Abstract
Arginine-rich dipeptide repeat proteins (R-DPRs) are highly toxic proteins found in patients with C9orf72-linked amyotrophic lateral sclerosis and frontotemporal dementia (C9-ALS/FTD). R-DPRs can cause toxicity by disrupting the natural phase behavior of RNA-binding proteins (RBPs). Mitigating this abnormal phase behavior is, therefore, crucial to reduce R-DPR-induced toxicity. Here, we use FUS as a model RBP to investigate the mechanism of R-DPR-induced aberrant RBP phase transition. We find that this phase transition can be mitigated by Kapβ2. However, as a nuclear import receptor and phase modifier for PY-NLS-containing RBPs, the function of WT Kapβ2 could lead to undesired interaction with its native substrates when used as therapeutics for C9-ALS/FTD. To address this issue, it is crucial to devise effective strategies that allow Kapβ2 to selectively target its binding to the R-DPRs, instead of the RBPs. We show that NLS-binding deficient Kapβ2W460A:W730A can indeed selectively interact with R-DPRs in FUS assembly without affecting normal FUS phase separation. Importantly, Kapβ2W460A:W730A prevents enrichment of poly(GR) in stress granules and mitigates R-DPR neurotoxicity. Thus, NLS-binding deficient Kapβ2 may be implemented as a potential therapeutic for C9-ALS/FTD.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are two neurodegenerative diseases which are considered to belong to a common spectrum. Pathologically they are both characterized by the cytoplasmic mislocalization and aggregation of RNA-binding proteins (RBP) with low complexity domains (LCDs), including TAR DNA-binding protein of 43 kDa (TDP-43) and Fused in sarcoma (FUS)1. The most common genetic cause of ALS and FTD is an aberrant G4C2 (GGGGCC) hexanucleotide repeat expansion present in the first intron of the C9ORF72 (C9) gene2,3. RNA transcripts containing the sense and antisense expanded sequence are translated through repeat-associated non-AUG translation (RAN-T), resulting in 5 different dipeptide repeat proteins (DPRs; poly-GP, poly-GA, poly-GR, poly-PA, and poly-PR)4. Accumulation of DPRs is one of the three non-mutually exclusive mechanisms proposed for C9-mediated toxicity, besides loss-of-function due to reduced expression of the C9orf72 protein and gain-of-function of toxic C9-RNA repeat foci5. All five DPRs are found in aggregates in C9 patients, whereas arginine-containing poly-GR and poly-PR peptides (hereafter referred to as R-DPRs) are the most toxic DPRs based on studies in different cellular and animal models4,6,7,8,9,10. Therefore, it is important to develop therapeutic strategies to mitigate R-DPR-induced neuronal toxicity.
One mechanism through which R-DPRs induce toxicity is by engaging LCD-containing RBPs to disrupt their normal phase behavior. For example, R-DPRs can promote liquid-liquid phase separation (LLPS) of LCD-containing RBPs, including FUS11, TIA112, NPM113, and TDP-4314,15, and thus disturb the dynamics and functions of the membrane-less organelles formed by these RBPs, such as stress granules (SGs) and nucleoli11,12,13. Another mechanism for R-DPR-induced toxicity is through induction of nucleocytoplasmic transport defects, which could result from (1) R-DPRs being phase modifiers of the FG-Nups in the nuclear pore, (2) sequestration of the nuclear transport machinery and cargo through direct binding to R-DPRs or aberrantly formed SGs, or (3) both15,16,17. It is proposed that the combined effect of dysregulated nucleocytoplasmic transport and RBP phase behavior leads to the mislocalization and co-aggregation of TDP-43 with R-DPRs, which is a key pathological feature in C9-ALS/FTD4,14. However, the molecular mechanism by which R-DPRs induce LLPS and aggregation of TDP-43 and other RBPs is unclear.
Because aberrant aggregation and phase transition of RBPs is a hallmark of ALS and FTD, an attractive therapeutic approach would be to mitigate these transitions. Indeed, restoring the functional phase of an RBP by nuclear import receptors (NIRs) or bait RNA oligonucleotides can rescue the toxicity caused by RBP aggregates18,19,20,21,22. For example, Kapβ2, a NIR of PY-NLS-containing proteins, can mitigate FUS aggregation and associated toxicity. Therefore, we hypothesize that mitigating aberrant RBP phases induced by R-DPRs could also rescue R-DPR toxicity. Interestingly, a large subset of the R-DPR interactome consists of RBPs with nuclear localization signals (NLSs), which also represent cargos of nuclear import receptors4,12,23. Moreover, another large subset of the R-DPR interactome is nuclear import receptors4,12,23. In addition, several genetic screens have identified nuclear import receptors as modifiers of R-DPR toxicity12,24,25,26. Indeed, we recently demonstrated that the nuclear import receptor Kapβ2 can rescue poly(GR)-induced neurotoxicity in CNS tissue27, suggesting that nuclear import receptors are prime candidates to mitigate aberrant RBP phase transitions caused by R-DPRs. However, the mechanism underlying these rescue activities of Kapβ2 against poly(GR)-induced neurotoxicity remains to be elucidated. In addition to its role as a nuclear import receptor, wild type (WT) Kapβ2 is a potent phase modifier of FUS and other PY-NLS-containing RBPs. However, this could lead to unintended interactions with its physiological substrates when developed as therapeutics for C9-ALS/FTD. Therefore, it is essential to develop Kapβ2 variants that do not interact with native substrates of Kapβ2. These variants have the potential to be used as therapeutics for C9-ALS.
In this study, using a model RBP (i.e., FUS), whose LLPS and aggregation can be induced and enhanced by R-DPR, we investigated the mechanism of R-DPR-induced RBP aggregation and LLPS. We aimed to understand the mechanism behind the R-DPR-induced aberrant RBP phase transition and develop strategies to mitigate the aberrant phase transition caused by R-DPRs. Our results show that Kapβ2 potently prevents and reverses R-DPRs-induced aggregation of FUS and TDP-43. However, due to its localization to SGs through binding to SG proteins such as FUS, Kapβ2 does not prevent R-DPR’s interaction with SGs. To develop R-DPRs specific chaperone, we demonstrated that NLS-binding-deficient Kapβ2W460A:W730A selectively extracts R-DPRs from FUS assemblies without affecting normal FUS LLPS in vitro, and prevents recruitment of poly(GR) in SGs in cells. More importantly, Kapβ2W460A:W730A counteracts neuronal toxicity caused by poly(GR). Based on our findings, Kapβ2 lacking NLS-binding has the potential to be utilized as a therapeutic option for the treatment of C9-ALS/FTD. Notably, this alternative approach would avoid any unintended interference with the native transport cargoes of WT Kapβ2 in cells.
Results
R-DPRs enhance aggregation and liquid-liquid phase separation of FUS
Previous studies have shown that R-DPRs can enhance the phase separation of the PrLD (Prion-like domain) of FUS11. To understand how R-DPRs affect the aggregation and LLPS of full-length FUS, we added R-DPRs into assays that favor formation of FUS fibrils (Fig. 1A–E) or FUS liquid droplets (Fig. 1F)18. To promote fibril formation, 5 µM of GST-tagged FUS (GST-TEV-FUS) was incubated with 1.6 µg TEV protease, which cleaved off the GST, eliciting rapid assembly of fibrils that are resistant to SDS treatment at a concentration of 0.01% or Sarkosyl treatment at a concentration of 0.04% (Supplementary Fig. 1A) and bear close ultrastructural resemblance to those that accumulate in disease (Fig. 1A)18,28. Under these conditions, fibrillization is the dominant pathway and is detectable as an increase in turbidity after a lag time. The addition of 10 µM of (GR)20 or (PR)20 reduced the lag time before aggregation and increased turbidity at the end of the aggregation, while the arginine-free DPR (GA)20 had no effect (Fig. 1B). Moreover, in the presence of (GR)20 or (PR)20, the size of FUS aggregates was significantly increased (Fig. 1C, D). We found that the concentration of R-DPRs is critical in promoting aggregation. For example, for 5 μM FUS, 5 μM or 10 μM, but not 1 μM R-DPRs, enhanced its aggregation (Fig. 1Band Supplementary Fig. 1B). Moreover, the effect of R-DPRs also depends on their length. For example, the addition of (GR)8 or (PR)8 also shortens the lag time of FUS aggregation, but to a lesser extent as compared to (GR)20 or (PR)20 (Fig. 1Eand Supplementary Fig. 1C). Thus, poly(GR) or poly(PR), but not poly(GA), promotes full length FUS aggregation.
A Schematic of the fibrillization and imaging assay. At 0 h, GST-TEV-FUS was incubated with 10 µM DPRs (i.e., (GA)20, (GR)20, or (PR)20) in the presence of TEV protease. Turbidity measurement at 395 nm was used to assess fibrillization. At the endpoint, 20 nM of TAMRA-(GR)20 or TAMRA-(PR)20 were added for visualization and image quantification purpose. Images were taken in both the DIC channel and the fluorescence channel. B GST-TEV-FUS (5 μM) was incubated with buffer, 10 μM of (GA)20, (GR)20, or (PR)20, in the absence or presence of TEV protease (16 μg/ml). Aggregation was assessed by turbidity measured at 395 nm. Solid lines are normalized mean. Dotted lines of corresponding colors represent ± SEM (n = 3 independent experiments). C Representative images of FUS aggregates formed in the presence or absence of the indicated R-DPR at T = 0 and T = 3 h of aggregation. Samples were supplemented with 20 nM of TAMRA-(GR)20 or TAMRA-(PR)20 before imaging for visualization and quantification. Scale bars, 5 μm. D Quantification of the images collected in (C) showing the integrated area of FUS aggregates. Data shown are mean ± SEM. N = 10 images per condition. An unpaired Student’s t-tests were used to compare different conditions, ****p ≤ 0.0001. E GST-TEV-FUS (5 μM) was incubated with different concentrations (1 μM, 5 μM, and 10 μM) of (GR)8 or (PR)8 in the absence or presence of TEV (16 μg/ml). Turbidity at 395 nm was used to assess aggregation. Solid lines are normalized mean. Data shown are mean ± SEM (n = 3 independent experiments). F Representative images of FUS droplets acquired using DIC and fluorescence microscopy. GST-TEV-FUS (10 μM) was incubated with buffer or 10 μM (GR)20 or (PR)20 in the absence of TEV protease for 3 h. Samples were supplemented with 20 nM of TAMRA-(GR)20 or (TAMRA)-(PR)20 for quantification of the fluorescence channel. Scale bars, 5 μm. G Quantification of the fluorescence images collected in (F) showing the distribution of FUS droplet size. Number of droplets quantified in each condition (n) is indicated in the figure. The bar represents the average droplet size ± SEM. An unpaired Student’s t-tests were used to compare different conditions, ****p ≤ 0.0001. H Quantification of the fluorescence images collected in (F) showing the integrated area of the FUS droplets in each condition. Data shown are mean ± SEM. N = 10 images per condition. An unpaired Student’s t-tests were used to compare different conditions; *p ≤ 0.05, **p ≤ 0.01. I Change of TAMRA anisotropy when 100 nM TAMRA-(GR)20 or TAMRA-(PR)20 binds to increasing concentrations of GST-TEV-FUS (0 μM to 7 μM) in assembly buffer (50 mM Tris-HCl pH 8, 20 mM Trehalose, 1 mM DTT, and 20 mM glutathione). Values represent means ± SEM (n = 4 (TAMRA-(GR)20), and 6 (TAMRA-(PR)20) independent experiments). Binding curves were fitted by Prism. The solid line represents the fit, and the fitted Kd was reported in the figure.
Interestingly, immediately upon the addition of (GR)20 or (PR)20, before the formation of FUS aggregates, we observed an increase in turbidity and formation of FUS assemblies that have similar morphology to FUS liquid droplets (T = 0 in Fig. 1B, C), suggesting that R-DPRs might promote FUS LLPS. To test whether R-DPRs promote FUS LLPS, we initiated FUS self-assembly under conditions that favor the formation of FUS liquid droplets by omitting TEV protease and increasing FUS concentration18. While (GR)20 and (PR)20 did not form liquid droplets under this buffer condition (Supplementary Fig. 1D), the R-DPRs significantly enhanced FUS LLPS, evidenced by the increased droplet size and overall phase-separated area (Fig. 1F–H). To rule out the possible effect of the GST tag, we also performed the experiment using MBP-tagged FUS-GFP, where the formation of FUS droplet was initiated by cleavage of the MBP tag29 (Supplementary Fig. 1E). Similarly, (GR)20 and (PR)20 induced an increase in droplet size and integrated phase-separated area of FUS-GFP (Supplementary Fig. 1E–G). Moreover, these FUS droplets are enriched with heterogeneously distributed R-DPRs (Supplementary Fig. 1E). In contrast, the distribution of FUS-GFP signal inside FUS droplet is homogeneous (Supplementary Fig. 1E). Interestingly, FRAP analysis of FUS droplets indicates that the presence of (GR)20 does not change the dynamics of FUS-GFP inside the droplet (Supplementary Fig. 1H-I). However, the phase formed by (GR)20 is less dynamic than FUS-GFP in the droplets, indicated by a longer half-time of recovery (Supplementary Fig. 1J, K). Anisotropy assays showed that both R-DPRs bind strongly to FUS with similar affinity (Fig. 1I). Therefore, our results show that (GR)20 and (PR)20 enhance the LLPS and aggregation of FUS by tightly binding to FUS, similar to R-DPRs binding induced LLPS of other RBPs that are stress granule components12.
