Abstract
The circadian clock orchestrates behavior and physiology through the oscillation of key clock proteins like PERIOD (PER). Here, we investigate the role of ubiquitin-specific peptidase 14 (USP14) in modulating PER stability and circadian rhythms in Drosophila. We find that overexpression of USP14 in clock cells reduces PER protein levels without altering its mRNA levels whereas USP14 knockdown increases PER protein levels, suggesting that USP14 regulates PER post-translationally. Interestingly, despite these alterations in PER levels, neither USP14 overexpression nor knockdown significantly impacts circadian behavioral rhythms, likely because of slight effects on PER levels in small ventral lateral neurons (sLNvs). Further analysis shows that USP14 physically interacts with Supernumerary Limbs (SLIMB), a protein involved in PER degradation. Moreover, reducing slimb expression mitigates the effects of USP14 on PER protein stability. Mass spectrometry identifies two ubiquitination sites on PER (Lys1117 and Lys1118) critical for its degradation. Expression of PER1117A, 1118A mutant in per01 background impairs circadian rhythm strength. In conclusion, this study demonstrates that Drosophila USP14 indirectly modulates PER protein stability by affecting SLIMB and highlights the critical role of specific ubiquitination sites on PER in maintaining circadian rhythms.
Similar content being viewed by others
Introduction
Circadian rhythms are endogenous oscillations of behavior and physiology with a period length of approximately 24 h. These rhythms are observed in most organisms and are driven by circadian clocks1,2. The circadian clock functions at the cellular level through a transcription-translation feedback loop involving mutually interacting positive and negative elements. In Drosophila, the positive components of the major feedback loop include the transcriptional activators CLOCK (CLK) and CYCLE (CYC), whereas the negative components are the transcriptional repressors PERIOD (PER) and TIMELESS (TIM). CLK and CYC form a heteromeric complex to activate the E-box-mediated transcription of per and tim; in a feedback connection, PER and TIM form complexes in the cytosol and translocate to the nucleus at a specific time of day to inhibit the transcriptional activity of the CLK/CYC complex. Among the clock proteins, the oscillation in PER protein levels primarily determines the duration (period) and phase of the circadian cycle3. The turnover of PER is regulated by post-translational modification; specifically, PER is phosphorylated by Doubletime (DBT)/Casein Kinase 1ε (CK1ε)4,5, ubiquitinated by E3 ubiquitin ligase Supernumerary Limbs (SLIMB), and subsequently degraded by the 26S proteasome6,7,8. The degradation of PER releases the repression of the CLK/CYC complex, thereby initiating a new cycle of transcriptional activation.
The ubiquitin-proteasome system (UPS), one of the principal protein quality control systems, is implicated in the regulation of circadian rhythms in various organisms, including mammals, Drosophila, and Neurospora6,7,9,10,11,12. FWD1, the Neurospora homolog of SLIMB, regulates the degradation of the circadian clock protein FREQUENCY, while β-TRCP, the mammalian homolog of SLIMB, targets mammalian PER (mPER) for degradation. Moreover, a recent study suggests that maintaining a balance between ubiquitination and deubiquitination is crucial for regulating the stability of clock proteins13. According to these findings, USP14 (a deubiquitinase) and β-TRCP function in opposition to regulating the stability of the mPER protein, as evidenced by disrupted behavior rhythms observed in β-TRCP1/2 mutant mice. However, the exact mechanism of PER degradation by SLIMB in Drosophila remains unknown.
To investigate the role for deubiquitinase in Drosophila, we analyzed the impact of USP14 on the stability of PER in Drosophila. Overexpressing USP14 in clock cells downregulated the levels of the PER protein without affecting TIM, the other main clock protein. In addition, we demonstrated that USP14 stabilized SLIMB, which is required for USP14 to regulate the stability of PER. We also identified the putative ubiquitination residues in PER through mass spectrometry and found that mutations in the lysine residue to alanine at positions 1117 and 1118 rendered the protein resistant to USP14-mediated protein degradation. These results collectively indicate that Drosophila USP14 regulates the degradation of PER.
