Abstract
Inevitable gefitinib resistance is the biggest bottleneck in current treatment and the mechanisms are not fully understood. Here, we observe that PFTK1 (also named CDK14) is significantly enhanced in NSCLC with gefitinib resistance. And the upregulation of PFTK1 is negatively associated with progression-free survival (PFS) in NSCLC patients who receive gefitinib treatment. Further study suggests that gefitinib can critically accelerate PFTK1 through suppressing its promoter methylation in a DNMT3B-dependent manner. Gain and loss of function assays demonstrate that desregulation of PFTK1 significantly enhances gefitinib resistance in NSCLC. PFTK1 interacts with LRP6 and activates Wnt/β-catenin signaling to attenuate gefitinib-induced cellular apoptosis. Moreover, FMF-04-159-2, a specific covalent inhibitor of PFTK1, can reverse the effect of PFTK1 on gefitinib resistance in vitro and in vivo. Consequently, these findings shed new light on the mechanism underlying gefitinib resistance, and suggest PFTK1 as a target for gefitinib treatment in NSCLC.
Similar content being viewed by others
Introduction
Lung cancer is one of the main causes of cancer-related mortality. Accounting for ~85% of all lung cancer is non-small cell lung cancer (NSCLC)1. Recently, several epidermal growth factor receptor (EGFR) tyrosine-kinase inhibitors (TKIs) have been developed to improve the prognosis of NSCLC patients whose tumors harbor EGFR-sensitizing mutations2,3. Among them, gefitinib, the first-generation EGFR-TKIs, is the standard treatment choice in China. Unfortunately, inevitable resistance develops and limits its clinical application.
Increasing studies have explored the mechanism of gefitinib resistance. Approximately 50% cases of acquired gefitinib resistance are due to a secondary T790M mutation in EGFR gene4. In addition, c-MET and HER2 gene amplification, K-Ras gene mutation, epithelial–mesenchymal transition (EMT), and cancer stemness were the main reasons for gefitinib resistance5,6,7. However, even 30% of NSCLC patients still suffered from gefitinib resistance due to unknown mechanisms8. Therefore, it is extremely urgent to discover new mechanisms of gefitinib resistance.
As we all know, gefitinib is highly specific for exon 19 deletion (del 19) and exon 21 mutation (L858R) of EGFR9. Thus, we constructed gefitinib-resistant cell lines from HCC827 and PC9, which harbor EGFR exon 19 deletion. Through transcriptome sequence and subsequent study, we confirmed that serine/threonine-protein kinase PFTAIRE 1 (PFTK1, also named as CDK14) played an important role in gefitinib resistance and provided evidence for using a PFTK1 inhibitor to reverse gefitinib resistance.
PFTK1 is a member of the cyclin-dependent kinase family. It is highly expressed in the pancreas, brain, heart, kidney, testis, and ovary but lowly expressed in the placenta, lung, and liver tissues10. Initially, PFTK1 was noted to specifically interact with cyclin D3 (CCND3) and form a ternary complex with the cell cycle inhibitor p21 (Cip1) in mammalian cells. Suppressing PFTK1 caused cell cycle arrest at the G1 stage11. Subsequently, PFTK1 was identified to interact with CCNY (Cyclin Y) in several studies12,13. CCNY enhanced PFTK1 kinase activity and recruited PFTK1 to the plasma membrane to phosphorylate low-density lipoprotein receptor-related protein 6 (LRP6)14. LRP6, a single-span transmembrane receptor, plays a key role in the transduction of the canonical Wnt/β-catenin signaling pathway15. Moreover, PFTK1 was reported to be significantly upregulated in esophageal squamous cell carcinoma (ESCC)16, hepatocellular carcinoma (HCC)17, prostate cancer (PCa)18. PFTK1 promotes cell proliferation, migratory properties, and chemotherapy resistance in these tumors. Little is known about the function of PFTK1 in gefitinib resistance.
In the current study, we set out to elucidate the mechanisms that PFTK1 mediates the resistance of NSCLC cells to gefitinib. This aim to identify a feasible therapy target and overcome gefitinib resistance in NSCLC.
Results
Upregulation of PFTK1 in gefitinib resistance NSCLC tissues and cells
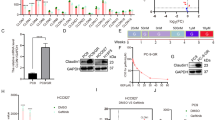
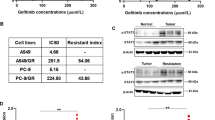
To investigate the mechanism of gefitinib resistance, two NSCLC cell lines HCC827 and PC9 (both harboring EGFR exon 19 deletion) were exposed to gefitinib for more than one year to establish gefitinib-resistant cell lines (HCC827/GR and PC9/GR). Strikingly, compared to parental cells, HCC827/GR cells showed an obvious EMT phenotype, whereas PC9/GR cells exhibited less likely EMT phenotype (Supplement Fig. 1a). Briefly, we conducted cell viability assays to measure the sensitivity of mentioned cells. The results demonstrated that HCC827/GR and PC9/GR cells were highly resistant to gefitinib in contrast with their respective parental cells (Fig. 1a, b). To confirm which gene contributed to gefitinib resistance in NSCLC, Gene Expression Microarray analysis was carried out in HCC827/GR and parental cells. Here, we focused on Wnt signaling pathway to discover its role in gefitinib resistance. The top 10 differentially expressed genes of Wnt signaling pathway were shown and measured by qRT-PCR assays. The results noted that PFTK1 was the most differentially expressed gene among these 10 genes (Supplement Fig. 1b, Fig. 1c). Therefore, we focused on PFTK1 and measured its protein expression. In line with this observation, western blot assay revealed that PFTK1 was significantly enhanced in HCC827/GR and PC9/GR cells comparing with HCC827 and PC9 cells, respectively (Fig. 1d). The results of qRT-PCR assays suggested that PFTK1 was remarkably enhanced in a time-dependent manner by gefitinib (Fig. 1e). Furthermore, we estimated the subcellular location of PFTK1 by immunofluorescence (IF) assays. And, the results suggested that PFTK1 was mainly expressed in the cytoplasm and nucleus (Fig. 1f). As shown in Supplement Fig. 1c, the results of Kaplan–Meier analysis suggested that PFTK1 was positively correlated with poor prognosis of NSCLC patients (p = 0.019). Then, we used the online software COMIP to evaluate the correlation between EGFR mutation and PFTK1 expression in LUAD from the TCGA databank. As indicated in Supplement Fig. 1d, PFTK1 was significantly enhanced in the EGFR mutation group (n = 60), compared to that in the EGFR wild-type group (n = 437). Subsequently, we measured PFTK1 expression in 6 pairs of sensitive and gefitinib-resistant tissues via qRT-PCR assays. The results suggested that PFTK1 was accelerated in gefitinib-resistant tissues than in paired sensitive tissues (Fig. 1g). In addition, immunohistochemistry (IHC) staining analysis was performed in 67 cases of NSCLC tissues with gefitinib treatment to determine PFTK1 expression (Fig. 1h, Supplement Fig. 1e). PFTK1 was markedly increased in 16 out of 19 cases of the resistant group. However, PFTK1 was inhibited in 72.92% of tissues of the 48 cases sensitive group. According to the clinical parameters, we observed that upregulation of PFTK1 was negatively associated with progression-free survival (PFS) in NSCLC patients who received gefitinib treatment (Fig. 1h). Together, these results proved that PFTK1 was highly expressed in NSCLC with gefitinib resistance. Patients with high PFTK1 have a poor response to gefitinib, indicating that PFTK1 may be used to predict the efficacy of gefitinib therapy in NSCLC patients.