R-DPRs engage the N-terminal PrLD to promote FUS LLPS and the C-terminal RGGs to promote FUS aggregation
To better understand how R-DPRs promote the aggregation or LLPS of FUS, we generated and tested a series of FUS deletion mutants (Fig. 2A). These mutants include the FUS PrLD (FUS1-214), FUS PrLD-RRM with the C-terminal RGGs deleted (FUS1-371), FUS ΔPrLD (FUS215-526), and FUS RRM-RGGs (FUS267-526). Consistent with previous results that interaction between N-terminal PrLD and C-terminal RGGs is important for FUS assembly28, these FUS variants did not assemble into either aggregates or liquid droplets even 100 minutes after cleaving the GST tag (Fig. 2B–E).
A A diagram illustrating the domain structure of WT FUS and truncation constructs used in this study. (B-E) Top: 5 μM GST-TEV-FUS1-371 (B), GST-TEV-FUS1-214 (C), GST-TEV-FUS215-526 (D), or GST-TEV-FUS267-526 (E), was incubated with buffer, 10 μM (GR)20, or (PR)20, in the absence or presence of TEV protease (16 μg/ml). The extent of aggregation was determined by turbidity measured at 395 nm. Solid lines are the average of raw turbidity data subtracted background turbidity of buffer. Dotted lines of corresponding colors represent ± SEM (n = 3-4 independent experiments). Bottom, the corresponding DIC and fluorescence microscopy images taken at the end of the reaction. R-DPR containing samples were supplemented with 20 nM of TAMRA-(GR)20 or (TAMRA)-(PR)20 for quantification of the fluorescence channel. Scale bars, 5 μm. F, G Quantification of the fluorescence images collected in (B–E, bottom) in the presence of R-DPRs showing the distribution of FUS assembly size (F), or circularity (G). The bar represents average ± SEM. The number of droplets quantified in each condition (n) is indicated in (G).
Interestingly, in the presence of R-DPRs, the N-terminal and C-terminal fragments of FUS show different assembly behavior. For the N-terminal fragments lacking the two C-terminal RGG domains (i.e., FUS1-214 and FUS1-371), the addition of (GR)20 and (PR)20 induces an immediate increase in turbidity (Fig. 2B, C, top). However, no further aggregation is observed, even at 100 minutes (Fig. 2B, C, top). When the FUS assembly was observed under the microscope, only small round condensates were present, indicating the formation of liquid droplets induced by R-DPRs (Fig. 2B-bottom, C-bottom, F, and G). On the other hand, for the C-terminal fragments (i.e., FUS215-526 and FUS267-526), the addition of (GR)20 and (PR)20 promoted aggregation by shortening the lag time and increasing the final turbidity of the aggregation (Fig. 2D, E). Imaging showed larger and more irregularly shaped FUS assemblies with low circularity in the presence of (GR)20 and (PR)20, indicating that R-DPRs induced these FUS constructs to form aggregates instead of liquid droplets (Fig. 2D–G). To determine whether R-DPRs can promote LLPS of C-terminal fragments in other conditions, we also imaged FUS C-terminal fragments in the presence of R-DPRs under droplet-forming conditions (i.e., without TEV), but did not observe any assembly, even after three hours (Supplementary Fig. 2A, B). To determine whether co-purified RNA (Supplementary Table 1) mediates the interaction between R-DPRs and FUS C-terminal fragments, we added RNase A into the assay and observed that even without RNA, R-DPRs promoted formation of FUS215-526 and FUS267-526 aggregates, although smaller than the aggregates formed without RNase A (Supplementary Fig. 2C). Therefore, these results indicate that R-DPRs interact with the N-terminal PrLD to promote FUS LLPS and the C-terminal RGGs-containing domains to promote FUS aggregation.
Next, we investigated whether the different effects of R-DPRs on the assembly of N-terminal and C-terminal FUS fragments were a result of different binding affinity. We used fluorescence anisotropy to measure the binding affinity of (GR)20 and (PR)20 to different FUS fragments. Our results showed that binding affinity of FUS fragments to R-DPRs seems to correlate with the length of the FUS fragment as shorter fragments show weaker binding (higher Kd) (Supplementary Fig. 2D–G). However, the effect of the R-DPRs on FUS assemblies does not correlate with their binding affinity. For example, both FUS1-371 and FUS215-526 bind R-DPRs with affinity in the 50-65 nM range (Supplementary Fig. 2E, F) but have different effects in promoting FUS assembly. While promoting LLPS but not aggregation of FUS1-371 (Fig. 2B), R-DPRs enhanced aggregation of FUS215-526 (Fig. 2D). Therefore, differences in binding affinity between R-DPRs and FUS regions cannot account for the disparate impact that R-DPRs have on FUS LLPS and aggregation.
Kapβ2 prevents and reverses the aggregation and liquid-liquid phase separation of wild-type FUS in the presence of arginine-rich dipeptide repeats
R-DPR-induced aberrant phase transition and aggregation of RBPs was proposed to be a mechanism of pathogenesis for C9-ALS/FTD4. Therefore, strategies that mitigate R-DPR-induced aberrant phase transition and aggregation of RBPs could rescue R-DPR toxicity. We and others have shown that nuclear import receptors function as potent chaperones and disaggregases to prevent and reverse LLPS and aggregation of the RBPs with the corresponding nuclear localization signal (NLS)18,19,20,21,22. Specifically, Kapβ2 (TNPO1) mitigates LLPS and aggregation of FUS by engaging the PY-NLS at its C-terminus. Therefore, we tested whether Kapβ2 mitigates FUS aggregation in the presence of R-DPRs.
For WT FUS, the addition of Kapβ2 effectively inhibited aggregation, even in the presence of (GR)20 or (PR)20 (Fig. 3A, B). Sedimentation analysis also confirmed the activity of Kapβ2 in preventing the self-assembly of FUS and R-DPRs (Supplementary Fig. 3A–D). In the absence of Kapβ2, after TEV cleavage, both FUS and R-DPRs are assembled into aggregates, which accumulate in the pellet fraction (Supplementary Fig. 3A–D). Addition of Kapβ2 leads to FUS and R-DPRs being retained in the supernatant fraction (Supplementary Fig. 3A–D). To investigate whether Kapβ2 can disassemble preformed co-aggregates of FUS and R-DPRs, we initiated FUS aggregation in the presence of (GR)20 or (PR)20, and then added Kapβ2 after aggregation had reached plateau. When added to pre-formed aggregates, Kapβ2 potently reversed FUS aggregation and reduced turbidity to baseline level (Fig. 3C, D).
A Schematic of the fibrillization inhibition reaction. At 0 h, GST-TEV-FUS fibrillization was initiated with or without R-DPRs, in the presence or absence of WT Kapβ2. Turbidity measurement was used to assess fibrillization. B GST-TEV-FUS (5 μM) fibrillization was initiated by adding TEV protease (16 μg/ml). The fibrilization reaction was incubated with 10 μM (GR)20 or (PR)20 in the presence or absence of 5 μM WT Kapβ2. Aggregation was assessed by turbidity measured at 395 nm. Solid lines are normalized mean. Dotted lines of corresponding colors represent ± SEM (n = 3-4 independent experiments). C Schematic of the fibrillization disaggregation reaction. GST-TEV-FUS was incubated with R-DPRs in the presence of TEV protease, and turbidity was measured to assess fibrillization. After 100 mins of aggregation, an equal volume of WT Kapβ2 or buffer was added, and turbidity was continually measured. D GST-TEV-FUS (5 μM) fibrillization was initiated as described in (C). After 100 mins, an equal volume of WT Kapβ2 (5 μM) or buffer was added, and turbidity was continually measured at 395 nm. Solid lines are normalized mean. Dotted lines of corresponding colors represent ± SEM (n = 3-4 independent experiments). E FUS-GFP LLPS was initiated by adding 3 C protease (18 μg/ml) into MBP-FUS-GFP (3 μM) incubated with buffer (50 mM KCl, 50 mM Tris pH 7.4, 0.5% Glycerol, and 1 mM DTT) or 3 μM (GR)20 or (PR)20, in the presence or absence of 6 μM WT Kapβ2. All R-DPRs-containing samples were supplemented with 100 nM labeled TAMRA-(GR)20 or TAMRA-(PR)20. Droplets were visualized after 2 hours of incubation. Scale bars, 5 μm. The figure on the right quantifies the fluorescence signal of the GFP channels and shows the integrated area of FUS droplets in each condition. Data shown are mean ± SEM. The number of images quantified in each condition (n) is indicated in the figure. An unpaired Student’s t-tests were used to compare different conditions; ****p ≤ 0.0001. F FUS-GFP LLPS was initiated as in (E). At 2 hours, 6 µM WT Kapβ2 was added to preformed FUS-GFP droplets, and the samples were imaged 2 hours later. All R-DPRs-containing samples were supplemented with 100 nM TAMRA-(GR)20, or TAMRA-(PR)20. Scale bars, 5 μm. The figure on the right quantifies the fluorescence signal of the GFP channels and shows the integrated area of FUS droplets in each condition. Data shown are mean ± SEM. The number of images quantified in each condition (n) is indicated in the figure. An unpaired Student’s t-tests were used to compare different conditions; ****p ≤ 0.0001.
Although co-aggregation of FUS and R-DPRs has not been observed in ALS/FTD patients, expression of R-DPRs can disrupt the dynamics of membrane-less organelles such as stress granules, which contain phase-separated FUS12. Therefore, we next investigated whether Kapβ2 maintains its activity in regulating FUS phase behavior in the presence of R-DPRs. Kapβ2 completely inhibited the formation of FUS droplets in vitro both in the presence and absence of R-DPRs, as indicated by the significant reduction of phase-separated droplet area (Fig. 3E).
Next, we tested whether Kapβ2 can disassemble FUS/R-DPR co-condensates after R-DPRs had induced an aberrant FUS phase transition. Remarkably, the addition of Kapβ2 completely disassembled FUS/R-DPR co-droplets and restored both FUS and R-DPR to a diffusive state, indicated by reduced droplet area (Fig. 3F). Therefore, addition of Kapβ2 prevents and reverses aberrant FUS phase transition induced by R-DPR. It is unclear whether Kapβ2 dissolves R-DPR or FUS first. To address this experimentally, we imaged the disassembly of FUS/R-DPR droplets at various time points. At the initial time point (T = 10 minutes), we observed that some of the FUS phase was dissolved first (Supplementary Fig. 3E, F). Interestingly, the portion of the FUS phase that remained undissolved at 10 minutes colocalized with the R-DPR phase (Supplementary Fig. 3E, F). This suggests that FUS interacts with R-DPR in the secondary aberrant phase, disrupting normal FUS liquid-liquid phase separation (LLPS). We propose that in the presence of R-DPR, FUS-R-DPR interactions facilitate the formation of this secondary aberrant phase, which may act as a scaffold to recruit additional FUS protein. Consequently, when Kapβ2 is added, the recruited FUS is dissolved first, while the FUS that directly interacts with R-DPR in the secondary phase is dissolved later, at 2 hours, along with the R-DPRs (Fig. 3F).
Since impaired dynamics of membrane-less organelles, such as stress granules, has been proposed to be one of the mechanisms for R-DPR toxicity30, we next tested if Kapβ2 can prevent (GR)20 from entering the stress granules (SGs) to impair their dynamics. To this end, we employed a SG recruitment assay in semi-permeabilized HeLa cells31. To our surprise, Kapβ2 did not prevent recruitment of (GR)20 into SGs (Supplementary Fig. 3G). We reasoned it is because Kapβ2 itself is recruited into the SGs via binding to SG components such as FUS30. Indeed, immunofluorescence of Kapβ2 showed SG localization (Supplementary Fig. 3H). Therefore, to eliminate this unwanted interaction of Kapβ2 with its native substrates in cells, it is important to develop Kapβ2 variants that specifically engage R-DPR only. To do this, we investigated the mechanism and interaction of Kapβ2 with FUS and R-DPRs.
