (A) were detected by qRT-PCR. C The levels of the METTL3 and CDS2 proteins in METTL3 expression were knocked down by siRNA in HUVECs were detected by western blotting and quantification (right panel) of the western blot results were shown (n = 3 per group). D The protein levels of CDS2 in wild-type or catalytic mutant Mettl3-overexpressing HUVECs were measured by western blotting and quantification (right panel) of the western blot results were shown (n = 3 per group). E qRT-PCR analysis of Cds2 mRNA from RIP assays of HUVECs. RNA 820 enrichment was determined relative to the nontargeting IgG control (n = 3 per group). F Global profiling of m6A in HUVECs and modification sites of m6A on Cds2. G Integrative genomics viewer (IGV) plots show m6A peaks at Cds2 mRNAs in MeRIR-seq of HUVEC cells. Ranges of reads are indicated. H RNA pull-down assay was performed using potential wild-type binding sequences in Cds2 mRNA. METTL3 protein was detected by western blot analysis. GAPDH was used as a nonspecific control (n = 3 per group). I RNA pull-down assay was performed using Oligo 3 potential wild-type binding or mutated sequences (n = 3 per group). J Relative luciferase activities with in vitro-transcribed RNA segments in control and Mettl3 knockdown cells (n = 3 per group). K, L The levels of Cds2 expression were measured by qRT-PCR in Mettl3-overexpressing (K), METTL3 knockout (L), and their corresponding control HUVECs treated with actinomycin D (2 µg/mL) at the indicated time points (n = 3 per group). M The mRNA levels of Cds2 in Igf2bp3 knockdown HUVECs were detected by qRT-PCR. (N) RNA immunoprecipitation (RIP)-qPCR detection was employed to examine the enrichment of IGF2BP3 binding Cds2 m6A modification sites by utilizing IGF2BP3-specific antibodies and IgG control antibodies. O Immunoblot analysis was performed to detect CDS2/COX IV proteins in control cells, Mettl3 knockdown cells, Igf2bp3 knockdown cells, and Mettl3/ Igf2bp3 knockdown cells. β-ACTIN was used as a loading control (n = 3 per group). P–R The levels of METTL3 (P) and CDS2 (Q) expression were analyzed in 413 cases of GC and 32 cases of normal gastric mucosa using GEO data. (R) METTL3 expression were positively correlated with CDS2 expression in HUVECs (linear regression). The data are represented as mean ± SD of at least three independent experiments. **p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 are considered significant. MeRIP-seq m6A-modified RNA immunoprecipitation sequencing, RNA-seq RNA sequencing, GEO Gene Expression Omnibus.
m6A modifications, which typically occur within the RRACH (R = G or A, H = A, C, or U) consensus sequence, are vital for mRNA regulation. The MeRIP-seq results indicated that the ACUUACC motif was significantly enriched in immunopurified RNA following METTL3 overexpression, suggesting specific binding preferences. This was further corroborated by identifying key m6A modification sites on the Cds2 mRNA (Fig. 4F). Utilizing a sequence-based predictor, we identified potential METTL3 binding sites on Cds2 mRNA, predominantly located within the coding sequence and the 3′ untranslated region (UTR) (Supplementary Fig. 3A). We segmented these sequences into nine parts, including the 5′UTR, two coding region (CR) segments, and six 3′UTR segments (Supplementary Fig. 3B). RNA pull-down assays using in vitro-transcribed, biotinylated fragments of these segments demonstrated METTL3’s association, particularly with the CR-2 and 3′-1 fragments of Cds2 mRNA (Supplementary Fig. 3C). To further explore the functional link between METTL3 and Cds2 mRNA, we constructed pSI3-derived reporter cells carrying Cds2 mRNA fragments for siRNA inhibition luciferase assays. The assay results were consistent with the RNA pull-down analysis (Supplementary Fig. 3D). Based on the m6A peak pattern identified in Cds2 (Fig. 4G), we partitioned these sequences into three distinct regions—Oligo 1, Oligo 2, and Oligo 3. Conducting RNA pull-down analysis using biotinylated fragments transcribed in vitro from these segments revealed a notable association with METTL3, with Oligo of Cds2 mRNA exhibiting particularly strong correlation (Fig. 4H, I). To further delve into the functional interplay between METTL3 and Cds2 mRNA, we engineered reporter cells derived from pSI3 carrying Cds2 mRNA fragments for siRNA inhibition luciferase assays. The measurement results (Fig. 4J), align seamlessly with the findings from the RNA pull-down analysis, reinforcing the robust functional relationship between METTL3 and Cds2 mRNA. Additionally, we treated HUVECs with actinomycin D and observed that Cds2 mRNA levels remained stable with Mettl3 overexpression but decreased following Mettl3 knockout (Fig. 4K, L). These experiments collectively elucidate the intricate regulatory relationship between METTL3 and Cds2 mRNA, highlighting the role of specific m6A modifications in this context.
IGF2BP3, an m6A-binding protein, is essential for promoting the stability and storage of its target mRNAs in an m6A-dependent manner, a function that is especially prominent under both normal and stress condition22. In our study, we specifically designed two siRNAs targeting Igf2bp3 and confirmed their efficiency in achieving knockdown. Our findings indicated that silencing Igf2bp3 led to a significant reduction in the expression of Cds2 mRNA (Fig. 4M). Additionally, RIP-qPCR assays using an IGF2BP3-specific antibody demonstrated a marked enrichment of Cds2 mRNA compared to the IgG control antibody (Fig. 4N). Consistent with the effects observed upon Mettl3 silencing, the knockdown of Igf2bp3 led to a decrease in CDS2 protein levels. Intriguingly, we also observed a concurrent reduction in the mitochondrial protein COX IV (Fig. 4O). These findings suggest a complex interplay between IGF2BP3 and the regulation of both Cds2 mRNA and protein levels, with implications extending to mitochondrial function as reflected in the levels of COX IV.
