Abstract
Throughout evolution, addition of numerous cyanobacteria-derived subunits to the photosynthetic NDH-1 complex stabilizes the complex and facilitates cyclic electron transfer around photosystem I (PSI CET), a critical antioxidant mechanism for efficient photosynthesis, but its stabilization mechanism remains elusive. Here, a cyanobacteria-derived intermolecular salt bridge is found to form between the two conserved subunits, NdhF1 and NdhD1. Its disruption destabilizes photosynthetic NDH-1 and impairs PSI CET, resulting in the production of more reactive oxygen species under high light conditions. The salt bridge and transmembrane helix 16, both situated at the C-terminus of NdhF1, collaboratively secure the linkage between NdhD1 and NdhB, akin to a cramping mechanism. The linkage is also stabilized by cyanobacteria-derived NdhP and NdhQ subunits, but their stabilization mechanisms are distinctly different. Collectively, to the best of our knowledge, this is the first study to unveil the stabilization mechanism of photosynthetic NDH-1 by incorporating photosynthetic components into its conserved subunits during evolution.

Similar content being viewed by others
Introduction
Cyanobacteria, often referred to as blue-green algae, are widely recognized as the earliest organisms to produce oxygen through photosynthesis on Earth1,2,3. As a consequence, the Earth transitioned from a reduced to an oxidized state, a crucial development for the emergence of complex life4,5. On the contrary, the increase in oxygen levels on Earth inevitably results in the generation of reactive oxygen species (ROS) within cyanobacterial cells, particularly when exposed to environmental stresses. To cope with and defend against the environmental stresses that generate ROS, causing cellular damages6, cyanobacteria have to evolve new antioxidant mechanisms to minimize ROS production or scavenge ROS7. Among these mechanisms, cyclic electron transfer mediated by photosynthetic NDH-1 (NDH-CET) stands out as a crucial antioxidant mechanism. The ATP/NADPH ratio from linear electron transfer is around 1.298, whereas the Calvin-Benson-Bassham (CBB) cycle requires a ratio of 1.59. The lower ATP/NADPH ratio due to ATP shortcut slows down the CBB cycle, causing electron accumulation on the photosystem I (PSI) receptor side and leading to ROS generation through electron-oxygen reactions10. The NDH-CET not only generates additional ATP, but also enhances the ATP to NADPH ratio necessary for improving the CBB cycle, thus reducing ROS production11. As a crucial antioxidant mechanism, the accelerated NDH-CET in response to environmental stresses reduces ROS production, which is essential for the survival of cyanobacteria in aerobic environments12,13,14.
It is reasonable to postulate that maintaining the stability of photosynthetic NDH-1 architecture is a prerequisite for the NDH-CET activity15,16. Consequently, such stabilization plays a crucial role in facilitating the operation of this antioxidant mechanism in cyanobacteria. To adapt to aerobic environment, numerous cyanobacteria-derived subunits have been incorporated into the photosynthetic NDH-1 complex to stabilize its architecture during evolution. The photosynthetic NDH-1 complex is situated in the thylakoid membrane17 and comprises a catalytic domain, a hydrophilic connecting arm, and a membrane-embedded arm, all of which are stabilized by specific cyanobacteria-derived subunits: NdhO, NdhV and NdhS18,19,20,21,22; NdhM and NdhN22,23,24; and NdhL, NdhP and NdhQ25,26,27,28,29, respectively.
In addition to the conserved NDH-1L, three other types of photosynthetic NDH-1 complexes, namely NDH-1L’, NDH-1MS, and NDH-1MS’, have evolved exclusively in cyanobacteria to enable adaptation to various environmental stresses13,30,31,32,33. Among them, NDH-1MS and NDH-1MS’ are involved in CO2 uptake, while NDH-1L and NDH-1L’ are associated with cellular respiration34,35,36,37. All of these four complexes are involved in NDH-CET38 and share a common NDH-1M module, which comprises six cyanobacteria-derived subunits, namely NdhL, NdhM, NdhN, NdhO, NdhS, and NdhV, in addition to nine conserved components: NdhA–NdhC, NdhE, and NdhG–NdhK16,30,32,39. To carry out the NDH-CET function and prevent oxidative stress, the common NDH-1M module has to associate selectively with one of the four variable modules, each of which comprises distinct NdhD and NdhF subunits31,32,33. As a result, the connection between NdhD and NdhB is less stable, making it more susceptible to dissociation into the common NDH-1M module and the variable modules.
The conserved NDH-1L is regarded as the original photosynthetic NDH-1 complex and correspondingly, it may have encountered a broader range of environmental stresses and developed more stabilization mechanisms throughout its evolution. The NdhP and NdhQ subunits derived from cyanobacteria are exclusively found in the conserved NDH-1L, and their inclusion enhances the stability of the NdhD1-NdhB connection28,29. Furthermore, a cyanobacteria-derived intermolecular salt bridge between the two conserved subunits, NdhF1 and NdhD1, has been identified through sequence alignment and structural analysis13,40,41,42. However, the role of photosynthetic components that have been incorporated into the conserved NDH-1 subunits during evolution has not been investigated as of yet.
Here, our data demonstrate that the connection between NdhD1 and NdhB is stabilized by both the cyanobacteria-derived intermolecular salt bridge and the cyanobacteria-derived NdhP and NdhQ subunits, but their mechanisms of stabilization are entirely distinct. Our data further demonstrate that the disruption of their connection impairs NDH-CET activity and boosts ROS production in aerobic environment, particularly under conditions of high light irradiation.
Results
The Asp635-Lys166 salt bridge stabilizes photosynthetic NDH-1
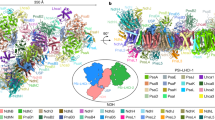
During evolution from archaea to cyanobacteria, a sequence of 25 amino acids has been added to the long horizontal helix of NdhF1 (Fig. 1a and Supplementary Fig. 1). Several recently published structures of photosynthetic NDH-1 have revealed the presence of a cyanobacteria-derived intermolecular salt bridge formed between Asp635, located on a long amphipathic helix of NdhF1, and Lys166 of NdhD1 (as depicted in Fig. 1b, c and Supplementary Fig. 2)13,40,41,42. Considering its position within photosynthetic NDH-1, it is plausible that the Asp635-Lys166 salt bridge contributes to the stabilization of the connection between NdhD1 and NdhB.