Kapβ2’s interaction with FUS and R-DPRs is tunable
The function of Kapβ2 as a phase modifier of FUS depends on its interaction with the PY-NLS18,20. To understand if this mechanism is preserved in the presence of R-DPRs, we tested Kapβ2 against FUSP525L which has a mutation in PY-NLS that disrupts Kapβ2 binding32,33. In the absence of R-DPRs, the activity of Kapβ2 in preventing FUSP525L aggregation was reduced (Fig. 4A, B), but not eliminated, with ~70% of FUSP525L remaining soluble (Fig. 4C, D, Supplementary Fig. 4A). Surprisingly, in the presence of (GR)20 or (PR)20, Kapβ2 is completely unable to inhibit FUSP525L aggregation (Fig. 4A, B), as almost 100% of FUSP525L was assembled in the pellet fraction (Fig. 4C, D). Instead, Kapβ2 inhibits the co-aggregation of R-DPRs into FUSP525L aggregates and chaperones R-DPRs, as ~85% of (GR)20 and (PR)20 remained in the soluble fraction (Fig. 4C, E). Similar results were observed when Kapβ2 was added to preformed R-DPR/FUSP525L co-aggregates, where Kapβ2 brought most (~80%) of R-DPR into the soluble fraction without reducing the portion of FUSP525L in the aggregated fraction (Supplementary Fig. 4B–D). These results indicate that in the presence of R-DPRs, Kapβ2 activity toward FUSP525L aggregation is further diminished, which could be explained by the combined impact of the PY-NLS mutation and the competitive binding of R-DPRs to Kapβ2. Indeed, anisotropy assays revealed that Kapβ2 binds to (GR)20 and (PR)20 with similarly strong binding affinity compared to its binding to WT FUS (Supplementary Fig. 4E–G).
A, B GST-TEV-FUSP525L (5 μM) fibrillization was initiated by adding TEV protease (16 μg/ml). Fibrilization reaction was incubated with 10 μM (GR)20 (A), or 10 μM (PR)20 (B), in the presence or absence of 5 μM WT Kapβ2. Aggregation was assessed by turbidity at 395 nm. Solid lines are normalized mean. Dotted lines of corresponding colors represent ± SEM (n = 3 independent experiments). C GST-TEV-FUSP525L (5 μM) fibrillization was initiated by adding TEV protease (16 μg/ml). Fibrilization reaction was incubated with buffer, 10 μM (GR)20, or 10 μM (PR)20, in the presence or absence of 5 μM WT Kapβ2. After 100 min of incubation, fibrillization was assessed by sedimentation assay. Pellet and supernatant fractions were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. A representative gel is shown. Quantification of the sedimentation gels in (C). The amount of FUS P525L (D), and R-DPR (E) in the pellet fraction was determined by densitometry. Values represent means ± SEM (n = 4 independent experiments). Unpaired Student’s t-tests were used to compare different conditions; ****p ≤ 0.0001. F FUS-GFP (3 μM) liquid droplets were formed in LLPS buffer (50 mM KCl, 50 mM Tris pH 7.4, 0.5% Glycerol, and 1 mM DTT) in the presence of 12 μM (GR)20. At 2 hours, 3 µM WT Kapβ2 was added, and the sample was then imaged continuously for 135 seconds. Representative images were shown. 100 nM TAMRA-(GR)20 was added for visualization of (GR)20. Scale bars, 5 μm. See also Supplementary Movie 1-3. G Quantification of images collected in (F) showing the changes in the enrichment factor (ratio of mean fluorescence intensity in liquid droplet to the mean fluorescence intensity in bulk) of FUS-GFP and TAMRA-(GR)20 after addition of Kapβ2. Values represent means ± SEM. 31 droplets were quantified. Change of anisotropy when 100 nM FUS-GFP (H), 100 nM TAMRA-(GR)20 (I), or 100 nM TAMRA-(PR)20 (J) binds to increasing concentrations of WT Kapβ2 or Kapβ2W460A:W730A (0 μM to 7 μM) in assembly buffer (50 mM Tris-HCl pH 8, 20 mM Trehalose, 1 mM DTT, and 20 mM glutathione). Values represent means ± SEM (n = 3-5 independent experiments). Binding curves were fitted by Prism. Solid line represents the fit and the fitted Kd was reported on the figure.
The further loss of Kapβ2 activity toward PY-NLS mutant FUS in the presence of R-DPRs is intriguing and indicates that R-DPRs influence Kapβ2 interaction with its native substrate by acting as a competitive binder. We hypothesized that by tuning the interactions between Kapβ2, R-DPRs, and WT FUS to favor Kapβ2 binding to R-DPRs, one could tune the activity of Kapβ2 to selectively engage R-DPRs in FUS/R-DPR droplets without affecting FUS LLPS. Indeed, when (GR)20 concentration was increased to 4-fold that of WT FUS in FUS/R-DPR droplets, which should promote Kapβ2:(GR)20 interaction, the addition of Kapβ2 selectively extracted (GR)20 from preformed FUS/(GR)20 droplets, whereas FUS droplets remain intact throughout the assay and the enrichment of FUS inside the droplet was not affected by the addition of Kapβ2 (Fig. 4F–G, Supplementary Movies 1–3). This selective activity is useful, as it is advantageous to preserve the functional phase separation of RBPs while depleting R-DPRs from these condensates in diseased conditions. Therefore, we next sought to achieve R-DPR-selective activity by designing a Kapβ2 variant that favors binding to R-DPRs but has a weakened binding affinity for FUS.
Kapβ2W460A:W730A shows enhanced R-DPR binding
To identify a Kapβ2 variant that can chaperon R-DPRs without interacting with FUS, we utilized the GalaxyPepDock docking server to identify the interacting surface between Kapβ2 and R-DPRs34. GalaxyPepDock predicts the 3D structure of protein-peptide complexes by combining information from structural databases and energy-based optimization34. Docking revealed that the binding pocket of poly(GR) and poly(PR) partially overlaps with the PY-NLS binding surface in Kapβ2 (Supplementary Fig. 5A–E), similar to a previous docking study26. This finding is also consistent with NMR data indicating that the poly(PR) binding site on Kapβ2 partially overlaps with the region of Kapβ2 that is responsible for PY-NLS recognition35. It is worth noting that the PY-NLS binding pocket can only accommodate a short stretch of GR/PR repeats (Supplementary Fig. 5C–E). For longer repeats, interactions with other regions of Kapβ2, such as the outer surface, should also be considered36. In addition, the unstructured acidic loop of Kapβ2, which is not included in the crystal structure (PDB ID: 4FDD) used for docking, could interact with RG-rich sequence31. Indeed, when we repeated the docking with an AlphaFold generated structure of full-length Kapβ2 (ID: AF-Q92973-F1), while both R-DPRs still docked into the PY-NLS binding pocket with slightly different orientation (Supplementary Fig. 5F–H), multiple interactions with the acidic loop were observed (Supplementary Fig. 5I–K). Interestingly, although both poly(GR) and poly(PR) occupy the PY-NLS-binding pocket, they engage different residues than those involved in FUS PY-NLS binding, making it possible to design mutations that specifically interrupt PY-NLS but not R-DPR binding. For example, Kapβ2W460A:W730A is a PY-NLS binding deficient mutant that is also inactive in chaperoning and transporting FUS (Supplementary Fig. 6A)18,37. Binding energy calculated using the docked structure of Kapβ2W460A:W730A:PY-NLS showed a 10% increase in binding energy compared to WT Kapβ2:PY-NLS (Supplementary Table 2). In contrast, binding energy only increased by 2% for poly(PR) and 5% for poly(GR) when Kapβ2W460A:W730A was used for docking instead of WT Kapβ2 (Supplementary Table 2). A smaller change in binding energy for the R-DPRs indicates that the W460A:W730A mutations disrupt the PY-NLS binding more than R-DPR binding38. We then measured the binding affinity between R-DPRs and Kapβ2W460A:W730A, as well as FUS and Kapβ2W460A:W730A. Interestingly, while FUS showed a reduced binding affinity for Kapβ2W460A:W730A compared to WT Kapβ2, both (GR)20 and (PR)20 showed enhanced binding affinity to Kapβ2W460A:W730A compared to WT Kapβ2 (Fig. 4H–J). As a result, while FUS and R-DPRs bind WT Kapβ2 with similar affinities, the affinity of R-DPRs for Kapβ2W460A:W730A is ~1.5 times that of FUS for Kapβ2W460A:W730A.
Kapβ2W460A:W730A preferentially removes R-DPR from co-aggregates and co-condensates of FUS/R-DPR
Next, we tested whether Kapβ2W460A:W730A exhibits selective binding activity toward R-DPRs in assemblies that have both FUS and R-DPRs. We first used a sedimentation assay to monitor the assembly of R-DPRs and FUS in the presence of Kapβ2W460A:W730A. Unlike WT Kapβ2, which retains both R-DPRs and FUS in the soluble fraction in this assay (Supplementary Fig. 3A–D), Kapβ2W460A:W730A selectively retains R-DPRs in the soluble fraction, whereas FUS remained in the pellet (Fig. 5A–D, Supplementary Fig. 6B–C). In addition, Kapβ2W460A:W730A remains in the soluble fraction with R-DPR, rather than binding to FUS in the pellet, confirming its selectivity for R-DPR (Fig. 5A, B).
A, B GST-TEV-FUS (5 μM) fibrillization was initiated by adding TEV protease (16 μg/ml). Fibrilization reactions were incubated with buffer, 10 μM (GR)20 (A), or 10 μM (PR)20 (B), in the presence or absence of 5 μM Kapβ2W460A:W730A. After 100 minutes of incubation, fibrillization was assessed by sedimentation assay. Pellet and supernatant fractions were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. A representative gel is shown. Quantification of the gel images in (A) and (B). The amount of FUS (C), or R-DPR (D) in the pellet fraction was determined by densitometry. Values represent means ± SEM (n = 3 independent experiments). Unpaired Student’s t-tests were used to compare different conditions. E FUS-GFP LLPS was initiated by adding 3 C protease (18 μg/ml) into MBP-FUS-GFP (3 μM) incubated with buffer (150 mM KCl, 50 mM Tris pH 7.4, 0.5% Glycerol, and 1 mM DTT), 3 μM (GR)20, or 3 μM (PR)20, in the presence or absence of 6 μM indicated Kapβ2 variant. All R-DPRs containing samples were supplemented with 100 nM TAMRA-R-DPR. Droplets were visualized after 2 hours of incubation. Scale bars, 5 μm. F Quantification of the GFP channel of the fluorescence images collected in (E) shows the integrated area of FUS droplets in each condition. Data shown are mean ± SEM. n = 29 images per condition. Unpaired Student’s t-tests were used to compare different conditions. (G) Quantification of the GFP channel of the fluorescence images collected in (E) shows the size of the droplets in each condition. Data shown are mean ± SEM. The number of droplets (n) quantified in each condition is indicated in the figure. Conditions with WT Kapβ2 were N/A because there were not enough droplets to be quantified. (H) Quantification of images collected in (E) showing the enrichment factor (ratio of mean fluorescence intensity in liquid droplets to the mean fluorescence intensity in bulk) of FUS-GFP and TAMRA-(GR)20 in each droplet. Data shown are mean ± SEM. The number of images (n) quantified in each condition is indicated in the figure. Conditions with WT Kapβ2 were N/A because there were not enough droplets to be quantified. Unpaired Student’s t-tests were used to compare different conditions; ****p ≤ 0.0001.