In our subsequent analysis utilizing the GEO database (GSE80178)23,24, we analyzed the mRNA levels of Mettl3 and Cds2 in skin tissues from DFU patients against those in normal skin tissue. This analysis demonstrated a notable reduction in the levels of both Mettl3 and Cds2 in DFU skin tissue (Fig. 4P, Q). Importantly, our investigation also uncovered a significant correlation between the expression levels of Mettl3 and Cds2 (R2 = 0.4723, p = 0.0046), indicating a potential interdependent regulatory mechanism. These findings are graphically represented in Fig. 4R. The observed correlation and differential expression in DFU tissue suggest a possible link between these genes and the pathophysiology of diabetic foot ulcers, underscoring their importance in the disease’s molecular landscape.
METTL3 increases the biological activity of HUVECs by upregulating CDS2 expression
To clarify the role of CDS2 in the energy metabolism of HUVECs, we designed two specific siRNAs targeting Cds2 and confirmed their knockout efficiency using qRT‒PCR and Western blot analysis (Fig. 5A). The depletion of Cds2 resulted in a significant impediment to angiogenesis, cell growth, and proliferation (Fig. 5B, C). Moreover, an assessment of ROS and ATP levels in Cds2-deficient cells revealed a substantial increase in ROS and a decrease in ATP levels, particularly noticeable after 4 and 8 h of cell starvation post-knockdown (Fig. 5D, E). Given that ATP metabolism predominantly occurs within mitochondria, we assessed the expression levels of mitochondrial marker proteins TOMM20 and OPA1. Interestingly, their expression levels were observed to decrease upon silencing of Cds2 (Fig. 5F). Employing MitoTracker staining, we observed a morphological transition in mitochondria to fragmented, dotted structures in HUVECs with Cds2 knockout, suggesting mitochondrial dysfunction (Fig. 5G). This observation was quantitatively supported by ImageJ analysis, indicating a significant reduction in mitochondrial area post-knockout (Fig. 5H). Further investigations with a structural illumination microscope (SIM) revealed significant alterations in the mitochondrial membrane state, including the loss of the inner membrane structure in Cds2-silenced cells, resulting in abnormal mitochondrial configurations (Fig. 5I). We also quantified these changes (Fig. 5J), which highlighted a substantial alteration in mitochondrial length following Cds2 knockout.
A The Cds2 knockdown efficiency was verified at the mRNA level (upper panel) and protein levels (down panel) in HUVECs by qRT-PCR and western blot assay, respectively (n = 3 per group). B Tube formation assay in HUVECs with Cds2 deficiency and corresponding control cells. The tubes were imaged (scale bars = 100 µm, upper panel) and quantified for tube formation and cell growth (down panel) (n = 3 per group). C EDU staining of HUVECs with Cds2 deficiency and from the corresponding control cells. Positive cells were imaged (scale bars = 100 µm, upper panel) and quantified (down panel) (n = 3 per group). D Cellular ROS levels of HUVECs with Cds2 deficiency and from the corresponding control cells (left panel). The ROS was quantified (right panel). (n = 3 per group). E Total cellular ATP levels were measured in HUVECs with Cds2 deficiency and from the corresponding control cell starved for the indicated times. (n = 3 per group). F The levels of the OPA1 and TOMM20 proteins in Cds2 expression were knocked down by siRNA in HUVECs were detected by western blotting and quantification (right panel) of the western blot results were shown (n = 3 per group). G, H HUVECs were transfected with Mito-DsRed for 15 min. The cells were fixed and immunostained with CDS2 antibody. Subsequently visualized using confocal microscopy. The average mitochondrial area was quantified (G) for each group using ImageJ software. (Scale bar: 100 µm) (n = 3 per group). I, J HUVECs were transfected with Mito-DsRed for 15 min. Then the structure of the mitochondrial inner membrane was photographed using Super Resolution Fluoroscopy Microscope (SIM). The average mitochondrial area was quantified (I) for each group using ImageJ software. (Scale bar: 100 µm). K Representative images (scale bars = 100 µm, left panel) and quantified for tube formation and cell growth (right panel) of the Tube formation assay of METTL3-overexpressing HUVECs transfected with the CDS2 siRNAs or their corresponding controls (n = 3 per group). L Representative wound images at d 0 and 14 during the healing process in animals treated with sense or AS-ODN. And KRT14 immunohistochemical staining of 14-day wound tissue. Wound diameters were measured on indicated days post-wounding from days 0–14 (the number of injections is 2) (n = 4 per group). M CDS2 protein in these wounds was analyzed by western blotting, and relative CDS2 expression was normalized to β-actin using ImageJ densitometry. N Observation of mitochondrial morphology (scale bars =1 µm or 500 nm, respectively) in mice skin tissue in animals treated with sense or AS-ODN using projection electron microscopy. O A laser Doppler imager show the blood flow perfusion in the wounds that were subjected to different treatments (upper panel) and Quantification of wound perfusion in the wounds (down panel) (n = 4 per group). The data are the means ± SD of at least three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 are considered significant.
As anticipated, the targeted application of specific siRNAs led to effective suppression of Cds2 expression in HUVECs overexpressing Mettl3. This downregulation of CDS2 significantly impacted cellular functions, notably inhibiting the enhanced angiogenesis and cell growth abilities that were initially induced by Mettl3 overexpression. These consequential effects on METTL3-induced HUVECs are clearly illustrated in Fig. 5K. This finding underscores the crucial role of CDS2 in supporting the angiogenic and growth-promoting activities of METTL3 in endothelial cells, highlighting the intricate interplay between these molecular factors.