Results
USP14 overexpression in clock cells reduces PER levels
In mammals, the degradation of mPER is regulated by a balance between ubiquitination and deubiquitination processes13. To explore whether Drosophila USP14 is involved in the degradation of circadian clock proteins, we overexpressed USP14 in clock cells using the tim (UAS)-gal4 driver. Flies were entrained under a standard 12 h/12 h light/dark (LD) cycle, where the zeitgeber time 0 (ZT0) corresponds to light-on and the zeitgeber time 12 (ZT12) to light-off. In control flies (tim>lacZ), newly synthesized PER was detected from ZT12 and reached peak levels by ZT24, followed by its degradation early in the day (Fgs. 1a, b). Notably, in USP14-overexpressing flies, PER levels were consistently lower throughout the day compared to controls. Interestingly, TIM protein levels remained unaffected between control and USP14-overexpressing flies (Fig. 1a, c). To determine whether USP14 influences the transcription of the per and tim genes, we measured the mRNA levels under the same conditions. No significant differences were observed between control and USP14-overexpressing flies (Fig. 1d, e), suggesting that USP14 primarily affects PER stability at the post-translational level rather than at the transcriptional level.
(a) Control (tim>lacZ) and USP14 OE (tim>Usp14-HA) flies were collected at the indicated times of the day (ZT). Head extracts were analyzed by immunoblotting with antibodies against PER, TIM, or α-Tub. (b and c) Quantification of PER (b) and TIM (c) protein levels was performed using the ImageJ software (NIH). All band intensities were normalized by the value of α-Tub band intensity. Values indicate mean ± SEM. p-values were obtained using Student’s t-tests. *p < 0.05, **p < 0.01. (d and e) Relative mRNA levels of period (d) and timeless (e) were determined by quantitative real-time PCR. All values were normalized by the values of rp49. (f) Free-running periods and rhythmicity (inside the bar) of the indicated genotype of flies are shown. Values indicate mean ± SEM (n = 16 ~ 62). p-values were obtained using one-way ANOVA followed by Tukey’s comparisons test. *p < 0.05, **p < 0.01, and ****p < 0.0001. (g) Brains of the control and USP14 OE flies were dissected at the indicated times of DD and stained with anti-PER (green) and anti-PDF (red) antibodies. Representative images of sLNvs are shown (scale bar: 10 μm). (h) Quantification of the PER fluorescence intensities of sLNvs. Values indicate mean ± SEM (n = 42 ~ 55). p-values were obtained using the Mann–Whitney test at each time point. *p < 0.05 and ****p < 0.0001.
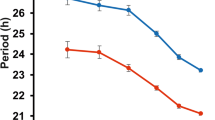
We next investigated whether USP14 overexpression impacts circadian rhythm by performing a circadian locomotor activity analysis (Fig. 1f and Supplementary Table 2). USP14 overexpression led to a statistically significant but minimal shortening of the circadian period compared to control flies (tim>lacZ), and the parental lines, questioning the biological significance of this effect. USP14 overexpression did not alter circadian rhythmicity of locomotor activity.
The Drosophila circadian network, which orchestrates daily rhythms, comprises pacemaker neurons, including the small and large lateral ventral neurons (sLNvs and lLNvs), lateral dorsal neurons (LNds), and posterior dorsal neurons 1 (DN1ps)14,15,16,17,18,19. Among these, the sLNvs are crucial for governing free-running rhythms20. To further assess the effect of USP14 on PER protein dynamics within pacemaker neurons, we measured PER protein levels in sLNvs throughout the day under constant darkness (DD) conditions. Both control and USP14-overexpressing flies displayed strong PER oscillation in the sLNvs (Fig. 1g, h). Although PER levels in the sLNvs of USP14 overexpressing flies were slightly reduced at CT28, they were comparable to control levels during the rest of the day (Fig. 1g, h). This may explain why circadian rhythms remained largely unaffected in USP14 overexpressing flies, despite the significant reduction in PER levels observed in whole-head extracts (Fig. 1a, b). To understand the discrepancy between PER levels in whole-head extracts and sLNvs, we performed immunostaining of the retina, as a significant portion of the whole-head extract originates from the eyes. Consistent with the immunoblotting results (Fig. 1b), PER intensities in the retina were markedly reduced in USP14-overexpressing flies compared to controls (Supplementary fig. 1). This suggests that the observed significant reduction in whole-head PER levels was primarily driven by changes in the retina.