We then tested the selective activity of Kapβ2W460A:W730A in preventing R-DPRs from forming FUS/R-DPRs co-condensates. Compared to the phase separation condition used in Fig. 3E, we increased the salt concentration from 50 mM to 150 mM to reduce non-specific interactions between proteins. Indeed, under this physiological salt concentration, the Kd of Kapβ2W460A:W730A to FUS-GFP (124.4±58.5 nM) shows a greater decrease compared to the Kd of Kapβ2 WT to FUS-GFP (39.6±17.7 nM) (Supplementary Fig. 6D), while both (GR)20 and (PR)20 showed enhanced binding affinity to Kapβ2W460A:W730A compared to WT Kapβ2 (Supplementary Fig. 6E, F). As a result, under physiological salt conditions, the affinity of R-DPRs for Kapβ2W460A:W730A is ~3 times that of FUS for Kapβ2W460A:W730A. The decrease in binding affinity for FUS:Kapβ2W460A:W730A is still less pronounced compared to the over 100-fold drop in hnRNP D PY-NLS binding when W460A or W730A mutation was introduced in Kapβ237, possibly due to the remaining interaction provided by the Kapβ2 acidic loop to the RGG rich region in full-length FUS31. Similar to our observation in 50 mM NaCl (Fig. 3E), in the presence of WT Kapβ2, co-condensation of FUS and R-DPRs was completely inhibited, and FUS could not form droplets (Fig. 5E). In contrast, when Kapβ2W460A:W730A was added to FUS/R-DPRs co-condensates, LLPS of WT FUS was maintained with the total integrated phase separated area comparable to the non-R-DPR condition (Fig. 5E, F). In addition, the enlarged size of FUS-GFP droplets caused by the addition of R-DPR was not observed in the presence of Kapβ2W460A:W730A (Fig. 5G). Remarkably, the preferred binding of Kapβ2W460A:W730A toward R-DPRs enabled it to selectively block R-DPRs from affecting FUS-GFP droplets, indicated by the disappearing of the heterogeneous phase formed by R-DPRs in FUS/R-DPR co-droplets and the decreased enrichment of TAMRA-DPR signal in these samples (Fig. 5E, H). For example, when quantified, the enrichment factor for (GR)20 in FUS-GFP droplets was significantly decreased from 7.8 ± 2.5 to 2.3 ± 0.5 (Fig. 5E, H). In contrast, the enrichment factor for FUS-GFP in the droplets remains unchanged with and without Kapβ2W460A:W730A (Fig. 5H). The selectivity of Kapβ2W460A:W730A toward (PR)20 in FUS/(PR)20 co-condensates is even more pronounced, as the enrichment factor for (PR)20 under this condition was 1.3 ± 0.1, indicating almost no enrichment of (PR)20 and near complete extraction from the FUS droplet by Kapβ2W460A:W730A (Fig. 5H). Thus, Kapβ2W460A:W730A binding to R-DPRs effectively inhibits the recruitment of R-DPRs into FUS droplets without affecting the normal phase separation of FUS. This effect is in stark contrast to WT Kapβ2, which completely inhibits normal phase separation of FUS (Fig. 5E-H).
We next investigated the ability of Kapβ2W460A:W730A to solubilize R-DPRs that have already assembled into FUS-GFP condensates. Unexpectedly, even with reduced binding to FUS, Kapβ2W460A:W730A was able to disassemble preformed FUS/R-DPR co-condensate completely (Supplementary Fig. 6G). It is possible that instead of simply being recruited into FUS droplets, R-DPR forms an important scaffolding structure for FUS/R-DPR co-condensates (Supplementary Fig. 3E, F). Therefore, when R-DPR is extracted by Kapβ2W460A:W730A, the droplets disassemble.
Kapβ2W460A:W730A effectively inhibits and reverses R-DPR-induced TDP-43 aggregation
In ALS/FTD patients, R-DPRs co-localize with TDP-43 aggregates39. A recent study showed that WT Kapβ2 can prevent poly(GR)-induced TDP-43 aggregation, even though TDP-43 is not a substrate for Kapβ215,18. We tested whether mutant Kapβ2 can also mitigate R-DPR-induced TDP-43 aggregation. Consistent with previous results, turbidity assays show that the addition of R-DPRs induces TDP-43 aggregation, with (PR)20 having a stronger effect than (GR)20 (Fig. 6A–D). Kapβ2W460A:W730A has a similar effect in inhibiting R-DPR-induced TDP-43 aggregation compared to WT Kapβ2 (Fig. 6A–D). Fluorescence images of TDP-43/R-DPR co-aggregates showed that the addition of R-DPRs induced the formation of large TDP-43 assemblies that co-localize with R-DPRs (Fig. 6E). When equimolar WT Kapβ2 was added, the size of TDP-43/R-DPR co-condensates was reduced relative to the aggregates that form in the absence of Kapβ2 (Supplementary Fig. 7A). When Kapβ2 was added at a ratio of 2:1 (Kapβ2:R-DPR), TDP-43/R-DPR co-condensates were inhibited entirely, and both TDP-43 and R-DPR remained diffuse (Fig. 6E). Kapβ2W460A:W730A showed similar results to WT Kapβ2 in these assays (Fig. 6E and Supplementary Fig. 7A). We then asked whether Kapβ2 can reverse R-DPR-induced TDP-43 aggregation by extracting R-DPRs from their co-aggregates. We found that even at a ratio of 2:1 (Kapβ2:R-DPR), Kapβ2 was not able to disassemble preformed R-DPR/TDP-43 co-aggregates (data not shown). To test whether a higher ratio of Kapβ2:R-DPR is more effective, we tested at a ratio of 4:1, but with lower concentrations of R-DPR (1 μM) and TDP-43 (1 μM), due to the limitation of the highest Kapβ2 concentration we can achieve without losing the protein. Even at lower concentrations, R-DPRs can induce aggregation of TDP-43 and co-aggregates with TDP-43 (Fig. 6F). Interestingly, at a ratio of 4:1 (Kapβ2:R-DPR), Kapβ2 WT and Kapβ2W460A:W730A were able to extract R-DPR into the soluble phase and at the same time, restore TDP-43 into a diffusive state (Fig. 6F). This activity of Kapβ2 is specific as BSA at the same concentration did not have an effect on R-DPR-induced TDP-43 aggregates (Supplementary Fig. 7B).
His-SUMO-TDP-43 (5 μM) incubated with buffer, 5 μM WT Kapβ2 or Kapβ2W460A:W730A, in the presence of 10 μM (GR)20 (A), or 10 μM (PR)20 (C). Aggregation was assessed by turbidity measured at 395 nm. Solid lines are normalized mean. Dotted lines of corresponding colors represent ± SEM (n = 3 independent experiments). Quantification of the area under the curve for the curves in (A) and (C) for (GR)20 (B), and (PR)20 (D), respectively. Color code is the same as in (A) and (C). Unpaired Student’s t-tests were used to compare different conditions. E Fluorescence images of His-SUMO-TDP-43 (2 μM) condensates formed in the presence or absence of the indicated R-DPR (2 μM) and the indicated Kapβ2 variant (4 μM). 200 nM His-SUMO-TDP-43-GFP and 100 nM TAMRA-R-DPR were added for visualization. Scale bar: 25 μm. F Indicated Kapβ2 variant (4 μM) was added into His-SUMO-TDP-43 (1 μM) condensates performed in the presence of the indicated R-DPR (1 μM). 2 hours after the addition of Kapβ2, fluorescence images of the samples were taken. 200 nM His-SUMO-TDP-43-GFP and 100 nM TAMRA-R-DPR were added for visualization. Scale bar: 25 μm.
Kapβ2W460A:W730A is more efficient in preventing poly(GR) recruitment into stress granules
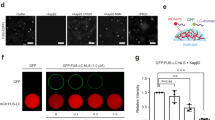
Having demonstrated that Kapβ2W460A:W730A selectively engages R-DPRs and effectively mitigates R-DPR-induced aberrant phase transition of FUS and TDP-43, which are both SG components, we next tested if Kapβ2W460A:W730A can prevent (GR)20 from entering SGs and impairing their dynamics. Compared to cells treated with WT Kapβ2, concomitant addition of Kapβ2W460A:W730A with (GR)20 resulted in reduced association of (GR)20 with SGs (Fig. 7A, B). This is consistent with a previous report that WT Kapβ2 is recruited into SGs, whereas Kapβ2W460A:W730A has a reduced tendency to be recruited30. Thus, Kapβ2W460A:W730A specifically prevents R-DPRs from interacting with SGs, while preserving physiological RBP granules.
A Representative images showing recruitment of TAMRA-(GR)20, in the presence of WT Kapβ2 or Kapβ2W460A:W730A, to G3BP1-positive stress granules in semi-permeabilized HeLa cells that were treated with sodium arsenite. Scale bar as indicated in the figure. B Quantification of the mean fluorescence intensity of TAMRA-(GR)20 in stress granules in the presence of WT Kapβ2 or Kapβ2W460A:W730A. The number of stress granules (n) quantified in each condition is indicated in the figure. Unpaired Student’s t-tests were used to compare different conditions; ****p ≤ 0.0001. C Co-immunoprecipitation (Co-IP) of (GR)50-GFP or (GA)50-GFP with V5-tagged WT Kapβ2 or Kapβ2W460A:W730A, from lysate of primary cortical neurons transduced with hsyn::(GR)50-GFP or hsyn::(GA)50-GFP, and hsyn::WT Kapβ2 or hsyn::Kapβ2W460A:W730A. Antibodies used for Co-IP and western blots are indicated. The intensity of the V5 and GFP bands was quantified, and the ratio of the IP band intensity to the input band intensity (normalized to vinculin) is shown in the bottom panel. D Representative confocal images of primary cortical neurons transduced with hsyn::(GR)50-GFP alone, or hsyn::(GR)50-GFP with hsyn::WT Kapβ2 or hsyn::Kapβ2W460A:W730A and treated with 0.5 mM sodium arsenite for 30 minutes. GFP signal is shown in green, TIAR signal is shown in Red, and nuclei are shown in blue. Scale bar = 10 µm. Shown on the right are the intensity profiles along the indicated lines across the stress granule. E Quantification of the confocal images collected in (D) showing relative fluorescence intensity of (GR)50-GFP in stress granules marked by TIAR. n = 3, m = 10. Parametric, unpaired Student’s t-tests were used to compare different conditions.
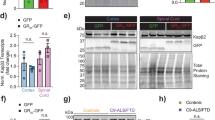
We extended our study to primary cortical neurons to investigate the selective interaction of Kapβ2W460A:W730A with polyGR in a disease relevant model. To achieve specific neuronal expression of the protein, we used constructs which express V5-tagged WT Kapβ2 or Kapβ2W460A:W730A under a human synapsin promoter. Upon viral transduction of V5-WT Kapβ2 or V5-Kapβ2W460A:W730A constructs, the two proteins were expressed at similar level in primary cortical neurons (Supplementary Fig. 8A, B). Exogenously expressed Kapβ2 localized both in the nucleus and in the cytoplasm as expected (Supplementary Fig. 8C)16. We then co-transduced the neurons with V5-WT Kapβ2 or V5-Kapβ2W460A:W730A, along with (GR)50-GFP or (GA)50-GFP as a control, all under the same human synapsin promoter and investigated their interaction in neurons. Both Kapβ2 variants co-immunoprecipitated with (GR)50-GFP, but not (GA)50-GFP (Fig. 7C and Supplementary Fig. 8D), consistent with our in vitro measurement. To investigate if Kapβ2 variants modulate the localization of poly(GR) in SGs, we stressed the primary neurons with sodium arsenite, which induced formation of SGs that contained (GR)50-GFP (Fig. 7D). Without (GR)50-GFP, expression of Kapβ2W460A:W730A does not change the SG dimension, number, and TIAR content per SG, compared to cells expressing WT Kapβ2 (Supplementary Fig. 8E–H). When co-expressed, the expression level of (GR)50-GFP is similar in WT Kapβ2- and Kapβ2W460A:W730A-expressing cells (Supplementary Fig. 8I), indicating that Kapβ2W460A:W730A does not influence (GR)50-GFP synthesis or degradation dynamics. Remarkably, Kapβ2W460A:W730A reduced the enrichment of (GR)50-GFP in TIAR-positive SGs, consistent with our observation in HeLa cells (Fig. 7D, bottom panel, and 7E). On the other hand, WT Kapβ2 failed to reduce the (GR)50-GFP SG enrichment (Fig. 7D, top panel, and 7E).