To assess the role of CDS2 in vivo, we utilized AS-ODN to specifically knockdown CDS2 expression in excisional cutaneous wounds (Fig. 5M). Starting from day 2 post-wounding, the wounds were treated daily with Cds2 AS-ODN. After a 14-day period, we found that wound healing was significantly delayed in the ASO-Cds2 group compared to the control group, and KRT14 expression was markedly reduced in the epithelium of the ASO-Cds2-treated group relative to the control group (Fig. 5L). Furthermore, to examine the mitochondrial morphology in the skin tissue of these mice, we employed transmission electron microscopy. Interestingly, in the Cds2 AS-ODN group, mitochondria exhibited a loss of their original rod-shaped structure (Fig. 5N). This alteration in mitochondrial morphology aligns with the observed repression of the angiogenic process in the ASO- Cds2 group, highlighted in Fig. 5O. These findings collectively suggest a critical role for CDS2 in both wound healing and maintaining normal mitochondrial morphology, with its knockdown leading to delayed healing and altered mitochondrial structure.
CDS2 interacts with OPA1 at the VAL-280 site
To elucidate how CDS2 impacts mitochondrial fusion, we hypothesized that CDS2 might influence mitochondrial morphology through its interactions with mitochondrial fusion proteins. To explore this, we employed Co-Immunoprecipitation followed by Mass Spectrometry (Co-IP-MS) and protein identification techniques to detect proteins physically interacting with CDS2. This analysis revealed a significant interaction between CDS2 and OPA1 (Fig. 6A–C). Intriguingly, we found that knocking down METTL3 led to a reduction in the interaction between CDS2 and OPA1, indicating a regulatory role of METTL3 in this interaction (Fig. 6D). Utilizing Protein-Protein Interaction Networks (PPI), we predicted the binding sites of CDS2 and OPA1 and identified that the interaction was strongest at the sites VAL-280, GLN-336, and ARG-331 (Fig. 6E). Single amino acid mutant analysis indicated that the CDS2-V280L mutant showed decreased interaction between OPA1 and CDS2, highlighting the essential role of valine 280 (V280) in this interaction (Fig. 6F, G). Interestingly, this binding at the VAL-280 site was notably weakened when METTL3 was knocked out (Fig. 6H), underscoring the interconnected roles of METTL3, CDS2, and OPA1 in mitochondrial dynamics.
A Co-IP Coomassie Bright Blue Dyeing Experiment. B Mass spectrometry analysis to obtain mitochondrial-related protein OPA1 with a high comprehensive score. C HUVECs were subject to immunoprecipitation (IP) with an anti-CDS2 antibody. The immunoprecipitation complex was probed via western blotting using an anti-OPA1 antibody (n = 3 per group). D HUVECs were treated with si-METTL3 and were then subjected to immunoprecipitation (IP) with an anti-CDS2 antibody. Co-IP of OPA1 was detected by western blotting using an anti-OPA1 antibody (n = 3 per group). E Protein-Protein Interaction Networks (PPI) predicted the binding sites of CDS2 and OPA1. F–H HUVECs transfected with CDS2-HA or CDS2 mutants for 24 h, followed by immunoprecipitation. Detection of CDS2 (wild type and mutants) was performed via Western blotting using an anti-HA antibody (n = 3 per group). I, J, and K Assessment of tubular structures and ROS in HUVECs under various conditions to evaluate cellular function and metabolism (n = 3 per group). L ATP assay conducted to assess the energy metabolism of HUVECs in different environmental conditions (n = 3 per group). M Western blot was utilized to detect the levels of proteins OPA1, METTL3, CDS2, and COX IV in HUVECs across various environments in each experimental group (n = 3 per group). N, O, and P Observation and analysis of the healing process and the expression of KRT14 in mouse back skin across different environments, accompanied by blood flow imaging to assess vascular changes (n = 4 per group). The data are the means ± SD of at least three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 are considered significant.
In our comprehensive study, we embarked on retrospective analyses through both in vivo and in vitro experiments to elucidate the therapeutic potential of Nr-CWS on HUVECs under various conditions. Initially, our in vitro exploration focused on assessing the effects of Nr-CWS on angiogenesis and ROS in HUVECs (Fig. 6I–L). Remarkably, we observed that the therapeutic efficacy of Nr-CWS was nullified in the context of silenced METTL3 and CDS2 expression. This finding was further supported by Western Blot experiments, which demonstrated that Nr-CWS did not induce a rebound in the mitochondrial inner membrane protein OPA1 when METTL3 and CDS2 were silenced (Fig. 6M).
Extending our investigation to in vivo models, we employed ASOs to silence METTL3 and CDS2 in mice (Fig. 6N–P). In these models, treatment with Nr-CWS resulted in a deceleration of wound healing and a reduction in angiogenesis efficiency (Fig. 6P). These in vivo findings corroborate our in vitro results, collectively suggesting that Nr-CWS enhances diabetic wound healing primarily by upregulating METTL3 expression. This upregulation appears to facilitate m6A methylation in HUVECs, thereby bolstering cellular energy metabolism and subsequently augmenting angiogenic capabilities.
Discussion
In this study, we elucidated that mitochondrial dysfunction in vascular endothelial cells, a critical factor in delayed wound healing, is primarily attributed to sustained damage from transient hyperglycemic exposure in MM conditions. m6A, the most prevalent mRNA and noncoding RNA modification in humans14,25, has recently been recognized as a crucial factor in the delayed healing of diabetic wounds26. We demonstrated that treatment with Nr-CWS significantly upregulates the methyltransferase METTL3 in HUVECs, thereby increasing the m6A modification levels. Mechanistically, Nr-CWS enhances METTL3 transcription through KLF9-mediated H3K27ac activation at the Mettl3 promoter. This activation results in enhanced m6A modification of Cds2 mRNA, with the m6A reader protein IGF2BP3 binding directly to the m6A site on the Cds2 mRNA, stabilizing it. The secreted CDS2 then interacts with OPA1, stabilizing mitochondrial morphology and boosting ATP production in HUVECs. This chain of molecular events fosters blood vessel formation in MM wounds, thus significantly promoting wound healing and improving the clinical prognosis of diabetic wounds.