a, b HCC827 and PC9 cells were treated with a stepwise dose escalation of gefitinib (ranging from 0.1 to 5 μM) over for 1 year to establish resistant cell lines HCC827/GR and PC9/GR. Cell viability assays were performed in parental and resistant cells. The IC50 values of each cell to gefitinib were evaluated (n = 3). **p < 0.01. c The mRNA levels of the top 10 differentially expressed genes in Wnt signaling pathway from Gene Expression Microarray analysis were determined by qRT-PCR assays (n = 3). **p < 0.01. d WB assays and densitometric analysis for the expression of PFTK1 were conducted in the mentioned cells (n = 3). **p < 0.01. e HCC827 and PC9 cells were treated with 1 μM gefitinib for 0 h, 6 h, 12 h, 24 h, and 48 h, respectively. The PFTK1 expression levels were measured via qRT-PCR assays (n = 3). f The subcellular location of PFTK1 was estimated in NSCLC cells via IF assays. The staining intensity of the nucleus and the cytoplasm were quantified by Image J software (n = 3). Scale bars: 40 μm. g Six pairs of sensitive and gefitinib-resistant NSCLC tissues were collected and detected for PFTK1 expression by qRT-PCR assays (n = 3). h The PFTK1 expression levels were measured in another cohort of 67 cases of NSCLC tissues with gefitinib treatment via IHC assays. Based on PFTK1 expression, the patients were divided into two groups. Kaplan–Meier analysis was performed to evaluate the relationship between PFTK1 expression and the PFS of these patients. **p < 0.01.
The Function of PFTK1 involved in gefitinib resistance of NSCLC
To verify the role of PFTK1 in gefitinib resistance, we restored PFTK1 in HCC827 and PC9 cells, and knocked down PFTK1 in gefitinib-resistant cell lines HCC827/GR and PC9/GR (Fig. 2a, b). It is found that over-expressing PFTK1 could significantly enhance cell resistance to gefitinib in HCC827 and PC9 cells (Fig. 2c). Conversely, inhibiting PFTK1 in HCC827/GR and PC9/GR cells did the opposite (Fig. 2d). HCC827 cells transfected with vector showed a slight decrease in cell colony formation ability with gefitinib treatment. However, cells transfected with PFTK1 showed a significant increase in cell colony formation ability with gefitinib treatment. Similar results were supported by PC9 cells (Fig. 2e, Supplement Fig. 2a). On the contrary, the knockdown of PFTK1 in gefitinib-resistant cells dramatically suppressed the cell colony formation ability with gefitinib treatment (Fig. 2e, Supplement Fig. 2b). Consistently, flow cytometry results suggested that both HCC827 and PC9 cells with restored PFTK1 showed a sharp decrease in apoptosis after gefitinib treatment, compared with each control group (Fig. 2f, Supplement Fig. 2c, d). To further investigate whether PFTK1 affects cell viability in vivo, cells (HCC827/Luc cells with restored PFTK1 or vector, HCC827/GR/Luc cells with shRNA targeting PFTK1 or control, PC9 cells with restored PFTK1 or vector, PC9/GR cells with shRNA targeting PFTK1 or control) were subcutaneously injected into BALB/c nude mice to establish a xenograft tumor model. The results of the luciferase image demonstrated that the PFTK1 overexpression group exhibited stronger luciferase activity and bigger tumor size with gefitinib treatment. However, knocking down PFTK1 got the opposite results (Fig. 2g, h, Supplement Fig. 2e). Collectively, the in vitro and in vivo results proved that PFTK1 was involved in the gefitinib resistance of NSCLC.
a HCC827 and PC9 cells were stably transfected with PFTK1 overexpression plasmid, b HCC827/GR and PC9/GR cells were transfected with shRNAs targeting PFTK1. a, b qRT-PCR and western blot analysis were conducted to measure the expression of PFTK1 in mentioned cells. Also, the densitometric analysis for the expression of PFTK1 was conducted in mentioned cells (n = 3). **p < 0.01. c, d Above cells were treated with gefitinib at the indicated concentration for 72 h, cell viability was evaluated by MTS assay (n = 3). **p < 0.01. e Cells (HCC827 and PC9 cells with restored PFTK1 and control group, HCC827/GR and PC9/GR cells with knocking down PFTK1 and control group) were seeded into a six-well plate, followed by 0.5 μM gefitinib treatment. Then, the cell clone numbers were estimated in each group (n = 3). **p < 0.01. f Cells were treated with 5 μM gefitinib for 72 h, and then the cell apoptosis was analyzed by flow cytometry (n = 3). **p < 0.01. g The workflow was showed. Tumor cells were cultured and subcutaneously inoculated into recipient nude mice. When the tumor volume was ~100 mm3, the mice were treated with gefitinib at a dose of 30 mg kg−1, daily for 20 days. The elements of this image were created from icofont website (https://www.iconfont.cn). h HCC827/Luc cells with restored PFTK1 and vector, HCC827/GR/Luc cells with shRNA targeting PFTK1 and control, were subcutaneously injected into BALB/c nude mice to establish a xenograft tumor model. These mice were treated with gefitinib. Luciferase activity was imaged and measured by In Vivo Xtreme from Bruker (n = 3). “gefitinib” was abbreviated as “Gef”. **p < 0.01.