Kapβ2W460A:W730A reduces (GR)100 aggregation and counteracts neuronal toxicity of poly(GR)
Finally, we asked whether by selectively engaging poly(GR), Kapβ2W460A:W730A can solubilize poly(GR) and reduce its neuronal toxicity. Because viral transduction of (GR)50-GFP does not lead to aggregation of (GR)50 without stress, we elected to transfect our primary neuron culture with (GR)100-mCherry, which spontaneously forms aggregates when expressed in absence of external stress (Fig. 8A). When Kapβ2W460A:W730A was co-expressed with (GR)100-mCherry, we observed localization of Kapβ2W460A:W730A to aggregates containing (GR)100 (Fig. 8A, arrow). Interestingly, co-expressing GFP-Kapβ2W460A:W730A with (GR)100-mCherry leads to a significant reduction in the number of (GR)100 aggregates (Fig. 8A, B). In the remaining (GR)100 aggregates in GFP-Kapβ2W460A:W730A expressing cells, although the size of the (GR)100 aggregates does not change (Fig. 8C), the mean mCherry fluorescence intensity in these aggregates is significantly reduced compared to the aggregates observed in GFP expressing cells, indicating less (GR)100 is assembled into aggregates. Finally, we monitored the survival of neurons by live cell imaging, which displayed overt survival deficits when expressing (GR)100, consistent with published results (Fig. 8E)10,40. Co-expressing GFP-Kapβ2W460A:W730A with (GR)100-mCherry leads to a modest, although significant, increase (9.76±1.61%) in neuronal survival rate after 5 days of transfection (Fig. 8E). To confirm the rescue of poly(GR) toxicity by Kapβ2W460A:W730A in an independent neuronal model of R-DPR toxicity, we treated primary neurons with synthetic (GR)20, which robustly induces neuronal death, compared to the nontoxic DPR (GR)8 (Fig. 8F, G). While addition of WT Kapβ2 or Kapβ2W460A:W730A does not change the viability of cells treated with (GR)8, suggesting that addition of both Kapβ2 does not affect neuronal viability, it significantly increased neuronal survival in (GR)20 treated cells (Fig. 8F). DIC images of the treated neurons confirmed the protective effect of Kapβ2W460A:W730A against neuron degeneration (Fig. 8G). For example, (GR)20 treated neurons exhibited retracted and fragmented neurites compared to (GR)8 treated neurons. Notably, when neurons were co-treated with (GR)20 and Kapβ2W460A:W730A, longer neurites were observed (Fig. 8G), indicating rescuing of neurodegeneration. Together, these data demonstrate that Kapβ2W460A:W730A can engage poly(GR) in neurons to reduce their aggregation formation and rescue their neuronal toxicity.
A Representative images of primary cortical neurons co-transfected with (GR)100-mCherry and GFP or GFP-Kapβ2W460A:W730A. GFP signal is shown in green, mCherry is shown in red, Tuj1 is shown in magenta, and nuclei are shown in blue. Scale bar = 10 μm. B Quantification of the number of (GR)100 aggregates per neuron in primary cortical neurons co-transfected with (GR)100-mCherry and GFP or GFP-Kapβ2W460A:W730A. The number of neurons quantified (n) in each condition is indicated in the figure. Data shown are mean ± SEM. Unpaired Student’s t-tests were used to compare different conditions; ***p ≤ 0.001. C Quantification of the size of (GR)100 aggregates formed in primary cortical neurons co-transfected with (GR)100-mCherry and GFP or GFP-Kapβ2W460A:W730A. The number of aggregates quantified (n) in each condition is indicated in the figure. Data shown are mean ± SEM. Unpaired Student’s t-tests were used to compare different conditions; ns: p > 0.05. D Quantification of the mean mCherry fluorescence intensity in each (GR)100 aggregates formed in primary cortical neurons co-transfected with (GR)100-mCherry and GFP or GFP-Kapβ2W460A:W730A. The number of aggregates quantified (n) in each condition is indicated in the figure. Data shown are mean ± SEM. Unpaired Student’s t-tests were used to compare different conditions; ***p ≤ 0.001. E Analysis of percentage of survival neurons 5 days after co-transfected with (GR)100-mCherry and GFP or GFP-Kapβ2W460A:W730A. N = 3. Unpaired Student’s t-test was used to compare different conditions. F Viability assay of primary cortical neurons treated with indicated R-DPR alone or with Kapβ2 WT or Kapβ2W460A:W730A. 5μM of DRAQ7, 2.5μM of the indicated R-DPRs and proteins were added into the culture media of treated cortical neurons. Cells positive for DRAQ7 fluorescence (observed in Cy5 channel) were counted 18 hours after treatment to assess the viability of the neurons. Data are represented as median ± S.E.M., n = 3, m (fields of view) = 5. Nested Student’s t-test was used to compare different conditions, ****p ≤ 0.0001. G The indicated R-DPRs (2.5μM) and Kapβ2 WT or Kapβ2W460A:W730A (2.5 μM) were added into the culture media of treated primary cortical neurons. DIC images of the treated primary cortical neurons were taken 18 hours after treatment to assess the morphology of the neurons. Scale bar = 20 μm.
Discussion
A proposed mechanism for R-DPR toxicity is by binding to LCD-containing RBPs, inducing their aberrant phase transition and aggregation11,12,13. Therefore, it is important to understand the mechanism of R-DPR induced RBP aberrant phase transition to develop strategies to mitigate this toxic effect. Here, we show that R-DPRs bind to the N-terminal LCD of FUS (i.e., PrLD) to induce its LLPS, whereas binding to the C-terminal LCD (i.e., the RGG domains) induces FUS aggregation. While WT Kapβ2 is effective in mitigating R-DPR-induced FUS aberrant phase transition, it also completely inhibits FUS LLPS which is important for FUS function in cells. Furthermore, FUS binding resulted in stress granule localization of Kapβ2 in cell, thus Kapβ2 could not prevent interaction of R-DPRs with stress granules. To eliminate this undesired Kapβ2 activity against FUS, we took advantage of the tunable interactions amongst Kapβ2, FUS, and R-DPRs, and used a known Kapβ2 mutant (i.e., Kapβ2W460A:W730A) with weakened binding to FUS. Interestingly, we find that Kapβ2W460A:W730A retains strong binding to R-DPRs (Fig. 4H–J and Supplementary Fig. 6E, F). As such, Kapβ2W460A:W730A selectively inhibits R-DPR assembling into FUS droplet without affecting normal FUS LLPS. In cells, Kapβ2W460A:W730A efficiently prevents the recruitment of (GR)20 into stress granules. Moreover, Kapβ2W460A:W730A reduced (GR)100 aggregate formation and counteracted poly(GR) toxicity in primary neurons. Our results demonstrate that nuclear import receptors are attractive candidates for maintaining functional RBP phase separation in the presence of R-DPRs. More importantly, the property of Kapβ2W460A:W730A to selectively inhibit R-DPRs from partitioning into FUS droplets without affecting normal FUS phase separation, and from enrichment into stress granules, makes it an ideal tool to mitigate toxicity induced by R-DPR in C9-ALS/FTD without the potential undesired interaction of WT Kapβ2 toward its native cargoes, such as FUS. The development of Kapβ2W460A:W730A extended the therapeutic potential of Kapβ2 beyond its initially proposed application on FUS-ALS/FTD, and demonstrated the potential of R-DPR-specific variant of Kapβ2 against the most common genetic form of ALS/FTD: C9orf72-ALS/FTD.
Although interacting with numerous LCD-containing RBPs, including FUS and TDP-43, R-DPRs have been found to co-aggregate with TDP-43, suggesting that R-DPRs affect LCD-containing RBPs differently. Indeed, our results showed that R-DPRs interact differently with the N-terminal and C-terminal LCDs of FUS. R-DPRs bind the N-terminal LCD of FUS, a PrLD, to promote phase separation. Conversely, R-DPRs interact with the C-terminal LCD, the RGG domains to promote FUS aggregation. Under the sticker-and-spacer model, LLPS of FUS is driven by the cation-π interaction between arginine and tyrosine stickers from the C-terminal RGG domains and N-terminal PrLD, respectively41. Indeed, without the C-terminal RGG, FUS PrLD has a low tendency to phase separate (Fig. 2C)41. R-DPRs could act as additional arginine stickers and thus re-establish the interaction between arginine and tyrosine to facilitate the LLPS of PrLD (Fig. 2B, C). On the other hand, a FUS construct containing the C-terminal two RGG domains (i.e., FUS267-526) are soluble on their own but aggregate rapidly in the presence of R-DPR, without forming droplets (Fig. 2E). TDP-43 also aggregates rapidly in the presence of R-DPRs (Fig. 6). Whereas the FUS PrLD is enriched with uncharged polar residues (e.g., tyrosine), the TDP-43 LCD is unique in that it is also enriched with hydrophobic residues42. Moreover, in contrast to the FUS PrLD which contains only 2.3% aspartic acid and glutamic acid residues, the full-length TDP-43 protein exhibits a higher abundance of negatively charged residues (11% consisting of aspartic acid and glutamic acid) (Supplementary Fig. 9A). These negatively charged residues could be shielded by arginines in R-DPRs, which could then increase the hydrophobicity of TDP-43 and induce aggregation43,44. Similarly, C-terminal fragments of FUS (i.e., FUS267-526) are more enriched in negatively charged residues than the N-terminal PrLD (Supplementary Fig. 9B). As a result, FUS267-526 has the tendency to form aggregates in the presence of R-DPRs (Fig. 2E). Alternatively, FUS267-526 aggregation in the presence of R-DPRs could be mediated or enhanced by the co-purified RNA. Indeed, the addition of RNase A reduced the size of the FUS267-526 aggregates formed, highlighting the contribution of RNA in the interaction between FUS and R-DPR. Further experiments are needed to understand the exact role played by RNA and whether the small aggregates are caused by remaining FUS-bound RNA that wasn’t digested by RNase A.
Kapβ2 is a potent chaperone and protein disaggregase for PY-NLS-containing proteins such as FUS, and it functions by interacting with the PY-NLS and arginine residues of the RGG domains of these proteins18,20,21,22,45. In the presence of R-DPRs, Kapβ2 is still able to completely prevent and reverse FUS aggregation. In this situation, Kapβ2 functions by binding to both PY-NLS and the R-DPRs, keeping both FUS and R-DPRs in the soluble fraction. Interestingly, Kapβ2 binds to FUS and (GR)20/(PR)20 with similar Kd (Fig. 1I, Supplementary Fig. 4E–G). As a result, the equilibrium of binding can be shifted to favor Kapβ2-R-DPR interactions by either increasing R-DPR concentration (Fig. 4F, G, Supplementary Movie 1–3) or by using a FUS mutant (i.e., FUSP525L) with disrupted Kapβ2 binding (Fig. 4A–E, and Supplementary Fig. 4). In cells, this could be further regulated by other factors that disrupt FUS-Kapβ2 binding such as methylation of FUS RGG domains46, which could drive binding of Kapβ2 to R-DPRs, potentially sequestering Kapβ2 and subsequently leading to a nucleocytoplasmic transport defect. It is important to emphasize that the role of Kapβ2 as a chaperone and disaggregase is influenced by the delicate balance between its interaction with its substrate, FUS, and with R-DPRs. Even minor alterations can lead to the specific targeting of Kapβ2 towards R-DPRs, resulting in selectivity in activity. For example, the Kd of Kapβ2 for FUS PY-NLSP525L is only ~6–9-fold higher compared to WT FUS PY-NLS45,47, yet Kapβ2 activity against FUSP525L aggregation is more significantly impaired in the presence of R-DPRs (Fig. 4A–D and Supplementary Fig. 4A–D)18. Factors other than the Kd might also contribute to selective Kapβ2 activity toward R-DPR in co-aggregates of R-DPR and FUS. One factor is that FUS is more aggregation-prone than R-DPRs, and, thus, more challenging to solubilize. The combined effect of lowered binding affinity and stronger aggregation tendency results in the selective activity of Kapβ2 toward R-DPR in these aggregation and disaggregation assays.
In the current study, we took advantage of this tunable activity of Kapβ2 in the presence of R-DPR by utilizing a Kapβ2 variant that preferentially engages R-DPRs without affecting the normal LLPS function of FUS. Kapβ2 has been identified as a potent modifier of R-DPR toxicity in several genetic screen studies using different model organisms. For example, deletion of the fly homolog of Kapβ2 (Trn) enhances R-DPR toxicity in fly models of C9-ALS12. Furthermore, overexpression of the yeast homolog of Kapβ2 (Kap104) in DPR-expressing yeast acts as a potent suppressor of toxicity24. Most recently, we showed that expressing Kapβ2 in poly(GR)-expressing primary mouse neurons reduces poly(GR) toxicity in these cells27. Therefore, Kapβ2 represents a potential therapeutic for C9-ALS. However, the function of WT Kapβ2 as a nuclear import receptor and phase modifier of PY-NLS-containing RBPs could result in potential unintended interaction with its native cargoes when considering its development as therapeutics for C9-ALS. Kapβ2W460A:W730A is a PY-NLS binding deficient mutant whose activity in transporting PY-NLS cargo and regulating FUS LLPS is abolished18,38,48. Interestingly, this critical role played by Trp residues in NLS cargo binding is shared by other nuclear import receptors. For example, W342/W864 are essential to stabilize the helical conformation of the IBB in the cargo-binding groove of Importin β149. However, our study showed that the critical Trp residues in Kapβ2 are not required for R-DPR binding (Fig. 4I, J). Further, we demonstrated that Kapβ2W460A:W730A can selectively inhibit the participation of R-DPR into FUS liquid droplets without affecting the assembly of FUS droplets. This selectivity of Kapβ2W460A:W730A makes it more advantageous as therapeutics for C9-ALS than WT Kapβ2. It is desired to inhibit the deleterious effect of R-DPR on the dynamics of phase separated RBP condensates, while preserving the functional LLPS of FUS and other PY-NLS containing RBP as it is beneficial for the cell. To this end, we showed that Kapβ2W460A:W730A is more efficient than WT Kapβ2 in preventing poly(GR) recruitment into G3BP1-marked stress granules. Since the interaction between Kapβ2W460A:W730A and R-DPRs is strong in different environments, we expect that the activity of Kapβ2W460A:W730A is more general and should also extend to stress granules marked by other markers such as TIA1 and FMRP, which often colocalize with G3BP1. Importantly, Kapβ2W460A:W730A has similar activity compared to WT Kapβ2 in inhibiting and reversing R-DPR-induced TDP-43 aggregation (Fig. 6), as it maintains strong binding to R-DPR. Thus, its function as an R-DPR sink/sponge is preserved.