a Sequence alignment of a long horizontal (LH) helix of NdhF1 between archaea and cyanobacteria. The LH helix sequence of NdhF1 from the archaea Pyrococcus furiosus (Pyro) was compared with its homologous sequences from the cyanobacteria Thermosynechococcus elongatus BP-1 (Thermo) and Synechocystis sp. stain PCC 6803 (Syn6803). The sequences were aligned using MAFFT version 7. Asterisks indicate identical amino acids; colons and dots indicate conserved amino acid substitutions. b The LH helix of NdhF1 is exhibited in the structures of NDH-1 from the archaea Pyro (PDB: 6U8Y) and the cyanobacteria Thermo (PDB: 6L7O) and Syn6803 (predicted by AlphaFold-Multimer). The LH helix of NdhF1 is highlighted in red and its other parts are displayed as orange; the NdhD1 subunit is highlighted in blue; other NDH-1 subunits are displayed as gray. c A close-up view of the black box in (b). In Syn6803, an intermolecular salt bridge is formed by Asp635 (D635) of a LH helix of NdhF1 with Lys166 (K166) of NdhD1. This salt bridge is formed in Thermo (Supplementary Fig. 2), but is absent in Pyro. Synechocystis 6803 cells were cultured under a light intensity of 40 µmol photons m−2 s−1 with 2% CO2 in air and harvested during the logarithmic growth phase for experiments (d–h). d, e The disruption of Asp635-Lys166 salt bridge had little, if any, effect on the protein abundance of total photosynthetic NDH-1 in the thylakoid membrane. d Lanes were loaded with thylakoid membrane proteins corresponding to 1 µg of chlorophyll a. In the lowest lane, ATPβ was used as a loading control. e Semi-quantitative analysis of thylakoid proteins. Immunoblot results were analyzed using ImageJ software. Protein levels in wild-type (WT) Synechocystis 6803 were set to 100%. f-h The disruption of Asp635-Lys166 salt bridge destabilizes photosynthetic NDH-1. f Thylakoid protein complexes isolated from the wild type Syn6803 and its mutants were separated by BN-PAGE. Thylakoid membrane extract corresponding to 9 µg of chlorophyll a was loaded onto each lane. g Protein complexes were electroblotted to a polyvinylidene difluoride membrane, and the membrane was cross-reacted with anti-NdhI, -NdhK, and -NdhM to probe the assembly of the NDH-1L and NDH-1M complexes. Red and blue arrows indicate the locations of the NDH-1L and NDH-1M complexes, respectively. h Semi-quantitative analysis of thylakoid proteins. Immunoblot results were analyzed using ImageJ software, with protein levels in WT set as 100%. Error bars represent the standard deviations of four independent measurements (n = 4); *** indicates P < 0.001.
To test this hypothesis, we disrupted the Asp635-Lys166 salt bridge by substituting Asp635 using Ala635 (Supplementary Fig. 3). Our data revealed that the disruption had little, if any, effect on the overall protein abundance of photosynthetic NDH-1 in the thylakoid membrane (as shown in Fig. 1d, e). However, it did result in the disassembly of approximately half of the NDH-1L complex into the common NDH-1M module (as depicted in Fig. 1f–h). It is worth noting that under typical growth conditions, i.e., at a light intensity of 40 µmol photons m−2 s−1 and with 2% CO2 in air, NDH-1L emerges as the predominant type of photosynthetic NDH-1. The other three variants—NDH-1L’, NDH-1MS, and NDH-1MS’—are difficult to detect though analysis of the results obtained from blue-native (BN)-PAGE combined with a proteome approach15, supplemented by a western blot method (Supplementary Fig. 4). Collectively, we propose that the cyanobacteria-derived Asp635-Lys166 salt bridge may contribute to the stabilization of the connection between NdhD1 and NdhB.
To validate our proposal, we assessed the in vitro and in vivo activity of NDH-CET using chlorophyll fluorescence analysis and P700 redox kinetics13,43. The reduction of plastoquinone was assessed by measuring the increase in chlorophyll fluorescence emitted from photosystem II (PSII), as shown in Fig. 2a. In vitro ferredoxin-dependent plastoquinone reduction was overwhelmingly driven by photosynthetic NDH-1, with only a marginal contribution from PGR5/PGRL1 (Supplementary Fig. 5). This observation aligns with the NDH-CET and PGR5/PGRL1-CET activity levels reported in cyanobacteria44,45,46,47. As anticipated, both the kinetics and the final reduced level were significantly lowered by the disruption of Asp635-Lys166 salt bridge (Fig. 2b). This conclusion was supported by data from in vivo postillumination rise in chlorophyll a fluorescence (Fig. 2c), re-oxidation rate of P700 by far-red light after termination of actinic light illumination (Fig. 2d) and re-reduction rate of P700+ in the dark after illumination with far-red light in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) (Fig. 2e). Collectively, we conclude that the cyanobacteria-derived Asp635-Lys166 salt bridge serves to stabilize photosynthetic NDH-1 and its disruption impairs NDH-CET activity.
a Schematic model of electron flow from reduced ferredoxin (Fd) by photosystem I (PSI) to plastoquinone (PQ) via photosynthetic NDH-1 in this assay. Synechocystis 6803 cells were cultured under a light intensity of 40 µmol photons m−2 s−1 with 2% CO2 in air and harvested during the logarithmic growth phase for experiments (b–e). b In vitro analysis of NDH-CET activity. Increases in chlorophyll fluorescence by addition of Fd under the illumination of actinic light (AL; 620 nm, 918 µmol photons m−2 s−1) were monitored in thylakoid membranes (80 µg chlorophyll a mL−1) of the wild type (WT) Synechocystis 6803 and its mutants. c–e In vivo analysis of NDH-CET activity. c Monitoring of NDH-CET activity using chlorophyll fluorescence analysis. The top curve shows a typical trace of chlorophyll fluorescence in the WT Synechocystis 6803. Cells were exposed to AL (620 nm; 45 µmol photons m−2 s−1) for 30 s. AL was turned off, and the subsequent change in the chlorophyll fluorescence level was monitored as an indication of NDH-CET activity. d Redox kinetics of P700 after termination of AL illumination (800 µmol photons m−2 s−1) under a background of far-red (FR) light. The cells were illuminated by AL supplemented with FR light to store electrons in the cytoplasmic pool. After termination of AL illumination, P700+ was transiently reduced by electrons from the PQ pool; thereafter, P700 was reoxidized by background FR light. The redox kinetics of P700 were recorded. The P700+ levels were standardized by their maximum levels attained by exposure to FR light. e Kinetics of the P700+ rereduction in darkness after turning off FR light in the presence of 10 µM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). The chlorophyll a concentration was adjusted to 20 µg mL−1 before measurement, and curves are normalized to the maximal signal.
The Asp635-Lys166 salt bridge prevents oxidative stress
To strengthen our conclusion, we measured the CO2 fixation activity and ROS levels under conditions of high light irradiation. As deduced from the uptake rate of CO2, the accumulation of the Rubisco large (RbcL) subunit, and the growth performance of cyanobacterial cells on plates, disrupting the Asp635-Lys166 salt bridge suppressed the CO2 fixation activity under high light conditions, despite a similar level of CO2 fixation activity compared to the wild-type (WT) strain under growth light conditions (Supplementary Fig. 6a–d). As expected, the cyanobacteria-derived Asp635-Lys166 salt bridge improves the Calvin-Benson-Bassham cycle during adaption of cyanobacteria to high light stress.
In line with the results of CO2 fixation, disrupting the Asp635-Lys166 salt bridge significantly elevated intracellular ROS levels under high light irradiation, despite similarly low ROS levels compared to the WT strain under growth light conditions (Supplementary Fig. 7a–d). This inference is based on the fluorescence intensity of 2’,7’-dichlorofluorescein (DCF), an indicator of ROS concentration48. Collectively, the cyanobacteria-derived Asp635-Lys166 salt bridge improves the performance of the Calvin-Benson-Bassham cycle and prevents oxidative stress under high light conditions.