PFTK1 activating the Wnt/β-catenin signaling pathway mediated gefitinib resistance in NSCLC
To address the underlying mechanism by which PFTK1 enhances gefitinib resistance, we analyzed the proteomics of HCC827 cells by restoring PFTK1 and the control group. Compared with control cells, 742 proteins were obviously upregulated, and 767 proteins were significantly downregulated in HCC827 cells with restored PFTK1 (Fig. 3a). Subcellular location analysis demonstrated that approximately 31.81% of the proteins were just located in nucleus. While 26.83% of the proteins were predominantly expressed in cytoplasm (Fig. 3b). As shown in Fig. 3c, the differentially expressed proteins of Wnt signaling pathways were exhibited via GO analysis. Among these, we focused on the higher upregulation of β-catenin caused by PFTK1. Previous studies have suggested that phosphorylation of LRP6 is crucial for Wnt/β-catenin signal transduction. Activating Wnt signaling promotes β-catenin to transfer into the nucleus and bind to members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family, recruit transcriptional co-activators including p300 and/or CREB binding proteins to drive a Wnt transcriptional program19. Therefore, we conducted a series of experiments to verify this. The results suggested that PFTK1 greatly enhanced the phosphorylation levels of LRP6 and β-catenin, and facilitated the nuclear translocation of β-catenin. While suppressing PFTK1 did the opposite (Fig. 3d, e). CO-IP assays suggested that PFTK1 can interact with both LRP6 and p-LRP6 in NSCLC cells (Fig. 3f). To further investigate, we employed the TOPFlash (a wild type of TCF/LEF-binding site)/FOPFlash (a mutant type of TCF/LEF-binding site) reporter system to determine the impact of PFTK1 on the transcriptional activity of β-catenin. The luciferase reporter gene assay suggested that PFTK1 markedly accelerated TOPFlash luciferase activity in NSCLC cells, while having no effect on FOPFlash activity (Fig. 3g). In addition, we used Adavivint (SM04690), an effective classical Wnt signaling pathway inhibitor, to address the function of the Wnt/β-catenin signaling pathway caused by PFTK1 in gefitinib resistance. It is noted that Adavivint reversed the effect of PFTK1 on TOPFlash luciferase activity (Fig. 3g). The results of cell viability assays suggested that restored PFTK1 enhanced gefitinib resistance in NSCLC cells, while Adavivint significantly reversed this effect (Fig. 3h). Finally, we demonstrated that PFTK1 promoted tumor growth with gefitinib treatment in vivo, compared to the control group. Expectedly, Adavivint reversed this effect (Fig. 3i). Taken together, these findings demonstrated that PFTK1 activated the Wnt/β-catenin signaling pathway to enhance the resistance of NSCLC cells to gefitinib.
a The proteomics analysis was carried out from HCC827/Vec and HCC827/PFTK1 cells. The scatter plot showed the log2 fold-change of the proteome dataset. Red dots represented significantly upregulated proteins, while green dots represented significantly downregulated proteins. b The subcellular localization of the above proteins was exhibited. c Through GO analysis, we displayed the differently expressed proteins in Wnt/β-catenin signaling. d In HCC827 and PC9 cells with increasing PFTK1, HCC827/GR and PC9/GR cells with PFTK1 knockdown, the protein levels of p-LRP6 and β-catenin were determined (n = 3). **p < 0.01. e IF assays were performed to measure the distribution of β-catenin. Scale bars: 40 μm. f PFTK1-flag overexpression plasmid was transfected into HCC827 cells, and CO-IP assays were accomplished. Since p-LRP6 and LRP6 were almost the same molecular weight, p-LRP6 was shown in a different gel. g TOPFlash and FOPFlash plasmids were transfected into HCC827 and PC9 cells with overexpression PFTK1, respectively. Ada (Adavivint), an effective classical Wnt signaling pathway inhibitor, was used in the PFTK1 group. Luciferase activity was detected in mentioned cells (n = 3). **p < 0.01. h HCC827 and PC9 cells with restored PFTK1 and control group, together with Ada treatment group were all applied to gefitinib at the indicated concentration for 72 h, and the cell viability of each group was evaluated by MTS assays (n = 3). **p < 0.01. i The workflow was shown. The elements of this image were created from icofont website (https://www.iconfont.cn). HCC827/Luc with PFTK1 and control cells were subcutaneously injected into BALB/c nude mice to establish a xenograft tumor model. In addition, half of the HCC827/PFTK1/Luc group was treated with Ada. All the animals were treated with gefitinib. Luciferase activity was imaged and measured by In Vivo Xtreme from Bruker (n = 2). **p < 0.01.
Decreasing DNMT3B inhibited PFTK1 via promoter methylation in NSCLC with gefitinib resistance
To gain insights into the mechanism of PFTK1 upregulated in gefitinib-resistant cells, we first estimated its promoter. We downloaded a 2500 bp promoter sequence of PFTK1 from UCSC and predicted its CpG island via the online software CpG finder. It is suggested that there was a CpG island (1449-2040 nt) in the promoter of PFTK1 (Supplement Fig. 3a). Subsequently, we assessed the relationship between the methylation level and mRNA expression level of PFTK1 in LUAD via the online databank cBioPortal. As shown in Supplement Fig. 3b, the PFTK1 expression level was negatively related to its promoter methylation in 370 cases of LUAD from TCGA data (r = −0.20, P = 3.34e-7). Then, the MethSurv databank noted that the higher the methylation level of PFTK1 was, the better the patients’ prognosis was (Supplement Fig. 3c). Therefore, we aimed to validate whether the upregulation of PFTK1 was accompanied by the reductions in DNA methylation levels during gefitinib treatment.