Although WT Kapβ2 can rescue neuronal toxicity of R-DPR, the mechanism of this rescue is not well understood. Several proposed mechanisms of R-DPR toxicity are relevant to the function of Kapβ2, including disrupted nucleocytoplasmic transport and disrupted phase separation of LCD-containing RBP. By abolishing Kapβ2 function as a nuclear import receptor and RBP chaperone but preserving its function as R-DPR sink/sponge, Kapβ2W460A:W730A serves as a valuable tool to understand which function of Kapβ2 is important for it to be a toxic modifier of R-DPR, and which one of the pathways contribute more to R-DPR toxicity. The study conducted has revealed that the mutant Kapβ2W460A:W730A can save cells from the harmful effects of certain disease related proteins. This indicates that the ability of Kapβ2 to act as a sink or sponge for these proteins is a crucial factor in modifying their toxicity. However, further research is needed to completely comprehend how Kapβ2 and its mutants’ function in the complex environment of neurons. For example, a full dose-response experiment would be helpful to understand whether higher expression levels of Kapβ2 can work better. In the future, combining Kapβ2W460A:W730A with other therapeutic strategies, such as the importin α/β complex against TDP-43 aggregation, could lead to more effective treatments for neurodegenerative disorders. Additionally, deciphering the structure of Kapβ2 in complex with toxic proteins could assist researchers in designing Kapβ2 variants that are more precise in their binding to these proteins.
Materials and Methods
Cells culture
HeLa cells were cultured in 4.5 g/L glucose Dulbecco’s Modified Eagle Medium (Corning) supplemented with 10% fetal bovine serum (Cytiva) and 1% penicillin/streptomycin (Corning) in a humidified incubator (37°C, 5% CO2/95% air).
Primary culture
Primary neurons were obtained from E16 rat pups’ cortices. Briefly, cortices were dissected, cleaned from the meninges and treated with 0.25% trypsin for 45 minutes at 37 °C under gentle shaking. The tissue was then homogenized and strained through a 70 mm strain. Cells were counted and plated on PDL-coated plates.
Nucleic acid constructs
Full-length wild-type FUS protein, and its mutants including FUSP525L, N-terminal fragments FUS1-214, FUS1-371, and C-terminal fragments FUS215-526, FUS267-526 were expressed from GST fusion constructs using pDuet vector containing a TEV cleavage site. N-terminal fragments FUS1-214 and FUS1-371 constructs were created by inserting stop codons into a GST-TEV-FUS plasmid using the following primers:
-
FUS1-214: CTATGGAGAGCAGGACCGTGGATAAT
-
FUS1-371:
-
Forward: CAAGGTCTCATTTGCTACTCGCTAGTAAGACTTTAATCGGGGTGGTGGC
-
Reverse: GCCACCACCCCGATTAAAGTCTTACTAGCGAGTAGCAAATGAGACCTTG
-
C-terminal fragments FUS215-526 and FUS267-526 were created by quikchange mutagenesis using the following primer:
-
FUS215-516:
-
Forward: AGAATCTTTATTTTCAGGGAGGCCGCGGCAGGGGTGGCAGT
-
Reverse: TCCCTGAAAATAAAGATTCTCGCTAGCCAG
-
FUS267-526:
-
Forward: AGAATCTTTATTTTCAGGGAGGCCCTCGGGACCAAGGA
-
Reverse: TCCCTGAAAATAAAGATTCTCGCTAGCCAG
Wild-type Kapβ2, TDP-43, and TDP-43-GFP proteins were expressed as his-SUMO fusion construct using pE-SUMOpro Amp vector (LifeSensors). NLS-binding deficient variant Kapβ2W460A:W730A protein was expressed as GST fusion construct using pGEX-TEV vector. FUS-GFP protein was expressed as MBP fusion protein using pMAL vector. All constructs were confirmed by DNA sequencing.
Protein expression and purification
All recombinant proteins were expressed and purified from E. coli BL21-CodonPlus (DE3)-RIL cells (Agilent). GST-TEV-FUS, GST-TEV-FUSP525L, GST-TEV-FUS1-214, GST-TEV-FUS1-371, GST-TEV-FUS215-526, and GST-TEV-FUS267-526 were purified as described previously28. Briefly, for all recombinant GST-TEV-FUS proteins, E. coli cells were induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 18 h at 15 °C. Bacterial cells were then harvested and lysed with sonication in PBS buffer (pH 7.4) supplemented with protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). Cell lysates were clarified by centrifugation, applied to glutathione sepharoseTM 4 fast flow beads (Cytiva, Sweden), and then eluted in a buffer containing 20 mM Tris pH 8, 20 mM trehalose, and 20 mM reduced glutathione. Purified FUS proteins were flash frozen and stored at −80 °C.
Wild type Kapβ2 protein was expressed and affinity purified using previously published methods with minor modifications48. Briefly, Kapβ2 expression was induced with 1 mM IPTG for 18 h at 15 °C. Subsequently, the cells were harvested, resuspended, and lysed by sonication in a buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 20% glycerol, 10 mM imidazole, 2.5 mM β-mercaptoethanol, and protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). Cell lysates were clarified by centrifugation, and the supernatant was incubated with HisPur™ Ni-NTA resin (Thermo Scientific, USA) pre-equilibrated in lysis buffer. The bound Kapβ2 protein was eluted with buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 20% glycerol, 200 mM imidazole, and 2.5 mM β-mercaptoethanol. The eluted Kapβ2 protein was buffer exchanged into buffer A containing 20 mM imidazole, 75 mM NaCl, 20% glycerol, and 2.5 mM β-mercaptoethanol and cleaved with Ulp1 protease. Cleaved Kapβ2 was further purified by ion-exchange chromatography (HiTrap Q HP, GE Healthcare). Purified Kapβ2 protein was snap frozen and stored at −80 °C.
Kapβ2W460A:W730A was purified as described18. Kapβ2W460A:W730A protein was induced in BL21 (DE3) RIL E. coli cells using 1 mM IPTG for 18-20 h at 25 °C. Cells were harvested, resuspended, and lysed with sonication in a buffer containing 50 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 20% glycerol, 2 mM DTT, 5 mM pepstatin and protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). Kapβ2W460A:W730A was purified using glutathione sepharoseTM 4 fast flow beads (Cytiva, Sweden) pre-equilibrated in the lysis buffer. Subsequently, bound Kapβ2W460A:W730A protein was eluted in a buffer containing 20 mM imidazole (pH 6.5), 75 mM NaCl, 1 mM EDTA, 2 mM DTT, 20% glycerol, and 20 mM reduced glutathione. The eluted Kapβ2 protein was cleaved with TEV protease, and further purified by ion-exchange chromatography (HiTrap Q HP, GE Healthcare).
His-SUMO-TDP-43 and His-SUMO-TDP-43-GFP protein was purified as described50. E. coli cells were induced with 1 mM IPTG for 18 h at 15°C. The cells were then harvested, resuspended, lysed, and sonicated in a lysis buffer containing 50 mM HEPES (pH 7.5), 2% TritonX-100, 300 mM NaCl, 30 mM imidazole, 5% glycerol, 2 mM β-mercaptoethanol, 5uM pepstatin, and protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). Cell lysates were clarified by centrifugation and the supernatant was then incubated with HisPur™ Ni-NTA beads (Thermo Scientific, USA) pre-equilibrated in lysis buffer. TDP-43 and TDP-43-GFP proteins were eluted using an elution buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 300 mM imidazole, 5% glycerol, and 5 mM DTT. The eluted proteins were dialyzed against a buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 5% glycerol, 5 mM DTT.
To overexpress and purify MBP-FUS-GFP protein, E. coli cells were induced with 1 mM IPTG for 18 h at 15 °C and harvested by centrifugation at 4000 rpm for 20 mins at 4 °C. The cell pellet was resuspended, lysed, and sonicated in lysis buffer containing 50 mM HEPES pH 7.4, 1.5 M NaCl, 10% glycerol, 2 mM DTT, 30 mM imidazole and protease inhibitor tablets (cOmplete, EDTA-free, Roche Applied Science). After lysis, the precleared and filtered supernatant was passed through a HisTrap FF Crude Column (Cytiva, Sweden) pre-equilibrated with lysis buffer using AKTA pure (General Electric company, USA). Further, FUS-GFP was eluted using a buffer containing 50 mM HEPES (pH 7.4), 1 M NaCl, 10% glycerol, 2 mM DTT, and 500 mM imidazole, using a linear gradient. The eluted protein was further purified over a Heparin column (Cytiva, Sweden).
Recombinant GST-Prescission Protease was expressed in Escherichia coli cells and induced with 0.5 mM IPTG for 18 h at 30 °C. Bacterial cells expressing Prescission Protease were harvested by centrifugation, resuspended in a buffer containing 20 mM Tris (pH 8), 250 mM NaCl, 1 mM EDTA, 5 mM BME, 1 mM DTT, and protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). Then, resuspended cells were lysed with sonication. Cell lysates were clarified by centrifugation, applied to glutathione sepharoseTM 4 fast flow beads (Cytiva, Sweden) pre-equilibrated in resuspension buffer, and then eluted in a buffer containing 25 mM Tris pH 8, 250 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF, and 20 mM reduced glutathione. Purified Prescission Protease were then dialyzed overnight at 4 °C into a buffer containing 25 mM Tris (pH 8), 200 mM NaCl, 1 mM EDTA, and 0.2 mM PMSF, then stored in 20% glycerol.
Reagents
Twenty repeats of PR/GR/GA peptides were synthesized by Peptide 2.0 (Chantilly, VA). Also, TAMRA-(PR)20 and TAMRA-(GR)20) were synthesized by peptide 2.0 (Chantilly, VA). Lyophilized powder was reconstituted in 1× PBS and snap frozen and stored at -80 °C.
FUS turbidity assay
To measure the assembly kinetics of WT and mutant FUS, the purified GST-TEV-FUS proteins (wild-type FUS, FUSP525L, FUS1-214, FUS1-371, FUS215-526, and FUS267-526) were thawed on ice and centrifuged at 15000 rpm for 10 min at 4 °C to preclear aggregates. FUS assembly was initiated by addition of 1.6 µg TEV protease to 5 μM GST-TEV-FUS in the presence of the indicated peptide or equivalent volume of 1× PBS as a control in FUS assembly buffer (50 mM Tris-HCl (pH 8), 20 mM trehalose, 1 mM DTT, and 20 mM glutathione) for a total reaction volume of 100 μL. Aggregation was monitored via turbidity measurements at 395 nm (OD395nm) using a Tecan Spark plate reader for 100 min. To examine the inhibitory potential of WT Kapβ2 toward R-DPRs-enhanced aggregation of wild-type FUS and FUSP525L, aggregation reactions were performed as described above with 5 μM WT Kapβ2 or with an equivalent volume of Kapβ2 buffer as a control. Turbidity data were normalized by first subtracting FUS+buffer only condition. The resulting absorbance was then normalized to the maximum turbidity of FUS aggregation in the presence of TEV alone to determine the relative extent of aggregation. The lag time of the aggregation turbidity assay was obtained by extrapolating the maximum derivative down to the intercept with the pre-transition base-line51.
Disaggregation assays were performed similarly, where GST-TEV-FUS or GST-TEV-FUSP525L was incubated with or without R-DPRs in the presence of TEV protease, and turbidity was measured to assess fibrillization. After 100 min of aggregation, an equal volume of WT Kapβ2 or buffer was added, and turbidity was continually measured. The absorbance was normalized to that of the fully assembled FUS fibrils before addition of Kapβ2 to determine the relative extent of disaggregation.
Sedimentation assay to measure FUS aggregation
For sedimentation analysis, samples were sedimented by centrifugation for 10 min at 15,000 rpm at 4 °C after aggregation or disaggregation assays were completed. Pellet and supernatant fractions were separated immediately. The pellet was resuspended in 100 µL of FUS assembly buffer, and both samples were mixed with 4X Laemmli reducing sample buffer. SDS-PAGE was performed on equal volumes of each fraction. Coomassie stain was used to visualize proteins. The relative abundance of the protein in each fraction was quantified using ImageJ software.