The salt bridge and TM16 of NdhF1 jointly stabilize photosynthetic NDH-1
Sequence alignment, combined with structural data, reveals that the conserved NdhF1 subunit contains 16 transmembrane (TM) helixes and an exceptionally long horizontal (LH) helix (Supplementary Fig. 8). Based on their positioning (Fig. 3a), we propose that the Asp635-Lys166 salt bridge, along with TM16 of NdhF1, collaboratively stabilizes the connection between NdhD1 and NdhB on the stromal side of the thylakoid, facilitated by the LH of NdhF1. To test this idea, we truncated both TM16 and the C-terminal tail of NdhF1 from Synechocystis sp. strain PCC 6803 (hereafter referred to as Synechocystis 6803) (Supplementary Fig. 9), which includes LH and TM16 (Supplementary Fig. 8). Consistent with the disruption of the Asp635-Lys166 salt bridge, truncating TM16 and the C-terminal tail of NdhF1 had little, if any, effect on the overall abundance of photosynthetic NDH-1 in the thylakoid membrane (Fig. 1d and 3b). However, it similarly resulted in the disassembly of approximately half of the NDH-1L complex into the common NDH-1M module, as depicted in Figs. 1f–h and 3c, d. This was corroborated by the results of their NDH-CET activity (Fig. 2 and Supplementary Fig. 10). Collectively, the cyanobacteria-derived salt bridge and TM16 of NdhF1 collaboratively stabilize the connection between NdhB and NdhD1 on the stromal side of the thylakoid, facilitated by the LH of NdhF1, akin to a “cramp iron” structure (as illustrated in Fig. 3e).
a The TM16 of NdhF1 is exhibited in the structure of photosynthetic NDH-1 and is highlighted in blue. Synechocystis 6803 cells were cultured under a light intensity of 40 µmol photons m−2 s−1 with 2% CO2 in air and harvested during the logarithmic growth phase for experiments (b–d). b The deletion of the TM16 of NdhF1 had little, if any, effect on the protein abundance of total photosynthetic NDH-1 in the thylakoid membrane. Lanes were loaded with thylakoid membrane proteins corresponding to 1 µg of chlorophyll a. In the lowest lane, ATPβ was used as a loading control. c, d The deletion of the TM16 of NdhF1 destabilizes photosynthetic NDH-1. c Thylakoid protein complexes isolated from the wild type Synechocystis 6803 and its mutants were separated by BN-PAGE. Thylakoid membrane extract corresponding to 9 µg of chlorophyll a was loaded onto each lane. d Protein complexes were electroblotted to a polyvinylidene difluoride membrane, and the membrane was cross-reacted with anti-NdhI, -NdhK, and -NdhM to probe the assembly of the NDH-1L and NDH-1M complexes. Red arrows indicate the locations of the NDH-1L complex with the full-length NdhF1 subunit and the NdhF1 truncation, while blue arrows denote the locations of the NDH-1M complex. e A model schematically represents the cramp iron-like structure. This structure consists of salt bridge, TM16 and a part of C-terminus of NdhF1 and stabilizes the connection of NdhD1 with NdhB.
In bacterial studies, it was found that the deletion of the C-terminal tail of NdhF1, and even its truncated TM16, led to the complete absence of respiratory NDH-149,50,51,52. Conversely, in cyanobacteria, deleting the entire NdhF1 resulted in the disassembly of approximately half of the NDH-1LΔNdhF1 complex into the common NDH-1M module (Fig. 3c, d). This NDH-1LΔNdhF1 complex comprises the common NDH-1M module and a partially compromised variable module, potentially containing the NdhD1, NdhP, and NdhQ subunits. In addition, the intermolecular salt bridge derived from cyanobacteria is not present in bacteria and archaea49,50,52,53 but is conserved in photosynthetic NDH-154,55 (Supplementary Figs. 11 and 12). Sequence alignment, along with structural data, demonstrates that TM16 of NdhF1 is also conserved in photosynthetic NDH-154,55 (Supplementary Fig. 13). Hence, the “cramp iron” mechanism has evolved exclusively to stabilize the connection between NdhD1 and NdhB in photosynthetic NDH-1, preventing oxidative stress necessary for the survival of cyanobacteria in aerobic environment and, ultimately, contributing to the evolution of complex organisms on Earth.
The salt bridge and NdhP, Q jointly stabilize photosynthetic NDH-1
Consistent with the disruption of the Asp635-Lys166 salt bridge, the deletion of individual NdhP and NdhQ subunits resulted in the disassembly of approximately half of the NDH-1L complex into the common NDH-1M module28,29. Given the similar destabilization outcomes and their positions (Fig. 4a, b), we postulate that they collaboratively contribute to the stabilization of photosynthetic NDH-1. To test this possibility, we substituted the Asp635 of NdhF1 using Ala635 in the NdhP or NdhQ-deletion mutant (Supplementary Fig. 14). Our data revealed that disrupting the Asp635-Lys166 salt bridge in the NdhP or NdhQ-deletion mutant had little, if any, effect on the total protein abundance of photosynthetic NDH-1 in the thylakoid membrane (Supplementary Fig. 15). However, it resulted in almost complete disassembly of the NDH-1L complex into the common NDH-1M module (Fig. 4c, d and Supplementary Fig. 16), just like the NdhP and NdhQ double deletion mutant29. Notably, the upper band observed in Fig. 4d does not correspond to NDH-1L and NDH-1L’ complexes, as it remains detectable in the NdhD1 and NdhD2 double deletion mutant. Furthermore, it is not associated with NDH-1MS or NDH-1MS’ complex, as antibodies against CupA and CupB failed to detect any signal (Supplementary Fig. 17). These findings suggest that the upper band represents a distinct entity, unrelated to the four known NDH-1 complexes, warranting further investigation. In conclusion, the Asp635-Lys166 salt bridge, along with NdhP and NdhQ, collectively contribute to the stabilization of photosynthetic NDH-1.
a The NdhP and NdhQ subunits are exhibited in the predicted structures of photosynthetic NDH-1 in Synechocystis 6803 by AlphaFold-Multimer. NdhP and NdhQ are highlighted in deep sky blue and red, respectively, and NdhD1 is displayed as blue. b A close-up view of the black box in (a) shows that the NdhP and NdhQ subunits exclusively interact with the NdhD1 subunit. Synechocystis 6803 cells were cultured under a light intensity of 40 µmol photons m−2 s−1 with 2% CO2 in air and harvested during the logarithmic growth phase for experiments (c, d). c, d The disruption of the salt bridge in the NdhP or NdhQ-deletion mutant destabilizes photosynthetic NDH-1. c Thylakoid protein complexes isolated from the wild type (WT) Synechocystis 6803 and its double mutants were separated by BN-PAGE. Thylakoid membrane extract corresponding to 9 µg of chlorophyll a was loaded onto each lane. d Protein complexes were electroblotted to a polyvinylidene difluoride membrane, and the membrane was cross-reacted with anti-NdhI, -NdhK, and -NdhM to probe the assembly of the NDH-1L and NDH-1M complexes. Red and blue arrows indicate the locations of the NDH-1L and NDH-1M complexes, respectively.