Briefly, we designed 5 sets of MSP primers by online software MethPrimer based on the 2500 bp promoter sequence (Supplement Fig. 3d). As a result, only primers of set 5 could amplify in MSP assays, we used this set of primers in the following study. We observed that the unmethylation status of the PFTK1 promoter was found in HCC827/GR and PC9/GR cells, while the methylation status of the PFTK1 promoter was exhibited in HCC827 and PC9 cells (Fig. 4a). Furthermore, we explored the methylation pattern of PFTK1 promoter from six patients with EGFR-mutant NSCLC, who underwent gefitinib treatment. The MSP results revealed that the PFTK1 promoter was unmethylated in all NSCLC tissues after gefitinib treatment, and the methylation status was present in these tissues before gefitinib treatment (Fig. 4b). DNA methyltransferases (DNMTs) could suppress the expression of target genes via hypermethylation of the CpG islands in the promoter regions20. Indeed, we used 5-Azacytosine (5-Aza, a typical DNMTs inhibitor) to treat NSCLC cells that contain methylation status of PFTK1 promoter. As expected, PFTK1 was enhanced by 5-Aza in a dose-dependent manner (Fig. 4c). Currently, the human genome encodes five DNMTs: DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L21. We performed qRT-PCR assays and found that only DNMT3B but no other DNMTs were repressed in gefitinib-resistant cell lines HCC827/GR and PC9/GR (Fig. 4d). Certainly, similar results were obtained via WB assays (Fig. 4e). A significantly negative expression relationship between DNMT3B and PFTK1 were disclosed by online databank TIMER (Supplement Fig. 3e). Subsequently, we restored DNMT3B in HCC827/GR and PC9/GR cells, and knocked down DNMT3B with shRNAs in HCC827 and PC9 cells (Supplement Fig. 3f). Both qRT-PCR and WB assays demonstrated that restored overexpression of DNMT3B obviously repressed PFTK1, while knocked down DNMT3B did the opposite (Fig. 4f, g). Moreover, a ChIP assay was performed to investigate the recruitment of DNMT3B to the promoter region of PFTK1. Increased occupancy of DNMT3B in the CpG region of PFTK1 was observed in parental cells by ChIP assay, which could be abolished in gefitinib-resistant cells (Fig. 4h). Moreover, we observed that DNMT3B was evidently repressed in NSCLC tissues with gefitinib resistance, compared to that of NSCLC tissues with gefitinib sensitive (Fig. 4i). As showed in Fig. 4i, DNMT3B was negatively related with PFTK1 in these NSCLC tissues with gefitinib treatment (r = −0.3792, p = 0.0016). Ultimately, we proposed that gefitinib could significantly suppress DNMT3B, and DNMT3B-regulated PFTK1 via enhancing methylation level of its promoter.
a MSP assays were performed to evaluate the methylation status of the PFTK1 promoter in gefitinib-resistant and parental cells. “Methylation” was abbreviated as “M”, “Unmethylation” was abbreviated as “U”. b Six pairs of NSCLC tissues [before and after gefitinib treatment, we abbreviated as sensitive (S) and resistance (R)] were collected, and the methylation status of the PFTK1 promoter was determined by MSP assays. c 5-Azacytidine (5-Aza), an inhibitor of DNMTs, was used to treat HCC827 and PC9 cells for 24 h. The expression of PFTK1 was measured by qRT-PCR assays (n = 3). **p < 0.01. d The relative expression levels of common DNMTs in gefitinib-resistant and parental cells were evaluated by qRT-PCR assays (n = 3). **p < 0.01. e The protein levels of DNMT3B in gefitinib-resistant and parental cells were assessed via WB assay (n = 3). **p < 0.01. f, g DNMT3B overexpression plasmid and control plasmid were transfected into HCC827/GR and PC9/GR cells (f). ShRNAs targeting DNMT3B were transfected into HCC827 and PC9 cells (g). WB and qRT-PCR assays were carried out to estimate the expression level of PFTK1 (n = 3). **p < 0.01. h The occupancy of DNMT3B in the promoter of PFTK1 among gefitinib-resistant and parental cells was valued via ChIP assay (n = 3). **p < 0.01. i The DNMT3B expression in 67 cases of NSCLC tissues with gefitinib treatment was determined by IHC assay. The relationship between DNMT3B and PFTK1 was analyzed. Scale bars: 40 μm (**p < 0.01).
Pharmacological inhibition of PFTK1 reversed the gefitinib resistance in NSCLC
To explore the potential of PFTK1 as a therapeutic target in NSCLC, we focused on FMF-04-159-2 (FMF), a specific covalent inhibitor of PFTK1. As shown in Fig. 5a, we observed the binding site of FMF on PFTK1 via MOE software. qRT-PCR and WB assays suggested that FMF neither regulated the mRNA nor the protein expression level of PFTK1 (Fig. 5b). The colony formation assays showed that FMF effectively inhibited the proliferation of cells with gefitinib treatment (Fig. 5c). The similar results were obtained by cell viability assay (Fig. 5d). In addition, it is indicated that the cell apoptosis was significantly increased in the presence of FMF by flow cytometry assays (Fig. 5e). WB analysis evaluated that FMF could remarkably suppress the phosphorylation of LRP6 and the total β-catenin (Fig. 5f). Meanwhile, we conducted the dual-luciferase reporter gene assays and found that the luciferase activity of TOPFlash but not FOPFlash were mainly reduced in FMF group, comparing with that in the control group (Fig. 5g). Next, we tested the effect of FMF on gefitinib resistance in vivo by subcutaneously injected cells into BALB/c nude mice. The results validated that FMF could manifestly inhibit tumor growth with gefitinib treatment (Fig. 5h). Together, all these results confirmed that FMF targeting PFTK1 evidently attenuated the Wnt/β-catenin signaling to enhance the sensitivity of NSCLC cells to gefitinib.