Recent studies have underscored the critical role of m6A modification in the progression of various refractory wounds27, highlighting the diverse biological functions it plays in these contexts. Among the m6A modulators, METTL3 has been extensively studied in a range of cell types for its impact on wound healing processes26,28. In our research, we present a novel finding: Nr-CWS activates METTL3 transcription through the KLF9-H3K27ac pathway, leading to an increase in METTL3 expression in HUVECs under MM conditions. This activation results in an enhanced level of m6A modification in HUVECs. Furthermore, our data indicate a significant association between low METTL3 expression and poor prognosis in diabetic wounds, suggesting a pivotal role for METTL3 in modulating wound healing processes. This discovery offers a new perspective on the molecular mechanisms underlying diabetic wound healing and presents potential therapeutic targets for improving clinical outcomes.
Using RNA-seq and MeRIP-seq, we discovered that Cds2 is a pivotal downstream target of METTL3 in HUVECs. CDS2, functioning as CDP-diacylglycerol synthase, is instrumental in converting PA to CDP-DAG, a precursor for phosphatidylinositol and phosphatidylglycerol synthesis in mammals29. Notably, Cds2 knockdown has been linked to abnormal mitochondrial morphology in diverse cell types, including H9c2 cardiomyocytes and Drosophila sperm30, as well as mitochondrial dysfunction in liver cells31. Our Co-IP-MS and Protein Identification studies further revealed that CDS2 interacts with OPA1, specifically binding at the VAL-280 site and inducing OPA1 expression. This finding emphasizes the essential role of mitochondria in cellular metabolism, ATP production, and programmed cell death, highlighting the importance of their proper function and quality control32,33,34. Additionally, we discovered that Nr-CWS can increase the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in HUVECs (Supplementary Fig. 4A), thereby enhancing the cells’ ATP production, maximal respiration, spare respiratory capacity, and non-mitochondrial oxygen consumption (Supplementary Fig. 4B–E). The observed changes in OCR indicators align with our observations of mitochondrial alterations, which may be related to the disruption of mitochondrial inner membrane proteins, leading to a diminished capacity for aerobic respiration in cells35,36. This mechanism warrants further investigation. Mitochondria must operate properly, and their quality control needs to be carefully regulated to support these various cellular functions37,38. Given OPA1’s role in maintaining mitochondrial crista structure39 and the observed cell damage from OPA1 deficiency40, our data suggest that CDS2 might act as an activator in certain conditions to regulate cell energy metabolism. Importantly, we found a positive correlation between the expression of METTL3, CDS2, and OPA1 in MM tissues, indicating the clinical significance of the METTL3/CDS2 axis in enhancing mitochondrial morphology and angiogenesis in MM wounds. These insights provide a novel regulatory model for the role of METTL3 in diabetic wound progression.
Our study illuminates the significant therapeutic role of Nr-CWS in treating MM ulcers. We have elucidated that Nr-CWS mechanistically enhances the METTL3/CDS2/OPA1 axis, thereby promoting wound healing in diabetic conditions through improved mitochondrial morphology and angiogenesis (Fig. 7). Crucially, we found that METTL3 expression is markedly reduced in MM tissues, correlating with a poorer prognosis in patients with diabetic ulcers. This observation positions METTL3 not only as a key biomarker but also as a potential therapeutic target for diabetic ulcers and metabolic memory wounds. By targeting the METTL3/CDS2/OPA1 pathway, new avenues for effective treatment strategies in diabetic wound care could be developed, offering hope for improved clinical outcomes in this challenging area of medicine.
Nr-CWS reduces the elevated levels of KLF9 protein induced by a high-glucose environment, thereby enhancing H3K27ac at the transcription initiation site, which leads to increased expression of METTL3. METTL3, through its interaction with IGF2BP3, elevates the m6A methylation level of Cds2 mRNA, subsequently stabilizing the mitochondrial inner membrane protein OPA1. This restoration of mitochondrial function enhances the metabolic capacity of HUVECs, thereby accelerating wound healing. (Image produced by BioRender).
Materials and methods
Cell culture and treatment
Human umbilical vein endothelial cells (HUVECs) ordered from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Nanjing Keygen Biotech Co., Ltd.), containing 10% fetal bovine serum (FBS). Cells were incubated in media containing either 30.5 nmol/L high glucose (HG) or 5.5 nmol/L normal glucose (NG) (Supplementary Fig. 1A), along with an osmotic control (30.5 nmol/L mannitol). For the Nr-CWS treatment, HUVECs were exposed to Nr-CWS at a concentration of 10 μg/L for 24 h before proceeding with the experiments.
Tube formation assay
Matrigel (BD Biosciences) was added to 48-well plates at 200 μl per well, incubating for 30 min at 37 °C in a 5% CO2 atmosphere. The pretreated HUVECs were trypsinized using 0.125% trypsin, with the trypsinization process terminated by adding DMEM supplemented with 10% FBS. Subsequently, 3 × 105 cells were plated on the Matrigel in endothelial cell basal medium-2 and incubated for 6 h, followed by fluorescent staining with yellow calcein (Beyotime). The vascular network of the HUVECs was then acquired under a microscope (Olympus). The total tube length, number of connections, and percentage of tube area were quantified using image analysis software (ImageJ).