To confirm our conclusion, we assessed NDH-CET activity in vitro and in vivo through chlorophyll fluorescence and P700 analyses13,43. Compared to their individual deletion mutants, disrupting the Asp635-Lys166 salt bridge in the NdhP or NdhQ-deletion mutant significantly diminished NDH-CET activity. This is evident from the experimental data of in vitro ferredoxin-dependent plastoquinone reduction activity (Fig. 5a and Supplementary Fig. 18), the in vivo postillumination rise in chlorophyll a fluorescence (Fig. 5b)28,29, re-oxidation rate of P700 by far-red light after the termination of actinic light illumination (Fig. 5c)28,29, and the re-reduction rate of P700+ in the dark after illumination with far-red light in the presence of DCMU (Fig. 5d)28,29. The physiological data provide further support for our conclusion that the Asp635-Lys166 salt bridge, along with NdhP and NdhQ, jointly stabilizes the connection between NdhD1 and NdhB in photosynthetic NDH-1.
Synechocystis 6803 cells were cultured under a light intensity of 40 µmol photons m−2 s−1 with 2% CO2 in air and harvested during the logarithmic growth phase for experiments (a–d). a In vitro analysis of NDH-CET activity. Increases in chlorophyll fluorescence by addition of Fd under the illumination of actinic light (AL; 620 nm, 918 µmol photons m−2 s−1) were monitored in thylakoid membranes (80 µg chlorophyll a mL−1) of the wild type (WT) Synechocystis 6803 and its mutants. b–d In vivo analysis of NDH-CET activity. b Monitoring of NDH-CET activity by chlorophyll fluorescence. Experimental procedure as in Fig. 2c. a.u., Arbitrary units. c Redox kinetics of P700 after termination of AL illumination (800 µmol photons m−2 s−1) under a background of far-red (FR) light. The cells were illuminated by AL supplemented with FR light to store electrons in the cytoplasmic pool. After termination of AL illumination, P700+ was transiently reduced by electrons from the PQ pool; thereafter, P700 was reoxidized by background FR light. The redox kinetics of P700 were recorded. The P700+ levels were standardized by their maximum levels attained by exposure to FR light. d Kinetics of the P700+ rereduction in darkness after turning off FR light in the presence of 10 µM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). The chlorophyll a concentration was adjusted to 20 µg mL−1 before measurement, and curves are normalized to the maximal signal.
The salt bridge and NdhP, Q jointly prevent oxidative stress
To confirm our conclusion further, we assessed CO2 fixation activity and ROS levels under high light irradiation. Compared to their individual deletion mutants, disrupting the Asp635-Lys166 salt bridge in the NdhP or NdhQ-deletion mutant significantly aggravated the suppression of the CO2 fixation activity under high light conditions. This effect was observed despite a similar level of CO2 fixation activity under growth light conditions when compared to the WT strain (Supplementary Figs. 19 and 20). Hence, the Asp635-Lys166 salt bridge, along with NdhP and NdhQ, collaboratively enhances the Calvin-Benson-Bassham cycle during the adaption of cyanobacterial cells to high light stress.
Consistent with the CO2 fixation results, disrupting the Asp635-Lys166 salt bridge in the NdhP or NdhQ-deletion mutant significantly raised intracellular ROS levels under high light irradiation. This effect was observed despite a similarly low level of ROS under growth light conditions compared to the WT strain, as indicated by DCF fluorescence levels (Supplementary Figs. 21 and 22). Collectively, the Asp635-Lys166 salt bridge, along with NdhP and NdhQ, jointly stabilizes photosynthetic NDH-1 and mitigates oxidative stress under high light conditions.
Discussion
It has been proposed that NDH-1L is a main type of photosynthetic NDH-1 during the early evolution of cyanobacteria32,56. The NDH-1L consists of the common NDH-1M module and a variable module that contains NdhD1 and NdhF1 subunits31,32,33. The connection between NdhB and NdhD1, situated between the common and variable modules, is particularly susceptible to dissociation. As a result, to facilitate the newly-developed antioxidant mechanism and adapt to aerobic environment, two cyanobacteria-derived subunits, NdhP and NdhQ, were incorporated into the photosynthetic NDH-1L complex during the early evolution of cyanobacteria. These subunits play a crucial role in stabilizing the connection between NdhD1 and NdhB.
Structural data show that the NdhP and NdhQ subunits are present in NDH-1L but absent in NDH-1MS13,57. Our functional data further demonstrate that NdhP and NdhQ collectively enhance the stability of the connection between the common module NDH-1M and the variable module containing NdhD1 and NdhF1 subunits28,29. In line with this structural and functional evidence, the NDH-1MS complex, as compared to the NDH-1L complex, readily disassembles into the common module NDH-1M and the variable module NDH-1S, which contains NdhD3 and NdhF315,58,59. This is supported by amino acid interaction analysis using RING v3.0, revealing that NDH-1L possesses a greater number of binding sites at the interface of NdhD with NdhB compared to NDH-1MS (Supplementary Fig. 23 and Tables 2 and 3)13,57. Together, NdhP and NdhQ reinforce the connection between NdhB and NdhD1 by increasing the binding sites at their interface (Supplementary Fig. 23 and Tables 2 and 3).
Prior to the incorporation of NdhP and NdhQ into photosynthetic NDH-1, we observed the addition of photosynthetic components to its 11 conserved subunits, including NdhF1 (Fig. 1a and Supplementary Fig. 1), during the early evolution of cyanobacteria. This was determined through sequence alignment and structural analysis. Whether they can indeed stabilize photosynthetic NDH-1 remains a mystery. Here, our data revealed that a cyanobacteria-derived intermolecular salt bridge, formed between the two conserved subunits NdhF1 and NdhD1, stabilizes the connection of NdhD1 with NdhB (Figs. 1, 2 and Supplementary Figs. 6, 7). To the best of our knowledge, this study is the first to explore the role of photosynthetic components incorporated into conserved NDH-1 subunits during evolution, opening a new avenue for investigating the stability mechanisms of photosynthetic NDH-1 over evolutionary time.
Here, we demonstrate that the cyanobacteria-derived intermolecular salt bridge, TM16, and LH of NdhF1 create a cramp iron-like structure at the stroma side of the thylakoid (Fig. 3a, e), essential for stabilizing the connection between NdhB and NdhD1 in photosynthetic NDH-1 (Fig. 3c, d). Indeed, it is quite intriguing that the disruption of this cramp iron-like structure does not cause an upward shift in the remaining NDH-1L band, whereas the deletion of NdhP and/or NdhQ does induce such a shift (Figs. 1f, g, 3c, d, and 4c, d). This serves as a reminder that NdhP and NdhQ play a role in augmenting the binding sites between NdhB and NdhD1, potentially stabilizing the conformation of NdhD1 and enhancing the cohesive interaction between NdhB and NdhD1, resembling a “molecular glue”. Collectively, the mechanism by which this salt bridge stabilizes NDH-1L is fundamentally distinct from that of NdhP and NdhQ, despite their collaborative role in stabilizing the interaction between NdhD1 and NdhB. To our knowledge, the “cramp iron” stabilization mechanism of photosynthetic NDH-1 has been first developed in cyanobacteria and is conserved among photosynthetic organisms.