Detergent solubility assay
FUS aggregation was initiated by the addition of 1.6 µg TEV protease to 5 μM GST-TEV-FUS in assembly buffer (50 mM Tris-HCl (pH 8), 20 mM trehalose, 1 mM DTT, and 20 mM glutathione) for a total reaction volume of 100 μL. After 100 minutes, aggregates were incubated with SDS or N-Lauroylsarcosine (sarkosyl; Sigma-Aldrich, 61747) for 2 minutes at room temperature. The insoluble fraction was separated by centrifugation for 10 minutes at 21,130 g at room temperature. The supernatant (soluble) fraction was transferred to a clean tube and denatured by adding 4x Sample Buffer and heating 5 minutes at 99°C. The SDS or sarkosyl-insoluble pellet was resuspended in 4x Sample Buffer, heated, and analyzed by gel electrophoresis and Coomassie staining.
Phase separation of GST-FUS
All the proteins used were centrifuged at 15,000 rpm for 10 min at 4 °C prior to the reaction to remove any preformed aggregates. Protein concentration was then measured via Bradford assay (Biorad). To image WT FUS liquid droplets, GST-TEV-FUS (10 µM) was incubated in the presence or absence of R-DPR (10 µM) in FUS assembly buffer (50 mM Tris-HCl pH 8, 20 mM Trehalose, 1 mM DTT, and 20 mM glutathione) for 3 hours at room temperature. Before imaging, 20 nM of TAMRA-(GR)20 or TAMRA-(PR)20 were added for visualization and image quantification purposes. The concentration of the added TAMRA-(GR)20 or TAMRA-(PR)20 (20 nM) was carefully chosen to make sure they do not affect FUS condensate. Protein samples were imaged at 0 and 3 hours by differential interference contrast (DIC) and fluorescence microscopy.
Phase separation of MBP-FUS-GFP
Purified wild-type MBP-FUS-GFP was thawed on ice and centrifuged for 10 min at 15000 rpm at 4 °C to preclear aggregates. Phase separation of MBP-FUS-GFP (3 µM) was initiated by the addition of 3 C protease (Prescission Protease, 18 μg/ml) to cleave off the MBP tag in LLPS buffer (50 or 150 mM KCl, 50 mM Tris pH 7.4, 0.5% Glycerol, and 1 mM DTT). Phase separation of FUS-GFP was performed in the presence or absence of the indicated R-DPR (3 µM).
To examine the inhibitory and reversal effects of WT Kapβ2 or Kapβ2W460A:W730A on R-DPR-enhanced LLPS of FUS-GFP, WT Kapβ2 or Kapβ2W460A:W730A (6 µM) was added to reactions containing FUS-GFP (3 µM) and R-DPR (3 µM) at the time 3 C was added (for inhibition), or 2 hours after 3 C was added (for reversal). Droplets were visualized by DIC and fluorescence microscopy at indicated times. Each droplet experiment was performed with three separate batches of purified protein. All R-DPR containing samples were supplemented with 100 nM of TAMRA-(GR)20 or TAMRA-(PR)20 for visualization. Droplets were visualized by DIC and fluorescence microscopy at 0 and 2 hours. The integrated droplet area, droplet size, and enrichment factor was measured using ImageJ.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments of the FUS-GFP/TAMRA-DPR co-droplets were conducted using a Nikon Ti Microscope with a Galvano scanner and with an 60X objective. Images were taken every second. Bleaching was performed by applying 25% 488 laser power to the selected ROI and fluorescence was recorded for 180 seconds. Data were normalized on the unbleached area and on the background using the online tool easyFRAP43 and then plotted using GraphPad Prism 9.0. A one-way ANOVA test was applied to analyze the differences between the curves.
TDP-43 turbidity assay
Purified His-SUMO-TDP-43 was thawed and precleared of aggregates by spinning at 15000 rpm for 10 minutes at 4 °C before measuring aggregation kinetics. Protein concentration was measured by Bradford, and TDP-43 was then diluted to a final concentration of 5 μM in PBS (pH 7.4), in the presence or absence of 5 μM indicated R-DPR, and 5 μM WT Kapβ2 or Kapβ2W460A:W730A. Aggregation was monitored via turbidity measurements at 395 nm (OD395nm) using a Tecan Spark plate reader.
Formation of TDP-43 condensates
Purified His-SUMO-TDP-43 protein was thawed on ice and precleared of aggregates by centrifuging for 10 mins at 15,000 rpm at 4 °C. For inhibition assay, TDP-43 (2 μM) was combined with 2 μM unlabeled (GR)20 or (PR)20, and 4 μM or 2 μM of WT Kapβ2 or Kapβ2W460A:W730A in PBS. All samples were supplemented with 100 nM of TAMRA-(GR)20 or TAMRA-(PR)20 and 200 nM of His-SUMO-TDP-43-GFP. After 1 hour of incubation, all the samples were imaged using DIC and fluorescent microscopy. For reversal assay, 4 μM WT Kapβ2, Kapβ2W460A:W730A, BSA, or buffer was added into His-SUMO-TDP-43 (1 μM) condensates preformed in the presence of the indicated R-DPR (1 μM). 2 hours after the addition of Kapβ2, fluorescence images of the samples were taken. 200 nM His-SUMO-TDP-43-GFP and 100 nM TAMRA-R-DPR were added for visualization.
Fluorescence anisotropy
All fluorescence anisotropy experiments were conducted in FUS assembly buffer or LLPS buffer as indicated in the Figure legend, with a total volume of 100 μL. Both TAMRA-R-DPR and FUS-GFP concentrations were fixed at 100 nM and anisotropy measurements were run in a Nunc MicroWell 96-well plate (Thermo Scientific, USA) using a TECAN Spark plate reader. For TAMRA fluorescence, excitation and emissions were set at 520 nm/20 nm and 580 nm/20 nm, respectively. For GFP fluorescence, excitation and emissions were set at 450 nm/25 nm and 510 nm/25 nm, respectively.
Computational analyses of protein structure and docking
In previous studies, several structures of human Kapβ2 in complex with different proteins have been solved20,38,47,52,53,54,55,56. In this study, we used a 2.3-Å crystal structure of Kapβ2 bound to the PY-NLS of FUS (PDB ID: 4FDD)47. The different chains in the 4FDD structure have been labeled from A to B, where chain A is Kapβ2 and chain B is PY-NLS of FUS47. Using PyMol, we removed the PY-NLS of FUS (chain B) from the 4FDD structure and saved separately the chain A of WT Kapβ2. Next, chain A of WT Kapβ2 was redocked with PY-NLS of FUS, as well as two test peptides (i.e., (GR)15 and (PR)15) using GalaxyPepDock docking server, which performs protein-peptide docking based on interaction similarity34. As this server only allows docking of peptides of up to 30 amino acids in length, we used (GR)15 and (PR)1534. GalaxyPepDock predicts 3D protein-peptide complex structures from an input protein structure (as a PDB file) and peptide sequence (in FASTA format) by combining information from structural databases and energy-based optimization34. The top ten models were downloaded, and the top model was used to generate the docking pose and analyzed for energy calculations using the HawkDock web server34,57.
Next, we introduced two point mutations, W460A and W730A, in Kapβ2 (chain A of the 4FDD structure) by using the Swiss PDB viewer58 to generate 4FDD-WWAA. Similar to the previous steps, PY-NLS of FUS (chain B) was removed from the 4FDD-WWAA structure and saved separately as chain A of Kapβ2W460A:W730A. Chain A of Kapβ2W460A:W730A was redocked with PY-NLS of FUS, as well as two test peptides (i.e., (GR)15 and (PR)15) using GalaxyPepDock docking server34 and the top one model was used for energy calculations using the HawkDock web server57.
This crystal structure of WT Kapβ2 (PDB ID: 4FDD) does not include the region of AA 337-36747. Therefore, we repeated docking using an AlphaFold predicted structure of full-length Kapβ2 (ID: AF-Q92973-F1). AlphaFold is an AI system developed by Google DeepMind and EMBL-EBI that predicts a protein’s 3D structure from its amino acid sequence59. It regularly achieves accuracy competitive with experiments. AF-Q92973-F1 structure was used to dock PY-NLS of FUS and test peptides (GR)15 and (PR)15 using GalaxyPepDock docking server as described previously.
Semi-permeabilized cell assay
Semi-permeabilized cell assays were conducted as previously described60, with slight modifications. HeLa cells were grown on coverslips coated in poly-D lysine (PDL). Cells were stressed with 500 mM sodium arsenite for 1 hour at 37 °C followed by permeabilization for 10 minutes with 25 μg/mL digitonin diluted in permeabilization buffer (20 mM HEPES, pH 7.5, 110 mM KOAc, 5 mM Mg(OAc)2, 0.5 mM EGTA, 250 mM sucrose, and protease inhibitors (Halt Protease Inhibitor Cocktail, EDTA-Free, Thermo Scientific)) on ice. Cells were washed several times in transport buffer (20 mM HEPES (pH 7.3), 110 mM KOAc, 2 mM Mg(OAc)2, 5 mM NaOAc, 0.5 mM EGTA, 250 mM sucrose, and protease inhibitors (Halt Protease Inhibitor Cocktail, EDTA-Free, Thermo Scientific)) prior to blocking the nuclear pore with 0.2 mg/mL wheat germ agglutinin (WGA) incubation for 15 minutes on ice. TAMRA-(GR)20 (100 nM), recombinant WT Kapβ2 (200 nM) or Kapβ2W460A:W730A (200 nM) were diluted in transport buffer and directly added to permeabilized cells for 30 minutes following WGA treatment. Cells were fixed with 4% paraformaldehyde (PFA), immunostained against G3BP1 (rabbit anti-G3BP1; 1:500, Invitrogen #PA5-29455) overnight at 4°C. Secondary antibodies (goat anti-rabbit AlexaFluor 647 (1:1000, Invitrogen #A-21245)) were incubated for 1 hour at room temperature. Fixed cells were imaged with a LEICA DMI8 microscope using a 40X objective. Images were processed and quantified using Image J.
Viral production and cortical neurons transduction
HEK293T were seeded at 8×106 cells per 15 cm plate using normal HEK media (DMEM, 10% FBS, Penn/Strep/Glutamine). On the next day, cells were transfected with 8 μg of transfer plasmid (pLenti_hsyn::(GR)50-GFP, pLenti_hsyn::(GA)50-GFP, pLenti_hsyn::WT Kapβ2, or pLenti_hsyn::Kapβ2W460A:W730A), 16 μg of psPAX2, and 4 μg of pMD2.G using 84 μL PEI MAX (1 μg/μL). The mixture was incubated in 1 mL OptiMEM for 15 minutes at room temperature and then added to the cells. After 4 hours of incubation, cell media was replaced with fresh media. 48 hours after transfection, cell media was collected and centrifuged at 500 g for 10 minutes at room temperature to remove cell debris. Viral particles were then concentrated using Lenti-X Concentrator. Viral particles were resuspended in PBS, aliquoted, snap frozen and stored at -80 °C. Cortical neurons at 7 DIV were treated with 2 μL/125,000 cells of each virus. Transduction efficiency was assessed though epifluorescence microscope 48 hours after transduction.
Co-Immunoprecipitation and Western blot
5 million cortical neurons at 7 DIV were treated with indicated viruses. 48 hours after transduction, cells were then treated for 30 minutes with 2 mM DSP, washed twice with PBS, and harvested in lysis buffer. Lysates were then incubated with magnetic beads that were pre-coupled with GFP antibody (Abcam, catalog#: ab5449). After overnight incubation, beads were washed twice after which the protein was eluted, boiled, and loaded into the polyacrylamide gel (10%). After electrophoresis run, proteins were transferred on nitrocellulose membrane and probed for Vinculin (ThermoFisher, catalog#: 700062, 1000), GFP (Abcam, catalog#: ab5449, 1:1000) and V5 (Cell Signaling Technology, catalog#: 13202, 1:1000).