Edu assay
Edu cell proliferation staining was performed using an Edu kit (riboEDIT, EDU-Apollo567). Briefly, HUVECs (2 × 104 cells per well) were seeded in 12-well plates and incubated overnight. Then, cells were treated with Edu for 2 h, after which they were fixed for 15 min and then permeabilized with 0.3% Triton X-100 for another 15 min. Subsequently, the cells were incubated with the Click Reaction Mixture for 30 min at room temperature in the dark, followed by a 10-min incubation with DAPI for nuclear staining.
Measurement of cellular ROS
HUVECs were seeded in a 6-well plate at a density of 2 × 105 cells per well. For evaluating total cellular ROS, living cells were added 10 μmol/L carboxy-DCFDA (Beyotime), and after 20 min incubation, harvested cells were resuspended in 200 μL of buffer, followed by immediate analysis using flow cytometry. The fluorescence of carboxy-DCFDA was detected through the FITC channel.
Mouse experiments
Subject male BALB/c mice aged 6–8 weeks and purchased from Xuzhou Medical University, Jiangsu, China as Test Animals were used in four groups: control group, diabetes group, MM group, treatment group with Nr-CWS. Diabetic mice were conducted by administration of streptozotocin at 55 mg/kg using the Sigma–Aldrich. Control-mange diabetic forms showed rather poor glycemia with glycated hemoglobin levels ranging from 10 to 12%. To achieve this, a total of 1 insulin unit (0.75 U/kg; MedChemExpress) was administered every other day for 4 months. MM group retreated mice after two months of badly controlled hyperglycemia saw an improvement in glycaemic control with 8 units of insulin administered twice daily up to 5–7% of glycated hemoglobin. After creating a 1 cm round wound, mice in the MM group received Nr-CWS treatment for 7 days. In control normal mice of the same age groups and strains, there was no treatment given. The weights of mice were recorded twice a week, the blood glucose of mice was undertaken weekly and the levels of glycosylated hemoglobin were measured every 2 months. The Ethics Committee of Xuzhou Medical University approved the execution of all animal experimentations (Ethical code. 202306T032). All applicable ethical requirements concerning the use of animals have been fulfilled.
Induction of cutaneous wounds
In order to reproduce the biological events related to human wound healing, the splinted mouse wound model was used. The splinting of the wound ensures that the healing process is reliant on epithelialization, cell growth, and new blood vessels. A circular wound of diameter equal to 1 cm was inflicted on the dorsal side of the skins of the mice. And after the wound’s silicone ring was adhered to the wound surface and interrupted stitches were made crossing the ring41,42. ASO-Mettl3 and ASO-Cds2 (sequences shown in Supplementary Table), these ODNs were diluted in PBS, dibutyrate (Haixing Biosciences) (30 ng/ml), and applied onto the wounds, Sense-ODN was applied as control. All wounds were covered.
Immunohistochemistry and immunofluorescence
Mouse skin samples were embedded in optimal cutting temperature (OCT) compound (Servicebio, G6059-110ML) and cut into 10 μm thick sections using a cryostat. The sections were fixed in 4% paraformaldehyde, followed by permeabilization with a solution containing 0.3% Triton X-100 and 1% bovine serum albumin (BSA) in PBS. To block nonspecific binding, sections were incubated with 10% normal goat serum. For immunostaining, the sections were incubated overnight at 4 °C with Alexa488-conjugated anti-KRT14 antibody (1:50, Proteintech, 10143-1-AP).
Additional immunohistochemical analyses were performed on formalin-fixed, paraffin-embedded skin tissues. These tissues were dehydrated, embedded in paraffin, and sectioned at 5 μm thickness. Dewaxing was carried out using Histo-Clear, followed by rehydration in a graded ethanol series. Antigen retrieval was performed by placing the slides in Antigen Decloaker (Zhejiang Jinhua Kedi Instrument Equipment Co., Ltd) at 95 °C for 20 min to break formaldehyde crosslinks. After blocking with 10% normal goat serum, sections were incubated with primary antibodies specific for α-SMA (1:100, Proteintech, 14395-1-ap), CD31 antibodies (1:200, Servicebio, GB15063), and VE-cadherin (1:100, Proteintech, 66804-1-Ig). Following several washes, sections were incubated with secondary antibodies conjugated to Alexa488 (1:400, Servicebio, GB25303) or Alexa-594 (1:400, Jackson, 111-585-003) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Servicebio, G1012). Finally, sections were mounted and visualized under the mounted Camedia Master digital camera (Olympus).
Hematoxylin-eosin (H&E) staining and Masson staining
Mouse skin tissues were collected and immediately fixed in 4% paraformaldehyde for 24 h. After fixation, the tissues were embedded in paraffin and sectioned into 3 μm slices. The sections were stained with hematoxylin and eosin using the H&E High Definition Constant Staining Kit (Servicebio, G1076) and with the Masson Stain Kit (Servicebio, G1006), following the manufacturer’s protocol. Micrographs were then captured using an Olympus light microscope.
Dot blot assay
For the dot blot analysis, total RNA was isolated using the RNA Isolation Kit V2 (Nanjing Vazyme Biotech). The RNA was denatured by preheating at 100 °C for 3 min, then cooled on ice for 60 s before being spotted onto a Hybond-N+ membrane (LABSELECT) and fixed by UV crosslinking. The membrane was incubated with an m6A antibody (1:1000, Proteintech, 68055-1-Ig) overnight at 4 °C, Afterward, the membranes were incubated with an HRP-conjugated secondary antibody, and the signal was detected using the ECL system to measure m6A levels.