When cyanobacterial cells are exposed to high light, there is an increased rate of cyclic electron transfer facilitated by the photosynthetic NDH-1 complex. Disruption of cyanobacteria-derived intermolecular salt bridge can destabilize this complex, potentially prolonging the lifetime of a semi-quinone anion intermediate at the quinone reduction site40,42. These semi-quinone anion species have been proposed to produce ROS in both the cytochrome b6f (cytb6f) complex60 and mitochondrial complex I61. Consequently, we hypothesize that ROS may accumulate in the destabilized form of the photosynthetic NDH-1 complex, especially under high light conditions. In support of this hypothesis, the NdhF1D635A mutant exposed to high light exhibited greater degradation of the photosynthetic NDH-1 complex compared to when they were exposed to growth light28,29 (Fig. 1f–h; Supplementary Fig. 24).
In summary, our functional data presented here, combined with previously reported structural data, shed light on a photosynthetic NDH-1-mediated antioxidant defense mechanism that emerged during the early evolution of cyanobacteria. In this model, the connection between the common module NDH-1M and the variable module containing NdhD1 and NdhF1 is strengthened by the cramp iron-like structure, which includes a cyanobacteria-derived intermolecular salt bridge (highlighted in yellow in Fig. 6), as well as the cohesive interaction resembling “molecular glue” induced by NdhP and NdhQ at the interface between NdhD1 and NdhB (illustrated in blue and red lines in Fig. 6). The enhanced ATP production is facilitated by the stabilized photosynthetic NDH-1-mediated PSI CET (NDH-CET), which serves to maintain the ATP/NADPH ratio, optimize the Calvin-Benson-Bassham cycle and mitigate the ROS production under high light conditions (Figs. 2, 5 and 6 and Supplementary Figs. 6, 7, and 19–22). We posit that the functioning of this antioxidant mechanism is essential for the survival of early cyanobacteria in an aerobic environment, ultimately paving the way for the evolution of complex organisms on Earth.
The stability of the connection between NdhB and NdhD1 relies on the formation of an intermolecular salt bridge within the cramp iron-like structure, facilitated by the functional “molecular glue” provided by NdhP and NdhQ (see WT). In contrast, disruption of the salt bridge and deletion of NdhP or NdhQ destabilizes NDH-1L, leading to impaired NDH-CET, thereby suppressing the CBB cycle and increasing ROS production, especially under high light (see F1D635A/ΔP and F1D635A/ΔQ). CBB, Calvin-Benson-Bassham; NDH-CET, NDH-1-depedent cyclic electron transfer; ROS, reactive oxygen species; TM, thylakoid membrane.
Methods
Culture conditions
A glucose-tolerant strain of the wild-type (WT) Synechocystis 6803 and its mutants, ∆ndhF1, ndhF1∆C, ndhF1∆TM16, NdhF1D635A, NdhF1D635A/∆ndhP (F1D635A/∆P), NdhF1D635A/∆ndhQ (F1D635A/∆Q), ∆ndhP/Q (∆P/Q)29, ∆ndhD1/D2 (∆D1/D2)34, and ∆ndhB (M55)62, were cultured at 30°C in BG-11 medium, as described by Allen in 196863, with Tris-HCl buffering at a concentration of 5 mM and a pH of 8.0. The culture was aerated with a 2% (v/v) mixture of CO2 in air. The solid medium used was BG-11, which was supplemented with 1.5% agar. The mutant strains were cultured in the presence of specific antibiotics, while being exposed to illumination by fluorescence lamps at 40 µmol photons m–2 s–1.
Construction of mutants
The mutant strains of ∆ndhF1, ndhF1∆C, and ndhF1∆TM16 were constructed as follows. The upstream and downstream regions of ndhF1, ndhF1N+15TMs or ndhF1N+15TMs+LH (Supplementary Fig. 8) were amplified by PCR, creating appropriate restriction sites. A DNA fragment encoding a KamR or a SpR cassette was also amplified by PCR, creating the BamHI and SalI sites using appropriate PCR primers (Supplementary Table 1). These three PCR products were ligated into the MCS of pUC19 (see Supplementary Fig. 9a-d) and used to transform the wild-type cells of Synechocystis 6803 to generate the ∆ndhF1, ndhF1∆C or ndhF1∆TM16 mutant, respectively. Subsequently, these transformants were spread on agar plates containing BG-11 medium and kanamycin (10 µg mL–1) or spectinomycin (10 µg mL–1) buffered at pH 8.0, and the plates were incubated in 2% (v/v) CO2 in air under illumination by fluorescent lamps at 40 µmol photons m–2 s–1. The complete or partial mutation regions of the ndhF1 gene in the transformants was segregated to homogeneity through successive streak purification, as confirmed by PCR amplification (see Supplementary Fig. 9e) and reverse transcription (RT)-PCR analysis (see Supplementary Fig. 9f).
The construction of strain NdhF1D635A with a site-directed mutation proceeded as follows. The DNA fragment encoding a KamR cassette was amplified by PCR using primers NdhF1D635A-E and NdhF1D635A-F, while another DNA fragment containing a downstream region of the ndhF1 gene was amplified using primers NdhF1D635A-G and NdhF1D635A-H. Both resulting DNA fragments were seamlessly assembled into the pUC19 vector, creating the pUC-KamR-Down plasmid with BamHI and KpnI sites, using the pEASY ®-Uni seamless cloning and assembly kit (Transgene, CU101). Furthermore, the upstream region of the ndhF1 gene consists of two distinct DNA fragments, referred to as fragments far away from and close to the KamR cassette. Two DNA fragments were amplified by PCR for assembly into the pUC-KamR-Down plasmid. The first fragment, located far away from the KamR cassette, was amplified using primers NdhF1D635A-A and NdhF1D635A-B. The second fragment, situated close to the KamR cassette, was amplified using primers NdhF1D635A-C and NdhF1D635A-D. These fragments were seamlessly assembled, resulting in the creation of pUC-NdhF1D635A plasmid with PstI and BamHI sites, using the pEASY®-Uni seamless cloning and assembly kit (Transgene, CU101). Within the upstream region, the triplet 1903GAC1905, which encodes aspartic acid, was modified to GCC, encoding alanine, using PCR primers NdhF1D635A-B, and NdhF1D635A-C (see red marks in Supplementary Table 1). The accuracy of the base changes at the mutation site was confirmed by DNA sequencing of the upstream region. This construct was then introduced into the WT Synechocystis 6803 to produce the NdhF1D635A mutant strain. The ndhF1 gene was amplified by PCR from both the WT and NdhF1D635A mutant strains and the correctness of the base modifications at the mutation site was validated through DNA sequencing (Supplementary Fig. 3c).
The F1D635A/∆P and F1D635A/∆Q double mutants were constructed as follows. The upstream and downstream regions of ndhP and ndhQ were amplified by PCR, creating appropriate restriction sites. A DNA fragment encoding a KamR or SpR cassette was also amplified via PCR, thereby introducing XbaI and KpnI sites for ΔndhP construction, and BamHI and SacI sites for ΔndhQ construction, using the corresponding PCR primers (Supplementary Table 1). They were ligated into the MCS of pUC19 to generate the pUC-ΔndhP and pUC-ΔndhQ plastids. These two plastids were introduced into the NdhF1D635A mutant strain, resulting in the generation of F1D635A/∆P and F1D635A/∆Q, respectively.