USP14 knockdown in clock cells increases PER levels
Given that USP14 overexpression reduced PER levels, we investigated whether endogenous USP14 regulates PER stability by knocking down USP14 in clock cells using the tim (UAS)-gal4 driver. Under standard LD conditions, PER levels in USP14 knocked down (KD) flies were slightly elevated compared to control flies, an effect opposite to that observed with USP14 overexpression (Fig. 2a, b). The effective knockdown of Usp14 mRNA in USP14 KD flies was verified through quantitative real-time RT-PCR (Fig. 2c).
(a) Adult flies of the indicated genotypes were harvested at the indicated ZT, and head extracts were prepared for immunoblotting with antibodies against PER or α-Tub. (b) The protein level of PER in representative images was quantified. These experiments were conducted at least in triplicate. Values indicate mean ± SEM. p-values were obtained using Student’s t-tests. *p < 0.05. (c) The levels of Usp14 mRNA were quantitated in control (tim > d2) and USP14 KD (tim > d2, Usp14 Ri) flies through real-time quantitative RT-PCR. Values indicate mean ± SEM. p-values were obtained using Student’s t-tests. **p < 0.05 (d) Free-running periods and rhythmicity (inside the bar) indicated genotype of flies are shown. Error bars indicate mean ± SEM (n = 29 ~ 37). p-values were obtained using the one-way ANOVA followed by Tukey’s comparisons test. ****p < 0.0001. (e) Brains of the control and USP14 KD flies were dissected at indicated times of DD and stained with anti-PER (green) and anti-PDF (red) antibodies. Representative images of sLNvs are shown (scale bar: 10 μm). (f) Quantification of the PER fluorescence intensities of sLNvs. Values indicate mean ± SEM (n = 54 ~ 74). p-values were obtained using the Mann–Whitney test at each time point. **p < 0.01 and ****p < 0.0001.
To determine whether the altered PER levels in USP14 KD flies affect circadian rhythm, we performed circadian locomotor activity analysis. Similar to USP14 overexpression, USP14 knockdown did not result in significant changes in either circadian period or rhythmicity when compared to parental lines and the control (tim>d2) flies (Fig. 2d and Supplementary Table 2).
To further correlate the PER dynamics with circadian locomotor activity, we examined PER levels in sLNvs under the DD condition. Both control and USP14 KD flies exhibited strong PER oscillation in sLNvs neurons (Fig. 2e, f). However, PER levels in sLNvs of USP14 KD flies were modestly higher than those in controls throughout most of the day. The relatively small increase in PER levels in USP14 KD flies likely explains the lack of dramatic changes in circadian behavior. Nonetheless, the opposing effects observed in PER levels with USP14 overexpression and knockdown indicate that USP14 is involved in the regulation of PER stability.