Stress treatments and analysis
48 hours after transduction with the indicated viruses, neurons were treated with 0.5 mM of sodium arsenite for 30 minutes (water was used as control), fixed with 4% PFA immediately, and immunostained against TIAR (BD, catalog#: 610352, 1:750) and V5 (Cell Signaling Technology, catalog#: 13202, 1:1000) overnight at 4 °C. The day after, two PBS washes were applied and cells were incubated with secondary antibody (Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647, Thermo Fisher, A-21235, 1:1000) diluted in 0.1% BSA. After two additional washes, cells were incubated with Hoechst (1:3000) for 10 minutes. Cells were then left in PBS and imaged with Nikon A1R confocal microscope with a 60X objective. To quantify enrichment of (GR)50-GFP in TIAR marked stress granules, mean green ((GR)50-GFP) and red (TIAR) fluorescence intensity were measured inside each SGs and then the ratio of the mean green and red fluorescence intensity was calculated. For all SGs analysis region of interests (ROI) were designed around each SGs based on red fluorescence. Area, TIAR content and GR content were calculated inside each ROI. Number of SGs was manually counted using maximum intensity projection of Z-Stack images. Profile intensity of green and red fluorescence was calculated along a line crossing the SGs from side to side. More than 10 cells per experiment were analyzed and 3 independent experiments were conducted and analyzed. GR intensity inside Kapβ2+ cells was calculated inside ROIs traced based on Kapβ2 fluorescence in neurons.
Primary culture transfection and live imaging
After 7 days in culture, cells were transfected using lipofectamine (Invitrogen) following manufacturer instructions. The following quantities of plasmids were used: pCDNA3_hU6_Flag_GR100_mCherry (400 ng)10; pCDNA3_CMV_eGFP (400 ng); pCDNA3_CMV_eGFP_Kapβ2WWAA (400 ng). After transfection, cells were imaged each day with a Nikon Eclipse epifluorescence microscope at 20X. Cells that were double positive for GFP and mCherry were counted in each experiment. Data were graphed and analyzed with Prism GraphPad 9.0.
Immunofluorescence
Primary neurons plated on glass bottom plates were fixed with 4% PFA for 20 min at 37 °C. Permeabilization and blocking steps were performed using a permblock buffer (1% BSA, 10% FBS, and 0.01% Triton X100). Primary antibody was incubated overnight at 4 °C with gentle rocking (Tuj1, Proteintech, 1:2000). The day after, two PBS washes were applied and cells were incubated with secondary antibody diluted in 0.1% BSA (Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647, Thermo Fisher, 1:1000). After two additional washes, cells were incubated with Hoechst (1:3000) for 10 minutes. Cells were then left in PBS and imaged with Nikon A1R confocal microscope with a 60X objective. 20 neurons per condition were imaged using Z-Stacks of 1um. Images were composed as maximum intensity projections and ROIs on the red channel (GR100) were automatically detected through by the Nikon software. Each ROI is an aggregate since the detection method used the brightest spots to detect ROIs. The software then gives mean brightness, dimension and number of the ROIs. Results were plotted with Prism GraphPad 9.0. and statistical analysis was also carried out through Prism GraphPad 9.0.
Viability assay of cortical neurons treated with (GR)20 peptide
Cortical neurons at 7 DIV were treated with 5 μM of DRAQ7 and 2.5 μM (GR)20, in the presence or absence of 2.5 μM of the indicated Kapβ2 proteins. DRAQ7 fluorescence was acquired by epifluorescence microscopy 18 hours after treatment. DRAQ7 fluorescence was then semi-automatically quantified in each cell and graphed using Prism GraphPad 10.0.3.
Statistics and reproducibility
All experiments were performed at least 3 times (n ≥ 3). Quantification and statistics are described in the figure legends. Statistical analyses were performed using the GraphPad Prism (GraphPad Software, Inc.; La Jolla, CA, USA) as described in figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated in this study are available in the Article, Supplementary Information, and Source Data files. Source data are provided with this paper in “Supplementary Data”.
References
Portz, B., Lee, B. L. & Shorter, J. FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 46, 550–563 (2021).
DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011).
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011).
Odeh, H. M. & Shorter, J. Arginine-rich dipeptide-repeat proteins as phase disruptors in C9-ALS/FTD. Emerg. Top. Life Sci. 4, 293–305 (2020).
Gitler, A. D. & Tsuiji, H. There has been an awakening: Emerging mechanisms of C9orf72 mutations in FTD/ALS. Brain Res. 1647, 19–29 (2016).
Mori, K. et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013).
Mori, K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013).
Ash, PeterE. A. et al. Unconventional translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 77, 639–646 (2013).
Freibaum, B. D. & Taylor, J. P. The role of Dipeptide repeats in C9ORF72-related ALS-FTD. Front Mol. Neurosci. 10, 35 (2017).
Wen, X. et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014).
Boeynaems, S. et al. Phase separation of C9orf72 Dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e1045 (2017).
Lee, K.-H. et al. C9orf72 Dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e717 (2016).
White, M. R. et al. C9orf72 Poly(PR) Dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol. Cell 74, 713–728.e716 (2019).
Cook, C. N. et al. C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci Transl. Med. 12. https://doi.org/10.1126/scitranslmed.abb3774 (2020).
Hutten, S. et al. Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 33, 108538 (2020).
Zhang, K. et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e917 (2018).
Hayes, L. R., Duan, L., Bowen, K., Kalab, P. & Rothstein, J. D. C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. eLife 9, e51685 (2020).
Guo, L. et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with Prion-like domains. Cell 173, 677–692.e620 (2018).
Mann, J. R. et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e328 (2019).
Yoshizawa, T. et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e622 (2018).
Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and Arginine methylation. Cell 173, 706–719.e713 (2018).
Qamar, S. et al. FUS phase separation is modulated by a molecular Chaperone and methylation of Arginine Cation-π interactions. Cell 173, 720–734.e715 (2018).
Bozic, J. et al. Interactome screening of C9orf72 dipeptide repeats reveals VCP sequestration and functional impairment by polyGA. Brain 145. https://doi.org/10.1093/brain/awab300 (2021).
Jovičić, A. et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 18, 1226–1229 (2015).
Freibaum, B. D. et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015).
Boeynaems, S. et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 6, 20877 (2016).
Cicardi, M. E. et al. Kapβ2 is a modifier of the C9orf72-linked glycine-arginine dipeptide neurotoxicity. bioRxiv, 2022.2009.2030.510384. https://doi.org/10.1101/2022.09.30.510384 (2022).
Sun, Z. et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9, e1000614 (2011).
Bogaert, E. et al. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537.e524 (2018).
Zhang, Y.-J. et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142 (2018).
Bourgeois, B. et al. Nonclassical nuclear localization signals mediate nuclear import of CIRBP. Proc. Natl Acad. Sci. 117, 8503–8514 (2020).
Conte, A. et al. P525L FUS mutation is consistently associated with a severe form of juvenile Amyotrophic Lateral Sclerosis. Neuromuscul. Disord. 22, 73–75 (2012).
Zou, Z. Y. et al. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol. Aging 34, 1312.e1311–1318 (2013).
Lee, H., Heo, L., Lee, M. S. & Seok, C. GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 43, W431–W435 (2015).
Nanaura, H. et al. C9orf72-derived arginine-rich poly-dipeptides impede phase modifiers. Nat. Commun. 12, 5301 (2021).
Jafarinia, H., Van der Giessen, E. & Onck, P. R. Molecular basis of C9orf72 poly-PR interference with the β-karyopherin family of nuclear transport receptors. Sci. Rep. 12, 21324 (2022).
Imasaki, T. et al. Structural basis for substrate recognition and dissociation by human Transportin 1. Mol. Cell 28, 57–67 (2007).
Lee, B. J. et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558 (2006).
Saberi, S. et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 135, 459–474 (2018).
Verdone, B. M. et al. A mouse model with widespread expression of the C9orf72-linked glycine–arginine dipeptide displays non-lethal ALS/FTD-like phenotypes. Sci. Rep. 12, 5644 (2022).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of Prion-like RNA binding proteins. Cell 174, 688–699.e616 (2018).
Schmidt, H. B., Barreau, A. & Rohatgi, R. Phase separation-deficient TDP43 remains functional in splicing. Nat. Commun. 10, 4890 (2019).
Koulouras, G. et al. EasyFRAP-web: a web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 46, W467–W472 (2018).
Bentley, E. P., Frey, B. B. & Deniz, A. A. Physical chemistry of cellular liquid-phase separation. Chemistry 25, 5600–5610 (2019).
Gonzalez, A. et al. Mechanism of karyopherin-β2 binding and nuclear import of ALS variants FUS(P525L) and FUS(R495X). Sci. Rep. 11, 3754 (2021).
Dormann, D. et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 (2012).
Zhang, Z. C. & Chook, Y. M. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc. Natl Acad. Sci. USA 109, 12017–12021 (2012).
Fare, C. M., Rhine, K., Lam, A., Myong, S. & Shorter, J. A minimal construct of nuclear-import receptor Karyopherin-β2 defines the regions critical for chaperone and disaggregation activity. J. Biol. Chem. 299, 102806 (2023).
Koerner, C., Guan, T., Gerace, L. & Cingolani, G. Synergy of Silent and Hot Spot Mutations in Importin β Reveals a Dynamic Mechanism for Recognition of a Nuclear Localization Signal. J. Biol. Chem. 278, 16216–16221 (2003).
McGurk, L. et al. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e709 (2018).
Arosio, P., Knowles, T. P. & Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 17, 7606–7618 (2015).
Soniat, M. & Chook, Y. M. Karyopherin-beta2 recognition of a PY-NLS variant that lacks the Proline-Tyrosine Motif. Structure 24, 1802–1809 (2016).
Soniat, M. et al. Crystal structure of human Karyopherin beta2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2. J. Struct. Funct. Genomics 14, 31–35 (2013).
Cansizoglu, A. E., Lee, B. J., Zhang, Z. C., Fontoura, B. M. & Chook, Y. M. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14, 452–454 (2007).
Huber, F. M. & Hoelz, A. Molecular basis for protection of ribosomal protein L4 from cellular degradation. Nat. Commun. 8, 14354 (2017).
Chook, Y. M. & Blobel, G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 399, 230–237 (1999).
Weng, G. et al. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 47, W322–W330 (2019).
Guex, N. & Peitsch, M. C. SWISS‐MODEL and the Swiss‐Pdb Viewer: an environment for comparative protein modeling. electrophoresis 18, 2714–2723 (1997).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Hutten, S. & Dormann, D. A quantitative assay to measure stress granule association of proteins and Peptidesin semi-permeabilized human Cells. Bio Protoc. 10, e3846 (2020).
Acknowledgements
L.G. was supported by Dr. Ralph and Marian Falk Medical Research Trust, Frick Foundation for ALS Research, the National Institute of General Medical Sciences grant R35GM138109, and the National Institute of Neurological Disorders and Stroke grant RF1NS121143. D.T. was supported by NIH grants R21-NS090912 and RF1-AG057882, DoD grant AL220064, Muscular Dystrophy Association grant 628389, the Farber Family Foundation, and the Family Strong for ALS Foundation. G.C. is supported by grants R35GM140733 and R21NS128396. M.E.C was supported by DoD grant W81XWH2110134. C.M.F. was supported by NIH grants T32GM008275 and F31NS111870. J.S. is supported by grants from The Packard Center for ALS Research at Johns Hopkins, Target ALS, The Association for Frontotemporal Degeneration, the Amyotrophic Lateral Sclerosis Association, the Office of the Assistant Secretary of Defense for Health Affairs through the Amyotrophic Lateral Sclerosis Research Program (W81XWH-20-1-0242 and W81XWH-17-1-0237), and NIH grant R01GM099836.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.S., G.C., D.T., and L.G.; methodology, K.M.K., A.G., M.E.C., V.K., M.H., J.L.C., J.S., G.C., D.T., and L.G.; validation, K.M.K., A.G., M.E.C., V.K., and J.L.C.; formal analysis, K.M.K., A.G., M.E.C., V.K., J.L.C., D.T., and L.G.; investigation, K.M.K., A.G., M.E.C., V.K., and J.L.C.; resources, M.H., R.Y., C.M.F., J.S., G.C., D.T., and L.G.; writing – original draft, K.M.K., A.G., M.E.C., J.L.C., and L.G.; writing – review & editing, K.M.K., A.G., M.E.C., M.H., J.L.C., C.M.F., J.S., G.C., D.T., and L.G.; visualization, K.M.K., A.G., M.E.C., J.L.C., and L.G.; supervision, J.S., G.C., D.T., and L.G.; project administration, L.G.; funding acquisition, M.E.C., C.M.F., J.S., G.C., D.T., and L.G.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests, except for: J.S. is a consultant for Dewpoint Therapeutics, ADRx, and Neumora. J.S. a shareholder and advisor at Confluence Therapeutics.
Peer review
Peer review information
Communications Biology thanks Andrew Tosolini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Ibrahim Javed and Benjamin Bessieres.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, K.M., Girdhar, A., Cicardi, M.E. et al. NLS-binding deficient Kapβ2 reduces neurotoxicity via selective interaction with C9orf72-ALS/FTD dipeptide repeats. Commun Biol 8, 2 (2025). https://doi.org/10.1038/s42003-024-07412-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-07412-x