Western blot
HUVECs were lysed in RIPA buffer (NCM Biotech). Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and blocked with 5% skimmed milk. The membranes were then incubated with primary antibodies overnight at 4 °C. Afterward, an HRP-conjugated secondary antibody was applied, and the signal was detected using the ECL system (NCM Biotech). The following primary antibodies were utilized: KLF9 (1:2000, ABclonal, A7196), METTL3 (1:2000, Proteintech, 15073-1-AP), H3K27ac (1:1500, Hangzhou Huaan Biotechnology, HO0702), CDS2 (1:2000, Proteintech, 28407-1-AP), β-Actin (1:2000, Servicebio, AC230511049), COX IV (1:2000, ABclonal, A11631), TOMM20 (1:2000, ABclonal, A19403), OPA1 (1:2000, Proteintech, 27733-1-AP), DRP1 (1:2000, Hangzhou Huaan Biotechnology, HA500487), MFN2 (1:1500, Hangzhou Huaan Biotechnology, HA720073), Anti-Rabbit Recombinat (1:4000, Proteintech, RGAR001), Anti-Mouse Recombinat (1:4000, Proteintech, RGAR002) and HA (1:2000, ZENbio, FF0517).
siRNA transfection
Cells were seeded onto 6-well plates and maintained in DMEM media containing 10% FBS at 37 °C, under 5% CO2. When the cell confluence reached approximately 50–70%, we transfected siRNAs targeting Mettl3 (si-Mettl3), Cds2 (si-Cds2), Klf9 (si-Klf9) and Igf2bp3 (si-Igf2bp3) (The sequences are shown in Supplementary Table) were purchased from GenePharma, China. Transient transfection with Lipofectamine 2000 (Invitrogen, USA) was performed according to the manufacturer’s instructions. For each well, transfection with siRNA was performed using Lipofectamine 2000 in the medium without serum in DMEM for 50 nM of siRNA for 5 min at room temperature to form complexes. The siRNA-transfection reagent mixture was then added to the cells and the cells incubated at 37 °C for 8 h. Following this incubation, the medium was removed and replaced with fresh complete culture medium for an additional 24–48 h for the purpose of siRNA-mediated knockdown. A total of forty-eight hours post-transfection the silenced efficiency of Mettl3, Cds2, and Klf9 genes were evaluated by qRT-PCR gene expression analysis.
RNA sequencing and co-immunoprecipitation followed by mass spectrometry (Co-IP-MS)
The total RNA from HUVECs treated with normal control or MM was used subjected to RNA-seq, which was provided by Majorbio Biotech Company in Shanghai, China. Relevant data has been uploaded to the SRA database (PRJNA1165978). The protein solution obtained from the Co-IP experiment using the IP-grade CDS2 antibody (1:50, Proteintech, 28407-1-AP) was used for the Co-IP-MS test conducted by the ApexBio Technology in Wuhan, China (supplementary data 4).
RNA extraction and purification, reverse transcription, and quantitative real-time qPCR assessment
Cells were first lysed in an RNA Isolation Kit V2 (NCM Biotech) and then transcribed into cDNA using first-strand cDNA Synthesis kit (Med Chem Express). Real-time quantitative PCR was conducted using SYBR qPCR mix (TransGen Biotech) with the following thermocycling conditions: It included an initial denaturation step, at 95 °C for 5 min, and 45 cycles of amplification, comprising of denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s and extension at 72 °C for 60 s. Details of the primer sequences are presented in the Supplementary Table. Comparative gene expression was determined using the 2-ΔΔCt method.
Immunofluorescence microscopy
HUVECs were allowed to grow directly on a coverslip until they reached approximately 60% confluence. Following treatment, the cells were fixed with 4% freshly prepared formaldehyde for 15 min at 37 °C and washed three times with PBS to increase the accessibility of the antigen. The cells were then incubated with primary antibodies for 2 h, washed twice with PBS, and at last stained with a secondary antibody for 45 min. Cell images were obtained with LSM480 Zeiss confocal microscope developed from Carl Zeiss Jena, Germany.
RNA pull-down assays
PCR-amplified DNA fragments were used as the template for the synthesis of biotinylated RNA with the help of T7 RNA polymerase in the presence of biotin-UTP. Purified biotinylated transcripts at a concentration of 1 μg/ml were used to transcribe 30 mg of cell lysates for 30 min at room temperature. Pull-down assays were then performed using paramagnetic streptavidin-conjugated Dynabeads (Dynal), and the material was subjected to Western blot. In the RNA pull-down experiment, biotin-labeled RNA probe in which biotin was attached to its 5′terminal was used.
Oligo 1: CAGAUCAGCAGCUCCACAUCUUCAACACGCUGCGGUCUCAUCU
Oligo 2: GAUCGACAAAGGGAUGCUGACAUCCACCACAGAGGACGAGUAG
Oligo 3: GGGCCACCCAGGGCCAGGAGAACAGGAACAGAACUGAGCAGGGG
Oligo 3 (mut): GGGCCACCCAGGGCCAGGAGUUCAGGUUCAGUUCUGAGCAGGGG
Constructs and luciferase reporter assay
Cds2 mRNA 5′UTR fragments were cloned into a pSI3 promoter vector (Focus Biosciences) using HindIII and NcoI sites to build reporter vectors. Constructs and luciferase reporter assayThe same pSI3 promoter vector was used, but the CR and 3 UTR fragments were instead cloned into FseI/Xba I sites. The 293T cells were plated in a density of 3.5 × 104 cells per well in the two-well culture system. SiRNA targeting METTL3 was transfected into the cells The cells were transfected with each of the pSI3-derived reporters 24 h later, post-coculture, and incubated for another 48 h. The luciferase activities of Firefly and Renilla were detected using the Dual Luciferase Assay System (Beyotime) as instructed by the manufacturer, and the sequence was as follows:
UTR-F: TCTAGGCGATCGCTCGAGGGGCCACCCAGGGCCAGGAGAAC
UTR-R: TATTGCGGCCAGCGGCCTTCCTGGTTGTGCCTCCTTCCTC
mut-out-F: gggccacccagggccaggagaacaggagcaggggcaggucuccaag
RNA half-life analysis
To determine the half-life of Cds2 mRNAs, actinomycin D (final concentration 10 mg/ml, Med Chem Express) was added to the HUVEC cultures. After silencing for 48 h with siRNA, total RNA was harvested at 0, 2, 4, 6, 8, and 10 h after the addition of actinomycin D. Levels of mRNA at these time points were examined by qRT-PCR.