USP14 regulates the stability of SLIMB
Our results showed that the overexpression of a deubiquitinating enzyme decreased PER levels, while its knockdown increased PER levels (Figs. 1 and 2). This led us to hypothesize that the role of USP14 in PER stability might not be direct. Alternatively, USP14 might regulate the stability of a specific protein that is responsible for degrading PER. To investigate this hypothesis, we first focused on SLIMB, a member of the F-box/WD40 protein family within the ubiquitin ligase SCF complex that controls the degradation of hyperphosphorylated PER6,7. We gradually increased USP14 levels in Drosophila S2 cells and then examined the stability of the SLIMB protein. We found that SLIMB protein levels increased with increasing levels of USP14 (Fig. 3a, b). To further determine the function of USP14 on SLIMB stability, we performed chase experiments using cycloheximide (CHX) to inhibit de novo protein synthesis. Our observation indicates that the degradation rate of SLIMB was delayed when USP14 was overexpressed (Fig. 3c, d). We also found that USP14 and SLIMB physically interacted, as determined by co-immunoprecipitation assays (Fig. 3e). Consistently, the ubiquitin-mediated protein degradation of SLIMB was reduced by USP14 overexpression (Fig. 3f). The phosphorylation of PER leads to its rapid degradation by the ubiquitin-proteasome pathway7; therefore, to mimic the animal clock, we induced the expression of dbt-V5 using the copper-inducible metallothionein (pMT) promoter that triggers the phosphorylation of PER. Similar to native PER protein mobility (Fig. 1a), the exposure of CuSO4 in Drosophila S2 cells led to a progressive decrease in PER mobility (Fig. 3g). Considering that USP14 indirectly regulates the protein degradation of PER through SLIMB, we then tested the stability of PER in the absence of SLIMB. To knock down the slimb gene in Drosophila S2 cells, we incubated the cells with a double-stranded RNA (dsRNA) against slimb, which resulted in an approximately 60% decrease in slimb mRNA levels as compared to control cells (Fig. 3h). Notably, the effect of USP14 on wild-type PER protein stability almost disappeared in slimb knockdown S2 cells (Fig. 3g, i), indicating that USP14 accelerates the degradation of the PER protein by enhancing the stability of SLIMB.
a Immunoblotting of the SLIMB protein according to increasing levels of USP14. b The normalized band intensities of the gels were quantified, and the data are shown in the graph. c SLIMB degradation was delayed by the overexpression of Usp14. HA-tagged SLIMB and Myc-tagged Usp14 were transiently cotransfected into S2 cells. Then, chase experiments were performed at the indicated time points after the addition of 1 μg/mL cycloheximide (CHX) at time zero. α-Tub was used as a loading control. d Quantification of SLIMB-3HA signals in (c) normalized to those of endogenous α-tubulin. e Co-immunoprecipitation of USP14 and SLIMB. Drosophila S2 cells were transfected with indicated plasmids, and whole-cell lysates were subjected to co-immunoprecipitation. f Effect of USP14 overexpression on ubiquitin-mediated degradation of SLIMB. USP14 fused with EGFP and three HA-tagged SLIMB were overexpressed in S2 cells and then incubated with 5 μM of proteasome inhibitor (MG132) for 12 h before lysis. Whole-cell lysates were subjected to immunoprecipitation with an anti-HA antibody, followed by immunoblotting with an anti-EGFP, anti-HA, and anti-ubiquitin antibody. g–i dsRNA against slimb was incubated on days 1 and 3. At day 4, dsRNA-treated S2 cells were transfected with the indicated plasmids. DBT was induced by adding 500 μM CuSO4 for the indicated times. Proteins (g) or RNA (h) were extracted and subjected to immunoblotting (g) or RT-qPCR (h). (h) The levels of slimb mRNA were quantitated in control (-) and slimb knockdown S2 cells. (i) Normalized PER levels at steady state (0 h after CuSO4 treatment) according to slimb knockdown. Notably, the effect of USP14 overexpression in wild-type PER protein degradation almost disappeared in slmb-downregulated S2 cells. p-values were determined using Student’s t-test. *p < 0.05, **p < 0.01.