a The 2D and 3D view of molecular docking was showed by MOE output. b–e HCC827/GR and PC9/GR cells were co-treated with FMF and gefitinib. The expression levels of PFTK1 (b), the cell colony-forming ability (c), the cell viability (d), and the cell apoptosis (e) were measured in each group (n = 3). **p < 0.01. f, g HCC827/GR and PC9/GR cells were treated with FMF, WB assays (f) and dual-luciferase reporter gene assays (g) were carried out to measure the activity of Wnt/β-catenin signaling pathway (n = 3). **p < 0.01. h Illustration for CDX development. The elements of this image were created from icofont website (https://www.iconfont.cn). HCC827/GR cells with luciferase were subcutaneously injected into nude mice. When tumor size reached 100 mm3, FMF and gefitinib were used daily to treat these nude mice by oral gavage for 20 days. Luciferase activity was imaged and measured by In Vivo Xtreme from Bruker (n = 3). **p < 0.01. i A schematic diagram depicts how PFTK1 upregulated by gefitinib actives Wnt/β-catenin signaling pathway to promote gefitinib resistance in NSCLC. We drew it on Figdraw (https://www.figdraw.com/).
Discussion
In our country, gefitinib is the most common drug in NSCLC with EGFR-mutant. Understanding and discovering the mechanisms of gefitinib resistance is critical for improving the therapeutic efficacy and survival rate of patients with NSCLC. As shown in Fig. 5i, we highlighted that PFTK1 was associated with gefitinib resistance. We observed that PFTK1 was significantly upregulated by decreasing DNMT3B in gefitinib-resistant NSCLC cells and clinical samples. Mechanistically, PFTK1 interacting with LRP6 accelerated the activity of Wnt/β-catenin signaling to confer gefitinib resistance. In addition, pharmacological inhibition of PFTK1 by FMF manifestly enhanced the sensitivity of NSCLC cells to gefitinib in vitro and in vivo. Therefore, PFTK1 might be given as a potential therapeutic strategy for overcoming resistance to gefitinib in NSCLC.
The past few years have seen an increase in studies that assess the functional role of PFTK1 in cancers. It is reported that PFTK1 was aberrantly expressed in several cancers and played an important role in cancer progression22,23. The expression level of PFTK1 was positively correlated with the poor prognosis of patients17. Recently, PFTK1 has been found to be associated with chemotherapy resistance16. Here, we found that PFTK1 was significantly increased in gefitinib-resistant NSCLC cells and clinical samples. And, we observed that the high expression of PFTK1 was correlated with PFS and gefitinib resistance in EGFR-mutant NSCLC patients. Functional investigations revealed that knocking down PFTK1 resensitized resistant NSCLC cells to gefitinib in vitro and in vivo. Consistently, FMF (a specific PFTK1 inhibitor) treatment significantly overcame gefitinib resistance and showed an obvious effect on the inhibition of gefitinib-resistant NSCLC cells. These data suggested that targeting PFTK1 can be a novel strategy to overcome gefitinib resistance in lung cancer.
Increasing studies have demonstrated that Wnt/β-catenin signaling pathway played an important role in EGFR-TKI resistance24. It is reported that inhibition of β-catenin enhances the anticancer effect of irreversible EGFR-TKI in EGFR-mutant NSCLCs25. Notably, knockdown of β-catenin decreased the expression of SOX2 and reduced the stemness of CSC properties and osimertinib resistance in NSCLC26. Combining the results of proteomics analysis and transcriptome sequence in gefitinib-resistant and parental cells, we proposed Wnt/β-catenin signaling as a potential downstream factor contributing to the effects of PFTK1. As discovered, CDK14/cyclin Y could phosphorylate LRP5/6 to constitutively activate Wnt/β-cantenin signaling during mitosis15,27. Consistent with this, we performed CO-IP and WB assays together with TOPFlash/FOPFlash reporter assays and revealed that PFTK1 interacted with LRP6 to trigger the Wnt/β-catenin signaling and confer gefitinib resistance in NSCLC. Similarly, a recent study reported that CDK14 was found to be elevated in triple-negative breast cancer (TNBC). CDK14 knockdown or pharmacological inhibition by FMF suppressed the progression and metastasis of TNBC via attenuating Wnt/β-catenin signaling28.
Aberrant DNA methylation is one of the most important epigenetic alterations in eukaryotic genomes29. The majority of methylation marks are present at 5mC and are often presented in CpG island30. For the first time, we predicted that there was a CpG island in the PFTK1 promoter by bioinformatics analysis. MSP assays combining with ChIP assays and several other experiments verified that PFTK1 was regulated by DNMT3B through modulating the methylation status of PFTK1 promoter. DNMT3B, a famous DNMTs, functions in the transcriptional repression of many target genes31,32. As revealed, DNMT3B altered multiple pathways including STAT3, NFκB, PI3K/Akt, β-catenin, and Notch signaling, which were critical for cancer cell survival, apoptosis, proliferation, invasion, and colonization33. Besides, several studies disclosed the regulation pattern of PFTK1 in cancers. It was suggested that hsa_circ_102229 directly targeted miR-152-3p as a ceRNA to regulate PFTK134. Coincidentally, HIF-1α regulated the expression of LINC01355 (Hypoxia Yield Proliferation Associated LncRNA, HYPAL) in GC cells and tissues. HYPAL served as a ceRNA of miR-431-5p to regulate CDK14 expression35. Through microarray analyses and other experiments, androgen receptor (AR) was identified as a candidate of dual-specificity tyrosine-regulated kinase 2 (DYRK2)-dependent transcription factors regulating CDK14 to promote cancer cell proliferation and invasion in vitro and in vivo36.