Human skin tissue samples
Human clinical samples of skin tissues were stored from the patients at the Affiliated Hospital of Xuzhou Medical University. All the samples were collected after seeking proper patient consent in addition to approval from the medical ethics committee of the affiliated hospital of Xuzhou Medical University (ethical code: XYFY2020-KL041-01). The research was performed in compliance with the Helsinki Declaration and internationally accepted ethical rights for health studies done on humans. H&E staining and RNA extraction were performed on the human samples as approved and in compliance with the guidelines. Proper ethical rules concerning the participation of human subjects in research were ensured to be adhered to.
ATP level determination
ATP content was determined by using the ATP bioluminescence assay kit (Beyotime Biotechnology) according to the instructions of the manufacturer.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the Chromatin Immunoprecipitation Kit following the protocol of the manufacturer (Bersin Bio™ Catalog No.Bes5001). The immunoprecipitation antibodies used were rabbit anti-H3K27ac (Hangzhou Huaan Biotechnology, HA600047) and Rabbit IgG (Proteintech, 30000-O-AP). qRT-PCR of immunoprecipitated DNA was performed using TOP Green Supermix (TransGen Biotech) and StepOne Real-Time PCR System (Applied Biosystems). Relative quantitation of the genes of interest was determined by the 2−ΔΔCT method to obtain the fold change of mRNA. The qRT-PCR primers are indicated in the Supplementary Table.
Seahorse measurement
OCR and ECAR were determined on the Seahorse XFe96 Analyzer produced by Agilent. HUVECs were divided into four groups and plated, at a density of 80,000 cells per well, into poly-L-lysine coated assay plates and allowed to attach overnight. The analysis was performed in Seahorse cell culture medium plus 10% FBS at the temperature of the room. IBMX was included in the cultures 30 min before performing the Seahorse analysis at the concentration of 0.1 mM. KH7 and H89 were administered to groups of cells 1 h before analysis at a final concentration of 10 µM. Oxygen consumption was measured every 6 min, with the following injections occurring after every four measurements: at 1 mM oligomycin, 1 mM FCCP, 2 mM rotenone, and 2 mM antimycin A. The oxygen consumption average per replicate between the four measurements obtained before the injections and the four measurements obtained afterward was calculated and respiratory parameters were determined.
Statistics and reproducibility
For all statistical tests, GraphPad Prism software, version 9.4.1 was used. In this study, data are expressed as mean ± SD. The number of independent replicates for each part of the experiment is noted in parenthesis in the legends to the figure Comparison between the means of the different groups and the control group was done using either the unpaired Student t-test or the one-way ANOVA with a post hoc test Bonferroni correction when necessary. Any statistically significant difference was determined at p < 0.05. The Western Blot data, along with other data were performed at least three times, and representative data are shown.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data have been submitted to the Sequence Read Archive (SRA), Accession to cite for these SRA data: PRJNA1165978. All source data are included in the “Supplementary data 1–5” file linked to this manuscript. Supplementary Fig. 5 lists all Western blot experiments in the manuscript in uncropped and unedited forms. The data which is used to substantiate the findings of this study are retrievable from the author at a reasonable request.
References
Boulton, A. J., Vileikyte, L., Ragnarson-Tennvall, G. & Apelqvist, J. The global burden of diabetic foot disease. Lancet 366, 1719–1724 (2005).
Nathan, D. M. et al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N. Engl. J. Med. 329, 977–986 (1993).
Zhao, J. et al. Transient high glucose causes persistent vascular dysfunction and delayed wound healing by the DNMT1-mediated Ang-1/NF-κB pathway. J. Invest. Dermatol. 141, 1573–1584 (2021).
Kowluru, R. A. Effect of reinstitution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes 52, 818–823 (2003).
Yao, Y. et al. Endothelial cell metabolic memory causes cardiovascular dysfunction in diabetes. Cardiovasc. Res. 118, 196–211 (2022).
Nathan, D. M. et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N. Engl. J. Med. 353, 2643–2653 (2005).
Aschner, P. J. & Ruiz, A. J. Metabolic memory for vascular disease in diabetes. Diabetes Technol. Ther. 14 (Suppl. 1), S68–S74 (2012).
Armstrong, D. G., Boulton, A. J. M. & Bus, S. A. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 376, 2367–2375 (2017).
Okonkwo, U. A. & DiPietro, L. A. Diabetes and wound angiogenesis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms18071419 (2017).
Yamakido, M. et al. Effect of N-CWS injection against influenza virus infection in the retired workers of the Okunojima Poison Gas factory. Hiroshima J. Med. Sci. 33, 547–551 (1984).
Li, Q., Guo, L., Wang, J., Tao, S. & Jin, P. Exosomes derived from Nr-CWS pretreated MSCs facilitate diabetic wound healing by promoting angiogenesis via the circIARS1/miR-4782-5p/VEGFA axis. Chin. J. Nat. Med. 21, 172–184 (2023).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Li, X., Ma, S., Deng, Y., Yi, P. & Yu, J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol. Cancer 21, 76 (2022).
Meyer, K. D. & Jaffrey, S. R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014).
Wei, W., Ji, X., Guo, X. & Ji, S. Regulatory role of N(6) -methyladenosine (m(6) A) methylation in RNA processing and human diseases. J. Cell Biochem. 118, 2534–2543 (2017).
Shen, F. et al. Decreased N6-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 100, E148–E154 (2015).