Identification of ubiquitination residue in the PER protein and its significance
To gain a more comprehensive understanding of the molecular mechanisms through which USP14 drives the degradation of the PER protein, we conducted mass spectrometry analyses to identify the post-translational modifications of PER. Head extracts at ZT20 were prepared from USP14-overexpressing flies and processed for immunoprecipitation followed by mass spectrometry (Fig. 4a). Consequently, we found three putative ubiquitination residues—K53, K1117, and K1118—in the PER protein. To determine whether those residues are required for the ubiquitination of PER, we generated various mutant constructs of PER: single mutant (PERK53A, PERK1117A, and PERK1118A), double mutant (PERK53A, K1117A, PERK53A, K1118A, and PERK1117A, K1118A), and triple mutant (PERK53A, K1117A, K1118A). When we tested the stability of PER mutant proteins, we found that the double mutation of the Lys-1117 and Lys-1118 residues into Ala rendered the PER protein resistant to protein degradation (Fig. 4b, c), which is validated using a chase experiment. The PER mutant with mutations at Lys-1117 and Lys-1118 residues exhibited greater resistance to protein degradation compared to the wild-type PER following DBT phosphorylation for 6 h (Fig. 4d, e). This suggests that these residues could potentially be targeted for ubiquitin conjugation. Consistently, the increased expression of SLIMB did not have a significant impact on the levels of PERK1117A, K1118A mutants, although there was a slight decrease. However, the levels of wild-type PER were noticeably reduced when SLIMB was co-expressed (Fig. 4f, g). To determine whether USP14 is involved in the stability of PER via the K1117 and K1118 residues, we co-expressed USP14 in combination with the PERWT or PERK1117A, K1118A mutants. The co-expression of UPS14 with PERWT accelerated the protein degradation of PERWT, similar to that of the native PER protein in fly heads (Fig. 1a). Furthermore, while USP14 affected the protein stability of wild-type PER, the effect of USP14 on PER protein stability was abolished in the PERK1117A, K1118A mutant protein (Fig. 4h).
a Schematic of the identification process of ubiquitination residues in Drosophila PER. Proteins were extracted from the fly heads at ZT20, and PER was isolated by immunoprecipitation. The purified PER proteins were used for the analysis of post-translational modifications by mass spectrometry, which revealed three putative Lys residues required for ubiquitin attachment. b, c S2 cells were co-transfected with the indicated plasmids, and DBT was induced by adding 500 μM CuSO4 for the indicated times. c Quantification of PERWT and PERK1117A,K1118A signals in (b) normalized to those of endogenous α-Tubulin. d Western blotting shows that PERK1117A,K1118A were longer-lived compared to PERWT after pre-treatment with 500 μM CuSO4. e Graphs show the rate of PER degradation. The PER level at 0 h chase in each panel of (d) was set to 1. f Anti-PER western blotting. Upon SLIMB overexpression, the levels of PERWT decreased, but the effect of SLIMB on PER stability was suppressed in PERK1117A, K1118A. g The normalized band intensities of gels in (f) were quantified and shown as a graph. (h) The impact of USP14 on PERWT was not observed in PERK1117A, K1118A. The experimental condition closely resembles condition (b). p-values were determined using Student’s t-test. *p < 0.05, **p < 0.01.
Finally, to evaluate the significance of the ubiquitination sites identified in PER, we expressed either wild-type PER (PERWT) or PER mutant (PERK1117A, K1118A) using the tim (UAS)-gal4 driver in a per-null background. We first analyzed molecular rhythms by performing western blotting analysis with head extracts. PERWT levels began to accumulate early in the night, peaking in the late night to early morning, before being degraded (Fig. 5a). This rhythmic accumulation is also evident in the quantitation graph (Fig. 5b). In contrast, PERK1117A, K1118A did not exhibit robust rhythms in protein levels. Specifically, the hyperphosphorylated isoforms of PER persisted (Fig. 5a), leading to significantly dampened molecular oscillations (Fig. 5b). This impaired degradation of hyperphosphorylated PERK1117A, K1118A is consistent with our findings in S2 cells (Fig. 4c). To further investigate the functional consequences of these mutations, we conducted circadian behavior analysis. Expression of PERWT under the tim (UAS)-gal4 driver successfully rescued the arrhythmic phenotype of per01 flies, although it slightly lengthened the circadian period to approximately 25.3 h. This lengthened periodicity could result from the elevated expression levels of PERWT under the tim (UAS)-gal4 driver compared to natural PER expression levels in wild-type flies. In contrast, flies expressing PERK1117A, K1118A exhibited statistically significant shortening of the period by 24.9 ± 0.21 h (Fig. 5d and Supplementary Table 2), although we do not consider it biologically significant. However, PERK1117A, K1118A did not effectively restore rhythmicity or rhythm strength compared to PERWT (Fig. 5c, e and Supplementary Table 2). Immunostaining of the sLNvs revealed pronounced oscillations of PERWT (left, Fig. 5f, g), whereas the oscillation amplitude of PERK1117A, K1118A was markedly reduced, with consistently higher levels than PERWT (right, Fig. 5f, g). These molecular rhythmic defects correlate with the impaired circadian behavior observed in PERK1117A, K1118A-expressing flies. Collectively, these results highlight the critical role of K1117 and K1118 residues in regulating PER protein stability and emphasize their importance in the maintenance of circadian rhythms in Drosophila.