To date, this study revealed a novel expression regulation of PFTK1 and elucidated a critical role of the PFTK1/β-catenin axis in patients with EGFR-mutant NSCLC with resistance to gefitinib. Based on this important finding, we suggest that targeting PFTK1 might serve as an effective strategy for overcoming acquired resistance to gefitinib in lung cancer.
Methods
Reagents, antibodies, and plasmids
gefitinib and adavivint (Ada, SM04690) were obtained from Selleck (Shanghai, China). FMF-04-159-2 (FMF, a PFTK1 inhibitor) and 5-Azacytidine (5-Aza) were obtained from MedChemExpress (New Jersey, USA). SYBR and iBright™ Prestained Protein Ladder were bought from Thermo Scientific (Waltham MA, USA). PFTK1 antibodies (ab224098, 1:50) and (21612-1-AP, 1:1000) were obtained from Abcam (Cambridge, UK) and Proteintech (Wuhan, China), respectively. Primary antibodies against β-actin (#3700, 1:8000), β-catenin (#8480, 1:1000), DNMT3B (#57868, 1:2000), p-LRP6 (#2568, 1:500), LRP6 (#3395, 1:500), and normal IgG (#2729, 1:500) were purchased from Cell signaling Technology (Boston, USA).
TOPFlash and FOPFlash plasmids were bought from Addgene (Mass, USA). Overexpression plasmids (PFTK1, DNMT3B, Luciferase), and shRNA knockdown plasmids targeting PFTK1 were bought from Genechem (Shanghai, China).
Cell culture
The human NSCLC cell lines HCC827 (RRID: CVCL_2063) and PC9 (RRID: CVCL_B260) with EGFR-activating mutation (deletion in exon 19) were bought from ATCC (Manassas, VA, USA). HCC827 and PC9 cells were treated with increasing concentrations (ranging from 0.1 to 5 μM) of gefitinib for 1 year to establish resistant cell lines HCC827/GR and PC9/GR. These cells were routinely cultured at 37 °C in a humidified incubator (5% CO2) in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). The short tandem repeat (STR) analysis was carried out to authenticate cells in the last three years by our lab. All cell lines were negative for mycoplasma contamination.
Briefly, HCC827 and HCC827/GR cells were transfected with Luciferase overexpression plasmids. G418 was used to generate Luciferase stable expression cells (named HCC827/Luc and HCC827/GR/Luc). Then, vector and PFTK1 overexpression plasmids were transfected into HCC827/Luc and PC9 cells, respectively. ShRNAs targeting PFTK1 and control plasmids were transfected into HCC827/GR/Luc and PC9/GR cells, respectively.
Clinical analysis
The study was approved by Guangzhou Institute of Cancer Research, the Affiliated Cancer Hospital, Guangzhou Medical University. All ethical regulations relevant to human research participants were followed. Six pairs of NSCLC tissues were collected before and after gefitinib therapy. The nineteen cases of NSCLC tissues with gefitinib resistance and forty-eight cases of NSCLC tissues with gefitinib sensitivity were contained in this cohort. All samples were collected with informed consent from the patients at the Guangzhou Institute of Cancer Research, the Affiliated Cancer Hospital, Guangzhou Medical University. Two pathologists independently identified the samples. The survival time of individuals with NSCLC was evaluated from the diagnosis date to the date of the last follow-up or death.
Tumor xenograft model and bioluminescence imaging
The animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Guangzhou Medical University. Standard animal care and laboratory guidelines were followed according to the IACUC protocol. We have complied with all relevant ethical regulations for animal use. Mice were housed in individually ventilated cages under standard room temperature (22 °C) and humidity (55%) and a 12/12 light/dark cycle. A total of 5 × 105 target cells were subcutaneously injected into Balb/C athymic nude mice (4 weeks, female, 18–22 g) from Guangdong Medical Laboratory Animal Center (Guangzhou, China) to generate xenograft tumors. Tumor volumes were assessed every 4 days by measuring the externally apparent tumors in two dimensions with calipers. The volume of the tumors was calculated using the formula: V = 2−1 LW2. When tumor size reached 100 mm3, the mice were treated with gefitinib (30 mg kg−1, daily for 20 days) or FMF (40 mg kg−1, daily for 20 days) by oral gavage. IACUC stipulated that the tumor volume can’t exceed 2000 mm3, and in none of the experiments were these limits exceeded. At the end of the experiment, mice were sacrificed with carbon dioxide. The tumors were collected, fixed, and embedded in paraffin. For imaging luciferase, we used In Vivo Xtreme from Bruker. Ten minutes before imaging, the mice were injected i.p. with 150 mg kg−1 D-luciferin (GoldBio).
RNA isolation and qRT-PCR
Total RNA was isolated from cells by an RNA extraction Kit (Omega, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized with a RevertAid First-Strand cDNA Synthesis Kit (Thermo Scientific). qRT-PCR assays were performed on a CFX96 Real-Time PCR System (Bio-Rad iQ5 program) using PowerUp SYBR Green Master Mix (Thermo Fisher). GAPDH was used as an internal control. The relative levels of gene expression were calculated by the 2−ΔΔCt method. Primer sequences were provided in Supplementary Table 1. All assays were conducted at least three times.
Bisulfite modification and MSP chain reaction
Genomic DNA was extracted from cells and fresh tissue samples by Tissue DNA Kit (Omega, #D3396). An EpiTect Bisulfite Kit (Qiagen, #59104) was applied to conduct the bisulfite modification of genomic DNA following the manufacturer’s instructions. MSP primers were designed by the online software MethPrimer and synthesized (BGI, Beijing, China) to detect unmethylated (U) and methylated (M) alleles. The MSP primers were listed in Supplementary Table 1. The expected sizes of U and M products were 136 and 132 bp, respectively.