RNA extraction and RT-PCR analysis
Total RNA was isolated and analyzed as described previously64. Reverse transcription (RT)-PCR was performed using the Access RT-PCR system (Promega) to generate products corresponding to ndhF1, ndhF1N+15TMs, ndhF1C, ndhF1TM16, ndhF1LH, ndhP, ndhQ and 16 S rRNA, with 0.5 µg of DNase-treated total RNA as starting material. RT-PCR conditions were 95 °C for 5 min followed by cycles of 95°C, 62°C, and 72 °C for 30 s each. The reactions were stopped after 25 cycles for 16 S rRNA, and after 35 cycles for ndhF1, ndhF1N + 15TMs, ndhF1C, ndhF1TM16, ndhF1LH, ndhP and ndhQ. The primers used are summarized in Supplementary Table 1.
Isolation of crude thylakoid membranes
The cell cultures (800 mL) were collected during the logarithmic growth phase (A730 = 0.6-0.8) and subjected to two washes with 50 mL of fresh BG-11 medium. Subsequently, thylakoid membranes were isolated in accordance with the method outlined by Gombos et al. 65, with some modifications as described below. The cells suspended in 5 ml of disruption buffer (10 mM HEPES-NaOH, 5 mM sodium phosphate, pH 7.5, 10 mM MgCl2, 10 mM NaCl, and 25% [v/v] glycerol) were supplemented by zirconia/silica beads and broken by vortexing 20 times at the highest speed for 30 s at 4 °C with 5 min cooling on ice between the runs. The crude extract was centrifuged at 5,000 × g for 5 min to remove the glass beads and unbroken cells. By further centrifugation at 20,000 × g for 30 min, crude thylakoid membranes were obtained from the precipitation.
Electrophoresis and immunoblotting
Blue native PAGE (BN-PAGE) of Synechocystis 6803 membranes was performed as described previously66 with slight modifications12,21,28,29,31,67,68,69,70,71. Isolated membranes were prepared for BN-PAGE as follows. Membranes were washed with 330 mM sorbitol, 50 mM Bis-Tris, pH 7.0, and 0.5 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO, USA) and resuspended in 20% (w/v) glycerol, 25 mM Bis-Tris, pH 7.0, 10 mM MgCl2, 0.1 units of RNase-free DNase RQ1 (Promega, Madison, WI, USA) at a chlorophyll a concentration of 0.3 mg mL−1, and 0.5 mM phenylmethylsulfonyl fluoride. The samples were incubated on ice for 10 min, and an equal volume of 3% n-dodecyl β-D-maltoside (DM) was added. Solubilization was performed for 40 min on ice. Insoluble components were removed by centrifugation at 18,000 × g for 15 min. The collected supernatant was mixed with a one-tenth volume of sample buffer, 5% Serva Blue G, 100 mM Bis-Tris, pH 7.0, 30% (w/v) Sucrose, 500 mM ε-amino-n-caproic acid, and 10 mM EDTA. Solubilized membranes were then applied to a 0.75 mm-thick, 5% to 12.5% acrylamide gradient gel (Hoefer Mighty Small mini-vertical unit, Hoefer, San Francisco, CA, USA). Samples were loaded on an equal chlorophyll a basis per lane. Electrophoresis was performed at 4 °C by increasing the voltage gradually from 50 up to 200 V during the 5.5 h run.
SDS-PAGE analysis of Synechocystis 6803 proteins from crude thylakoid membranes was conducted using a 12% polyacrylamide gel containing 6 M urea, following the procedure outlined by Laemmli in 197072. After electrophoresis, the proteins were visualized by Coomassie Brilliant Blue (CBB) staining.
For immunoblotting, the proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore, Bedford, MA, USA) and detected by protein-specific antibodies using an ECL assay kit (Amersham, Arlington Heights, IL, USA) according to the manufacturer’s protocol. An antibody against amino acids 452 to 509 of the NdhF1 subunit of Synechocystis 6803 (1000:1) was raised in our laboratory. This region corresponds to a hydrophilic segment located between TM13 and TM14 (Supplementary Fig. 8). To amplify this segment of the ndhF1 gene, primers were designed (listed in Supplementary Table 1) and the PCR products were ligated into vector pGEX-5X-1. Following amplification in Escherichia coli DH-5α. The construct was used to transform E. coli strain BL21(DE3)pLysS for expression. The resulting partial ndhF1 gene expression product from E. coli was purified and utilized as antigens for immunizing rabbits to generate polyclonal antibody. Antibodies against NdhA (1000:1), NdhI (1500:1), NdhK (1000:1), NdhM (1000:1), RbcL (1500:1) and ATPβ (1500:1) subunits of Synechocystis 6803 were raised previously in our laboratory68,71,73. Antibodies against CupA (2000:1) and CupB (2000:1) were provided from Professor Hualing Mi (Institutes of Plant Physiology and Ecology, Chinese Academy of Sciences).
In vitro assay of Fd-dependent PQ reduction
In vitro assay of Fd-dependent PQ reduction was performed as described previously13,45 with some modifications. In brief, the thylakoid membranes collected from the WT Synechocystis 6803 were suspended in buffer A (0.5 M sorbitol, 10 mM MgCl2, 10 mM NaCl, 10 mM HEPES, and 5 mM sodium phosphate, pH 7.5) at a final chlorophyll a concentration of 80 µg mL−1. Purified Fd (20 µM) was added, and an increase in chlorophyll fluorescence was recorded using a Dual-PAM-100 system (Walz).
Chlorophyll fluorescence and P700 analysis
The transient increase in chlorophyll fluorescence after actinic light had been turned off was monitored as described73. The redox kinetics of P700 was measured according to previously described methods19,21,28,29,67,68,69,70. Briefly, the levels of P700+ were assessed by monitoring absorbance at 820 nm using a Dual-PAM-100 instrument (Walz) equipped with an ED-101US/MD emitter-detector unit. P700 redox kinetics were tracked by turning off actinic red light (AL; 800 µmol photons m−2 s−1 for 30 s) while keeping far-red light (FR; >705 nm; 5.2 µmol photons m−2 s−1) on. The re-reduction of P700+ in darkness was measured with a Dual-PAM-100 (Walz) with an emitter-detector unit ED-101US/MD by monitoring absorbance changes at 830 nm and using 875 nm as a reference. Cells were kept in the dark for 2 min and 10 µM of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added to the cultures prior to the measurement. The P700 was oxidized by FR light with a maximum at 720 nm from a light-emitting diode (LED) lamp for 30 s, and the subsequent re-reduction of P700+ in the dark was monitored.
CO2 gas exchange kinetics measurement
Cells of WT and mutant strains were suspended in fresh BG-11 medium at a chlorophyll a concentration of 10 µg mL–1. The cell suspension was kept in the dark for 5 min and then illuminated with an intensity of 500 µmol photons m–2 s–1 for 5 min. CO2 gas exchange kinetics were monitored using an online membrane inlet mass spectrometer (MIMS, Omnistar, Pfeiffer Vacuum) and the CO2 uptake rate was subsequently calculated as described previously14,74.