a Flies expressing PERWT (w,per01;tim>perWT) and PERK1117A, K1118A (w,per01;tim > perK1117A,K1118A) were collected at the indicated times of the day (ZT). Head extracts were analyzed by immunoblotting with antibodies against PER or α-Tub. b Comparison of the levels of PER proteins between those presented in (a). Quantification of PER protein levels was performed using the ImageJ software (NIH). All band intensities were normalized by the value of α-Tub band intensity. Values indicate mean ± SEM. p-values were obtained using Student’s t-tests. *p < 0.05. The levels of mutant PER protein (PERK1117A, K1118A) were not significantly different throughout the day. c The locomotor activities of per01 (w,per01;tim > +), PERWT (w,per01;tim>perWT), and PERK1117A,K1118A (w,per01;tim>perK1117A,K1118A) flies are shown. Each panel represents the actogram of male flies for a given genotype during the 12 h/12 h LD cycle, followed by seven consecutive days of DD. The dotted red lines connect the evening peaks for each day of the experiments. d Free-running period and rhythmicity (inside the bar) of PERWT and PERK1117A,K1118A flies are shown. Error bars indicate mean ± SEM. p-values were obtained using the Mann–Whitney test; *p < 0.05. e Power of the rhythm strength of PERWT and PERK1117A,K1118A flies are shown. Error bars indicate mean ± SEM. p-values were obtained using the Student’s t-test; ****p < 0.0001. f, g Brains of the PERWT and PERK1117A,K1118A flies were dissected at the indicated times of DD and stained with anti-PER (green) and anti-PDF (red) antibodies. Representative images of sLNvs are shown (scale bar: 10 μm). g Quantification of the PER fluorescence intensities of sLNvs. Values represent mean ± SEM (n = 18 – 42). Asterisks indicate statistically significant differences between values at each time point using the Mann–Whitney test. ****p < 0.0001.
Discussion
The daily oscillation of PER protein levels is crucial for maintaining circadian rhythms in Drosophila, which is primarily governed by UPS-mediated regulation. The balance between ubiquitination and deubiquitination influences the physiological levels and functionality of proteins. The interplay between these processes has a substantial effect on the repetitive patterns of protein accumulation within the clock mechanism and establishes these post-translational modifications as a fundamental element in the rhythmic nature of the circadian clock. Herein, we suggest a relationship between clock protein PER, ubiquitinase SLIMB, and deubiquitinase USP14 in Drosophila. Unlike mammals, Drosophila USP14 did not have a direct effect on the PER protein. Alternatively, it enhanced the stability of SLIMB, thereby influencing PER function.
Our findings indicate that the overexpression of USP14 reduces PER levels in whole-head extracts (Fig. 1b). A reduction of repressor protein PER could theoretically derepress CLK-CYC, leading to increased levels of per and tim mRNA. However, our results demonstrated no difference in per and tim mRNA levels between control and USP14-overexpressing flies. CLK is present in limiting amounts and PER levels are significantly higher than those of CLK21. Thus, we reasoned that even a reduced amount of PER may still sufficiently repress CLK-CYC activity.