Yan, W., Lin, X., Ying, Y., Li, J. & Fan, Z. Specific RNA m6A modification sites in bone marrow mesenchymal stem cells from the jawbone marrow of type 2 diabetes patients with dental implant failure. Int. J. Oral. Sci. 15, 6 (2023).
Ren, Y. et al. m(6) A mRNA methylation: biological features, mechanisms, and therapeutic potentials in type 2 diabetes mellitus. Obes. Rev. https://doi.org/10.1111/obr.13639 (2023).
Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R. I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345 (2016).
Bai, Q. et al. Endothelial METTL3 (Methyltransferase-Like 3) inhibits fibrinolysis by promoting PAI-1 (plasminogen activator inhibitor-1) expression through enhancing Jun proto-oncogene N6-methyladenosine modification. Arterioscler. Thromb. Vasc. Biol. 41, 2877–2889 (2021).
Huang W. in Transcriptome Analysis of Gene Expression in Scramble and METTL3 Knockdown HUVEC Cell Lines https://doi.org/10.1161/ATVBAHA.121.316414 (2021).
Huang, H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Ramirez, H. A. et al. Staphylococcus aureus triggers induction of miR-15B-5P to diminish DNA repair and deregulate inflammatory response in diabetic foot ulcers. J. Invest. Dermatol. 138, 1187–1196 (2018).
Ramirez, H. A. et al. Genomic profiling of diabetic foot ulcers identifies miR-15b-5p as a major regulator that leads to suboptimal inflammatory response and diminished DNA repair mechanisms https://doi.org/10.1016/j.jid.2017.11.038 (2017).
Lan, Q. et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 79, 1285–1292 (2019).
Zhou, J., Wei, T. & He, Z. ADSCs enhance VEGFR3-mediated lymphangiogenesis via METTL3-mediated VEGF-C m(6)A modification to improve wound healing of diabetic foot ulcers. Mol. Med. 27, 146 (2021).
Luo, X. et al. Potential genetic therapies based on m6A methylation for skin regeneration: wound healing and scars/keloids. Front. Bioeng. Biotechnol. 11, 1143866 (2023).
Liu, Y., Gu, C., Li, X., Wang, T. & Yu, L. Involvement of METTL3/m(6)Adenosine and TGFβ/Smad3 signaling on Tenon’s fibroblasts and in a rabbit model of glaucoma surgery. J. Mol. Histol. 52, 1129–1144 (2021).
Heacock, A. M. & Agranoff, B. W. CDP-diacylglycerol synthase from mammalian tissues. Biochim. Biophys. Acta 1348, 166–172 (1997).
Laurinyecz, B. et al. Reduced expression of CDP-DAG synthase changes lipid composition and leads to male sterility in Drosophila. Open Biol. 6, 50169 (2016).
Xu, J. et al. Hepatic CDP-diacylglycerol synthase 2 deficiency causes mitochondrial dysfunction and promotes rapid progression of NASH and fibrosis. Sci. Bull. 67, 299–314 (2022).
Galluzzi, L., Kepp, O., Trojel-Hansen, C. & Kroemer, G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 111, 1198–1207 (2012).
Galluzzi, L. et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 15, 1113–1123 (2008).
Wallace, D. C., Brown, M. D., Melov, S., Graham, B. & Lott, M. Mitochondrial biology, degenerative diseases and aging. Biofactors 7, 187–190 (1998).
Madhu, V. et al. The mitophagy receptor BNIP3 is critical for the regulation of metabolic homeostasis and mitochondrial function in the nucleus pulposus cells of the intervertebral disc. Autophagy 19, 1821–1843 (2023).
Taylor, E. B. Functional properties of the mitochondrial carrier system. Trends Cell Biol. 27, 633–644 (2017).
Chen, H. & Chan, D. C. Mitochondrial dynamics-fusion, fission, movement, and mitophagy-in neurodegenerative diseases. Hum. Mol. Genet. 18, R169–R176 (2009).
Youle, R. J. & Narendra, D. P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011).
Frezza, C. et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 (2006).
Wai, T. et al. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 350, aad0116 (2015).
Dunn, L. et al. Murine model of wound healing. J. Vis. Exp. https://doi.org/10.3791/50265 (2013).
Galiano, R. D., Michaels, J. T., Dobryansky, M., Levine, J. P. & Gurtner, G. C. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen. 12, 485–492 (2004).
Acknowledgements
This research was funded by the Science and Technology SMEs Innovation Capacity Improvement Project of Shandong Province (Grant No. 2022TSGC1004), Key Medical Talents in Xuzhou, Jiangsu Province (Grant No. 2023214007), and National Natural Science Foundation of China (Grant No. 82072819).
Author information
Authors and Affiliations
Contributions
Jingyu Zhang: Writing - Original Draft, Formal Analysis, Visualization. Feifei Chen: Writing - Review & Editing, Conceptualization. Wuhan Wei: Investigation, Formal Analysis. Qianqian Ning: Data Curation (Sequencing Data and Public Database Analysis). Dong Zhu, Jiang Fan: Investigation, Visualization. Haoyu Wang: Animal Experimentation. Jian Wang: Methodology, Conceptualization. Aijun Zhang: Patient Recruitment. Peisheng Jin: Project Administration, Supervision. Qiang Li: Project Administration, Conceptualization, Validation, Funding Acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Subhadip Ghatak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Jesmond Dalli and Dario Ummarino. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, J., Chen, F., Wei, W. et al. Nr-CWS regulates METTL3-mediated m6A modification of CDS2 mRNA in vascular endothelial cells and has prognostic significance. Commun Biol 7, 1348 (2024). https://doi.org/10.1038/s42003-024-07047-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07047-y
This article is cited by
-
Targeting the NLRP3 in macrophages contributes to senescence cell clearance in radiation-induced skin injury
Journal of Translational Medicine (2025)