ROS measurement
Cells of WT and mutant strains are grown under growth light and high light were stained with fluorescence dye, 2’,7’-dichlorodi-hydrofluorescein diacetate (DCFH-DA) (Sigma-Aldrich) as described previously14,75,76. After the addition of 5 µM DCFH-DA (final concentration) to the cell suspension, the mixture was incubated in the dark on a shaker at room temperature for 1 h. Subsequently, intracellular ROS fluorescence was measured using an F4500 spectrofluorimeter (Hitachi) and analyzed by comparing the intensity of fluorescence peak at 520 nm.
Homology modeling of NDH-1 and its amino acid interaction analysis
The spatial structure of the NDH-1L complex from Synechocystis 6803 was modeled using AlphaFold-Multimer77,78. Amino acid interactions between NdhB and NdhD subunits were analyzed for both NDH-1L and NDH-1MS structures using the freely available RING v3.079 webserver, accessible at https://ring.biocomputingup.it/submit.
Statistics and reproducibility
A minimum of three independent experiments were conducted for all studies, with specific replicate numbers (n) provided in the figure legends. Statistical analyses were carried out using Windows Excel. Significance between two groups was assessed using a two-tailed Student’s t-test and a P-value <0.05 was considered statistically significant. The data were presented as the mean ± standard deviation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analyzed during this study have been included in both the published article and its supplementary information files. The numerical source data for all graphs are presented in the Supplementary Data 1. In addition, original uncropped blot/gel images, including the original size marker, have been made available in Supplementary Fig. 25. For more information, feel free to reach out to the corresponding authors upon a reasonable request.
References
Kasting, J. F. & Siefert, J. L. Life and the evolution of Earth’s atmosphere. Science 296, 1066–1068 (2002).
Lyons, T. W., Reinhard, C. T. & Planavsky, N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014).
Flamholz, A. & Shih, P. M. Cell biology of photosynthesis over geologic time. Curr. Biol. 30, R490–R494 (2020).
Brocks, J. J., Logan, G. A., Buick, R. & Summons, R. E. Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036 (1999).
Catling, D. C., Glein, C. R., Zahnle, K. J. & McKay, C. P. Why O2 is required by complex life on habitable planets and the concept of planetary “oxygenation time”. Astrobiology 5, 415–438 (2005).
Raymond, J. & Segre, D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767 (2006).
Latifi, A., Ruiz, M. & Zhang, C. C. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278 (2009).
Sacksteder, C. A., Kanazawa, A., Jacoby, M. E. & Kramer, D. M. The proton to electron stoichiometry of steady-state photosynthesis in living plants: a proton-pumping Q cycle is continuously engaged. Proc. Natl. Acad. Sci. USA 97, 14283–14288 (2000).
Walker, B. J., VanLoocke, A., Bernacchi, C. J. & Ort, D. R. The costs of photorespiration to food production now and in the future. Ann. Rev. Plant Biol. 67, 107–129 (2016).
Asada, K. & Takahashi, M. Production and scavenging of active oxygen photosynthesis. In Photoinhibition. Topics in photosynthesis. 9. (eds. Kyle, D. J., Osmond, C. B., & Arntzen, C. J.) 227–287 (Elsevier, 1987).
Kramer, D. M. & Evans, J. R. The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 155, 70–78 (2011).
Zhao, J., Gao, F., Fan, D. Y., Chow, W. S. & Ma, W. NDH-1 is important for photosystem I function of Synechocystis sp. strain PCC 6803 under environmental stress conditions. Front. Plant Sci. 8, 2183 (2018).
Zhang, C. et al. Structural insights into NDH-1 mediated cyclic electron transfer. Nat. Commun. 11, 888 (2020).
Zhao, J. et al. New insights into the effect of NdhO levels on cyanobacterial cell death triggered by high temperature. Funct. Plant Biol. 49, 533–541 (2022).
Zhang, P. et al. Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp. PCC 6803. Plant Cell 16, 3326–3340 (2004).
Ma, W. & Ogawa, T. Oxygenic photosynthesis-specific subunits of cyanobacterial NADPH dehydrogenases. IUBMB Life 67, 3–8 (2015).
Ohkawa, H., Sonoda, M., Shibata, M. & Ogawa, T. Localization of NAD (P) H dehydrogenase in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183, 4938–4939 (2001).
Battchikova, N., Zhang, P., Rudd, S., Ogawa, T. & Aro, E. M. Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 280, 2587–2595 (2005).
Battchikova, N. et al. Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 286, 36992–37001 (2011).
Chen, X., He, Z., Xu, M., Peng, L. & Mi, H. NdhV subunit regulates the activity of type-1 NAD(P)H dehydrogenase under high light conditions in cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 6, 28361 (2016).
Gao, F. et al. NdhV is a subunit of NADPH dehydrogenase essential for cyclic electron transport in Synechocystis sp. strain PCC 6803. Plant Physiol. 170, 752–760 (2016).
He, Z. & Mi, H. Functional characterization of the subunits N, H, J, and O of the NAD(P)H dehydrogenase complexes in Synechocystis sp. strain PCC 6803. Plant Physiol. 171, 1320–1332 (2016).
Prommeenate, P., Lennon, A. M., Markert, C., Hippler, M. & Nixon, P. J. Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J. Biol. Chem. 279, 28165–28173 (2004).
He, Z. et al. NdhM subunit is required for the stability and the function of NAD(P)H dehydrogenase complexes involved in CO2 uptake in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 291, 5902–5912 (2016).
Ogawa, T. Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol. 99, 1604–1608 (1992).
Nowaczyk, M. M. et al. NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 complex. Biochemistry 50, 1121–1124 (2011).
Schwarz, D., Schubert, H., Georg, J., Hess, W. R. & Hagemann, M. The gene sml0013 of Synechocystis species strain PCC 6803 encodes for a novel subunit of the NAD(P)H oxidoreductase or complex I that is ubiquitously distributed among cyanobacteria. Plant Physiol. 163, 1191–1202 (2013).
Zhang, J. et al. NdhP is an exclusive subunit of large complex of NADPH dehydrogenase essential to stabilize the complex in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 289, 18770–18781 (2014).
Zhao, J., Rong, W., Gao, F., Ogawa, T. & Ma, W. Subunit Q is required to stabilize the large complex of NADPH dehydrogenase in Synechocystis sp. strain PCC 6803. Plant Physiol. 168, 443–451 (2015).
Arteni, A. A. et al. Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim. Biophys. Acta 1757, 1469–1475 (2006).
Battchikova, N., Eisenhut, M. & Aro, E. M. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta 1807, 935–944 (2011).
Peltier, G., Aro, E. M. & Shikanai, T. NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu. Rev. Plant Biol. 67, 55–80 (2016).
Laughlin, T. G., Savage, D. F. & Davies, K. M. Recent advances on the structure and function of NDH-1: the complex I of oxygenic photosynthesis. Biochim. Biophys. Acta 1861, 148254 (2020).
Ohkawa, H., Pakrasi, H. B. & Ogawa, T. Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J. Biol. Chem. 275, 31630–31634 (2000).
Ogawa, T. & Mi, H. Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93, 69–77 (2007).