Western blot
Whole-cell lysates were prepared in RIPA buffer (Thermo Scientific) in the presence of Halt Protease and Phosphatase Inhibitor Cocktail (Pierce Chemical, Dallas, Texas, USA). Protein concentration was measured using a BCA Protein Assay Kit (Thermo Scientific). Thirty micrograms of lysates were resolved by SDS–polyacrylamide gel electrophoresis and then transferred to PVDF membranes (Millipore, Burlington Mass, USA). After blocking for nonspecific binding, membranes were incubated with specific primary antibodies overnight at 4 °C. Then, incubation with the specific HRP-conjugated antibody was performed. The signal was detected using an enhanced chemiluminescence western blot detection kit (Millipore). The densitometric analysis of all the western blot assays was conducted by Image J software. All assays were conducted at least three times.
Apoptosis assays
Target cells were seeded in a 6-well plate overnight. Cells were harvested after 72 h treatment with 5 μM gefitinib, incubated with Annexin V-APC and 7-AAD for 5 minutes, respectively. Furthermore, these cells were analyzed by flow cytometer (Becton Dickinson USA). All assays were conducted at least three times.
Cell viability assay
Cells were planted into 96-well plates and then incubated with different concentrations of gefitinib for 72 h. CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was used to test cell viability, following the manufacturer’s instructions.
Plate colony formation assay
A total of 500 target cells per well were seeded in triplicate in 6-well culture plates. These cells were treated with 0.1 μM gefitinib and cultured to form colonies. Two weeks later, these cells were fixed with methanol and followed by staining with 0.5% crystal violet. Finally, the plate was photographed, and the number of clones was calculated. All assays were conducted at least three times.
Co-immunoprecipitation (CO-IP) assay
Briefly, cells were lysed by RIPA buffer, and sonicated to get total proteins. Next, these proteins were incubated with anti-LRP6, anti-p-LRP6, and anti-IgG overnight at 4 °C, respectively. Then, these complexes were incubated with precleared protein AG-agarose beads (Millipore) for 2 h at room temperature. The reaction products were washed with lysis buffer three times and were subjected to western blot analysis for the potential proteins.
Chromatin immunoprecipitation (ChIP) assay
We performed ChIP assays with an EZ-ChIPTM chromatin immunoprecipitation kit (Millipore). At first, cells were cross-linked with 1% formaldehyde followed by glycine. Next, cells were lysed with lysis buffer containing Protease Inhibitor Cocktail II and sonicated on ice. 10% of the supernatant was saved as the input fraction. Then, the rest was used for immunoprecipitation with 5 µg of anti-DNMT3B antibody and anti-rabbit IgG, respectively. Finally, DNA was purified and quantified by qRT-PCR assay. IgG was included as a nonspecific control. The primers for ChIP-qPCR were shown in Supplementary Table 1.
Dual-luciferase report gene assay
Cells were seeded in 96-well plates at a density of 1 × 104 cells per well overnight. 3 ng Renilla luciferase reporter vectors and 30 ng luciferase reporter vectors (TOPFlash and FOPFlash plasmids, respectively) were co-transfected into the cells via Lipofectamine 3000 (Invitrogen). After 48 h, a dual-luciferase reporter assay system (Promega) was used to evaluate the luciferase activity. The firefly luciferase values were normalized to the Renilla luciferase values.
IF assay
Generally, cells were seeded onto the coverslips in a six-well plate overnight. The next day, cells on the coverslips were fixed with 4% paraformaldehyde, and permeabilized by 0.1% Triton X-100. Then, cells were incubated with the primary antibody for 1 h, followed by the secondary antibody. Finally, these cells were incubated with DAPI to stain the nucleus. Images were acquired on a confocal microscope (Zeiss, Germany). The staining intensity of the nucleus and cytoplasm was calculated by Imagine J software.
IHC assay
The tissues were fixed by formalin, embedded via paraffin, followed by xylene and grade ethanol. Further, the slides were immersed in 3% hydrogen peroxide and incubated with primary antibodies overnight. Moreover, the slides were incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, sc-2357, sc-516102). Finally, after the application of DAB chromogen, tissue sections were stained with hematoxylin. Images were captured with Panoramic 250 FLASH (3DHistech) and assessed with Case Viewer software.
Bioinformatics analysis
The promoter sequence (2500 bp) of PFTK1 was downloaded from UCSC, and potential CpG islands were predicted via CpG finder (http://www.softberry.com/berry.phtml?topic=cpgfinder&group=programs&subgroup=promoter). The MSP primers were designed by the online software MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The relationship between the methylation level of the PFTK1 promoter and its mRNA expression level in LUAD was evaluated by cBiorPortal (https://www.cbioportal.org/cbioportal). The online databank MethSurv (https://biit.cs.ut.ee/methsurv/) estimated that the higher the methylation level of the PFTK1 promoter was, the better prognosis of LUAD patients was. Moreover, the TIMER databank (https://cistrome.shinyapps.io/timer/) suggested that DNMT3B was negatively correlated with PFTK1 in LUAD. In addition, we also obtained the PFTK1 expression and EGFR mutation information from the online databank CAMOIP (https://portal.gdc.cancer.gov/).
Statistical analysis
Statistical analysis was performed by SPSS version 19.0 and GraphPad Prism 7. The results are presented as the mean ± SD of three or more experimental repeats. A χ2 test was carried out to analyze the relationship between gene expression levels. Survival analysis was implemented by Kaplan–Meier analysis. A comparison of the mean between the two groups was conducted by Student’s t test. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study are included in this published article (as well as in the accompanying Supporting Information). The data of Gene Expression Microarray analysis is available in GEO with accession number GSE278453. The proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD057124.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Thatcher, N. et al. gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet 366, 1527–1537 (2005).
Passaro, A., Janne, P. A., Mok, T. & Peters, S. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat. Cancer 2, 377–391 (2021).
Stewart, E. L., Tan, S. Z., Liu, G. & Tsao, M. S. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl. Lung Cancer Res. 4, 67–81 (2015).
Pao, W. et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2, e17 (2005).
Westover, D., Zugazagoitia, J., Cho, B. C., Lovly, C. M. & Paz-Ares, L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol. 29, i10–i19 (2018).
Chiu, C. F. et al. NF-kappaB-driven suppression of FOXO3a contributes to EGFR mutation-independent gefitinib resistance. Proc. Natl. Acad. Sci. USA 113, E2526–E2535 (2016).