Artier, J. et al. Modeling and mutagenesis of amino acid residues critical for CO2 hydration by specialized NDH-1 complexes in cyanobacteria. Biochim. Biophys. Acta 1863, 148503 (2022).
Walker, R. M., Zhang, M. & Burnap, R. L. Elucidating the role of primary and secondary sphere Zn2+ ligands in the cyanobacterial CO2 uptake complex NDH-14: the essentiality of arginine in zinc coordination and catalysis. Biochim. Biophys. Acta 1865, 149149 (2024).
Bernát, G., Appel, J., Ogawa, T. & Rögner, M. Distinct roles of multiple NDH-1 complexes in the cyanobacterial electron transport network as revealed by kinetic analysis of P700+ reduction in various Ndh-deficient mutants of Synechocystis sp. strain PCC6803. J. Bacteriol. 193, 292–295 (2011).
Birungi, M. et al. Possibilities of subunit localization with fluorescent protein tags and electron microscopy examplified by a cyanobacterial NDH-1 study. Biochim. Biophys. Acta 1797, 1681–1686 (2010).
Laughlin, T. G., Bayne, A. N., Trempe, J. F., Savage, D. F. & Davies, K. M. Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–414 (2019).
Schuller, J. M. et al. Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363, 257–260 (2019).
Pan, X. et al. Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase. Nat. Commun. 11, 610 (2020).
Munekage, Y. et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429, 579–582 (2004).
Mi, H., Endo, T., Schreiber, U., Ogawa, T. & Asada, K. Electron donation from cyclic and respiratory flows to the photosynthetic intersystem chain is mediated by pyridine nudeotide dehydrogenase in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 33, 1233–1237 (1992).
Mi, H., Endo, T., Ogawa, T. & Asada, K. Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 36, 661–668 (1995).
Yeremenko, N. et al. Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 46, 1433–1436 (2005).
Dann, M. & Leister, D. Evidence that cyanobacterial Sll1217 functions analogously to PGRL1 in enhancing PGR5-dependent cyclic electron flow. Nat. Commun. 10, 5299 (2019).
Keston, A. S. & Brandt, R. The fluorometric analysis of ultramicro quantities of hydrogen peroxide. Anal. Biochem. 11, 1–5 (1965).
Efremov, R. G., Baradaran, R. & Sazanov, L. A. The architecture of respiratory complex I. Nature 465, 441–445 (2010).
Efremov, R. G. & Sazanov, L. A. Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011).
Torres-Bacete, J., Sinha, P. K., Matsuno-Yagi, A. & Yagi, T. Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J. Biol. Chem. 286, 34007–34014 (2011).
Baradaran, R., Berrisford, J. M., Minhas, G. S. & Sazanov, L. A. Crystal structure of the entire respiratory complex I. Nature 494, 443–448 (2013).
Yu, H. et al. Structure of the respiratory MBS complex reveals iron-sulfur cluster catalyzed sulfane sulfur reduction in ancient life. Nat. Commun. 11, 5953 (2020).
Shen, L. et al. Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare. Nature 601, 649–654 (2022).
Su, X. et al. Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana. Mol. Plant 15, 454–467 (2022).
Brandt, U. Adaptations of an ancient modular machine. Science 363, 230–231 (2019).
Schuller, J. M. et al. Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I. Nat. Commun. 11, 494 (2020).
Herranen, M. et al. Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134, 470–481 (2004).
Zhang, P. et al. Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem. J. 390, 513–520 (2005).
Malone, L. A., Proctor, M. S., Hitchcock, A., Hunter, C. N. & Johnson, M. P. Cytochrome b6f–orchestrator of photosynthetic electron transfer. Biochim. Biophys. Acta 1862, 148380 (2021).
Cochemé, H. M. & Murphy, M. P. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283, 1786–1798 (2008).
Ogawa, T. A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 88, 4275–4279 (1991).
Allen, M. M. Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4 (1968).
McGinn, P. J., Price, G. D., Maleszka, R. & Badger, M. R. Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol. 132, 218–229 (2003).
Gombos, Z., Wada, H. & Murata, N. The recovery of photosynthesis from low-temperature photoinhibition is accelerated by the unsaturation of membrane lipids: a mechanism of chilling tolerance. Proc. Natl. Acad. Sci. USA 91, 8787–8791 (1994).
Kügler, M., Jänsch, L., Kruft, V., Schmitz, U. K. & Braun, H. P. Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth. Res 53, 35–44 (1997).
Dai, H. et al. Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J. 75, 858–866 (2013).
Gao, F. et al. The NDH-1L-PSI supercomplex is important for efficient cyclic electron transport in cyanobacteria. Plant Physiol. 172, 1451–1464 (2016).
Wang, X. et al. A cytoplasmic protein Ssl3829 is important for NDH-1 hydrophilic arm assembly in Synechocystis sp. strain PCC 6803. Plant Physiol. 171, 864–877 (2016).
Ran, Z. et al. Ssl3451 is important for accumulation of NDH-1 assembly intermediates in the cytoplasm of Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 60, 1374–1385 (2019).
Zhao, J., Gao, F., Zhang, J., Ogawa, T. & Ma, W. NdhO, a subunit of NADPH dehydrogenase, destabilizes medium size complex of the enzyme in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 289, 26669–26676 (2014).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Ma, W. & Mi, H. Expression and activity of type 1 NAD (P) H dehydrogenase at different growth phases of the cyanobacterium, Synechocystis PCC 6803. Physiol. Plant. 125, 135–140 (2005).
Beckmann, K., Messinger, J., Badger, M. R., Wydrzynski, T. & Hillier, W. On-line mass spectrometry: membrane inlet sampling. Photosynth. Res. 102, 511–522 (2009).
Rastogi, R. P., Singh, S. P., Häder, D. P. & Sinha, R. P. Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2’,7’-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem. Biophys. Res. Commun. 397, 603–607 (2010).
Ueno, M. et al. Moderate heat stress stimulates repair of photosystem II during photoinhibition in Synechocystis sp. PCC 6803. Plant Cell Physiol. 57, 2417–2426 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Evans, R. et al. Protein complex prediction with AlphaFold-multimer. bioRxiv. https://doi.org/10.1101/2021.10.04.463034 (2021).
Clementel, D. et al. RING 3.0: fast generation of probabilistic residue interaction networks from structural ensembles. Nucleic Acids Res. 50, W651–W656 (2022).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2023YFA0914600 to W.M.), National Natural Science Foundation of China (32170257 and 31770259 to W.M.), Shanghai Science and Technology Committee (22010503500 and 20ZR1441300 to W.M.).
Author information
Authors and Affiliations
Contributions
W.M. initiated and coordinated the project. M.Z. constructed and characterized the mutants and conducted physiological analyses. Y.J. performed biochemical analyses. Z.R. analyzed the NDH-1 structure. S.L. and T.X. identified the mutants. X.L. conducted in vitro ferredoxin-dependent plastoquinone reduction assays. All authors contributed to data interpretation. W.M., M.Z. and Y.J. collaborated on manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Mi Hualing and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, M., Jiang, Y., Ran, Z. et al. A cyanobacteria-derived intermolecular salt bridge stabilizes photosynthetic NDH-1 and prevents oxidative stress. Commun Biol 8, 172 (2025). https://doi.org/10.1038/s42003-025-07556-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07556-4