Zhao, H. et al. Apatinib plus gefitinib as first-line treatment in advanced EGFR-mutant NSCLC: the phase III ACTIVE study (CTONG1706). J. Thorac. Oncol. 16, 1533–1546 (2021).
Chia, P. L. et al. Expression of EGFR and conformational forms of EGFR in malignant pleural mesothelioma and its impact on survival. Lung Cancer 153, 35–41 (2021).
Yang, T. & Chen, J. Y. Identification and cellular localization of human PFTAIRE1. Gene 267, 165–172 (2001).
Shu, F. et al. Functional characterization of human PFTK1 as a cyclin-dependent kinase. Proc. Natl. Acad. Sci. USA 104, 9248–9253 (2007).
Zhao, X. et al. Cytoplasmic localization isoform of cyclin Y enhanced the metastatic ability of lung cancer via regulating tropomyosin 4. Front. Cell Dev. Biol. 9, 684819 (2021).
Kaldis, P. & Pagano, M. Wnt signaling in mitosis. Dev. Cell 17, 749–750 (2009).
Jiang, M., Gao, Y., Yang, T., Zhu, X. & Chen, J. Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1. FEBS Lett. 583, 2171–2178 (2009).
Davidson, G. et al. Cell cycle control of wnt receptor activation. Dev. Cell 17, 788–799 (2009).
Miyagaki, H. et al. Overexpression of PFTK1 predicts resistance to chemotherapy in patients with oesophageal squamous cell carcinoma. Br. J. Cancer 106, 947–954 (2012).
Leung, W. K. et al. A novel interplay between oncogenic PFTK1 protein kinase and tumor suppressor TAGLN2 in the control of liver cancer cell motility. Oncogene 30, 4464–4475 (2011).
Li, S. M. et al. The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J. Exp. Clin. Cancer Res. 37, 219 (2018).
Wang, T. et al. FTO-stabilized lncRNA HOXC13-AS epigenetically upregulated FZD6 and activated Wnt/beta-catenin signaling to drive cervical cancer proliferation, invasion, and EMT. J. BUON 26, 1279–1291 (2021).
Baubec, T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015).
Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 (2018).
Jiang, M. et al. Downregulation of PFTK1 inhibits migration and invasion of non-small cell lung cancer. Onco Targets Ther. 13, 9281–9289 (2020).
Zhang, W. et al. PFTK1 regulates cell proliferation, migration and invasion in epithelial ovarian cancer. Int. J. Biol. Macromol. 85, 405–416 (2016).
Yan, R. et al. Inhibition of DCLK1 sensitizes resistant lung adenocarcinomas to EGFR-TKI through suppression of Wnt/beta-Catenin activity and cancer stemness. Cancer Lett. 531, 83–97 (2022).
Togashi, Y. et al. Inhibition of beta-Catenin enhances the anticancer effect of irreversible EGFR-TKI in EGFR-mutated non-small-cell lung cancer with a T790M mutation. J. Thorac. Oncol. 10, 93–101 (2015).
Yi, Y. et al. P21-activated kinase 2-mediated beta-catenin signaling promotes cancer stemness and osimertinib resistance in EGFR-mutant non-small-cell lung cancer. Oncogene 41, 4318–4329 (2022).
Davidson, G. & Niehrs, C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell Biol. 20, 453–460 (2010).
Zhang, M. et al. CDK14 inhibition reduces mammary stem cell activity and suppresses triple negative breast cancer progression. Cell Rep. 40, 111331 (2022).
Baylin, S. B. & Jones, P. A. Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol. 8 https://doi.org/10.1101/cshperspect.a019505 (2016).
Endo, Y. et al. Clinicopathological impacts of DNA methylation alterations on pancreatic ductal adenocarcinoma: prediction of early recurrence based on genome-wide DNA methylation profiling. J. Cancer Res. Clin. Oncol. 147, 1341–1354 (2021).
Kausar, S., Abbas, M. N. & Cui, H. A review on the DNA methyltransferase family of insects: aspect and prospects. Int. J. Biol. Macromol. 186, 289–302 (2021).
Rhee, I. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556 (2002).
So, J. Y. et al. Induction of DNMT3B by PGE2 and IL6 at distant metastatic sites promotes epigenetic modification and breast cancer colonization. Cancer Res. 80, 2612–2627 (2020).
Du, C. et al. Hsa_circRNA_102229 facilitates the progression of triple-negative breast cancer via regulating the miR-152-3p/PFTK1 pathway. J. Gene Med. 23, e3365 (2021).
Piao, H. Y. et al. Hypoxia associated lncRNA HYPAL promotes proliferation of gastric cancer as ceRNA by sponging miR-431-5p to upregulate CDK14. Gastric Cancer 25, 44–63 (2022).
Imawari, Y. et al. Downregulation of dual-specificity tyrosine-regulated kinase 2 promotes tumor cell proliferation and invasion by enhancing cyclin-dependent kinase 14 expression in breast cancer. Cancer Sci. 109, 363–372 (2018).
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81872450), the Natural Science Foundation of Guangdong Province (2022A1515012162), the Science and Technology Program of Guangzhou (202102010086, 202201010839), and the plan on enhancing scientific research in GMU.
Author information
Authors and Affiliations
Contributions
X.J. and K.L. designed and wrote the manuscript, and Z.H. and D.L. revised it. J.T., P.C., and J.D. performed all the experiments. L.L., D.C., and J.Z. did the bioinformatics analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Valentina Masciale and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Silvia Belluti and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jia, X., Tian, J., Chen, P. et al. Methylation-modulated PFTK1 regulates gefitinib resistance via Wnt/β-catenin signaling in EGFR mutant non-small-cell lung cancer cells. Commun Biol 7, 1649 (2024). https://doi.org/10.1038/s42003-024-07339-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07339-3
This article is cited by
-
IRX3-CDK14 axis promotes glioblastoma progression by regulating LRP6-mediated canonical Wnt/β-catenin pathway
Cell Death & Disease (2025)