Although western blotting analysis of whole-head extracts revealed significant alterations in PER levels with USP14 manipulation, the impact on circadian rhythms was minimal, likely resulting from limited effects in sLNvs. Despite subtle alterations in PER levels within sLNvs following USP14 overexpression or knockdown, the consistent direction of PER stability changes underscored the role of USP14. However, these modest PER changes did not significantly affect circadian locomotor behavior, suggesting that although USP14 influenced PER stability, its effect may not be sufficient to noticeably alter circadian rhythms. Further sensitization by downregulating Usp14 in heterozygous slimb00295 loss of function mutants also failed to significantly affect circadian rhythms (Supplementary Fig. 2 and Supplementary Table 2). We hypothesize that SLIMB levels in clock neurons, including sLNvs, were sufficiently high to degrade PER, making USP14 manipulation less impactful on circadian rhythms compared to other cells.
As mentioned above, mPER degradation is mediated by two proteins: USP14 and β-TRCP, an ortholog of Drosophila SLIMB13. Notably, although USP14 functions as a deubiquitinating enzyme for the PER protein, the effect of USP14 in PER protein degradation differs between mice and flies, as mouse USP14 stabilizes the mPER protein whereas Drosophila USP14 accelerates PER protein degradation. The USP14 protein is highly conserved across species and has two conserved domains, UBQ and Peptidase_C19. Therefore, it is interesting that mouse USP14 and Drosophila USP14 have opposing effects on PER protein degradation. This discrepancy may be explained from an evolutionary standpoint as follows. In Drosophila, USP14 controls the stability of SLIMB, and SLIMB ubiquitinates PER; in other words, Drosophila USP14 goes through two steps to modulate PER protein stability. In contrast, mouse USP14 directly enhances the stability of PER. Moreover, the stability of mPER in mammals is directly regulated in the opposite direction by β-TRCP. Therefore, it is possible that during evolution, mammals evolved a direct way to more efficiently control the degradation of PER using two separate proteins (i.e., USP14 and β-TRCP) rather than going through two hurdles as in Drosophila.
In conclusion, our study demonstrates that USP14 regulates the degradation of the PER protein through SLIMB stabilization in Drosophila (Fig. 6).
Materials and Methods
Fly strains
All Drosophila stocks were raised on standard BDSC cornmeal containing 1.6% yeast, 0.9% soy flour, 6.7% cornmeal, 1% agar, and 7% light corn syrup at 25 °C. Genes in clock cells were misexpressed using the standard Gal4/UAS systems22. The generation of Tim (UAS)-gal4 flies has been previously described23. UAS-USP14-HA (F001032) and UAS-lacZ (BDSC1776) lines were obtained from Bloomington stock center and FlyORF, respectively.
Circadian rhythm analysis
Locomotor activity of individual flies was determined using the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA, USA). Young male flies in glass tubes containing 2% agar and 5% sucrose were exposed to a 12 L:12D cycle for four days and were then maintained in DD for seven days at 25 °C. The locomotor data analysis was performed using the FaasX software (Fly Activity Analysis Suite for Mac OS X), which was generously provided by Dr. Francois Rouyer (Centre National de la Recherche Scientifique, France). Periods were calculated for each fly using χ2 periodogram analysis, and the data were pooled to obtain an average value. Power was calculated by quantifying the relative strength of the rhythm during DD, and individual flies with a power of 10 or greater and a width of two or greater were considered rhythmic. Actograms exhibited double-plotted locomotor activities throughout the experimental period and were acquired using the Actogram J software24.
Plasmids and S2 cell culture
The act-gal4, uas-Usp14-HA, uas-egfp, pAct-per-V5, and pMT-dbt-V5 plasmids have been described previously7,23,25. PER mutants (K53A, K1117A, and K1118A) were generated using the QuickChange site-directed mutagenesis kit (Stratagene, San Diego, CA, USA). The sequences of the mutants were verified through DNA sequencing. Drosophila S2 cells were grown in Schneider’s Drosophila medium (Invitrogen, 21720, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Invitrogen, 16000-044) and 0.5% penicillin/streptomycin (Invitrogen, 15140-122). For transfection, the Effectene reagent was used following the manufacturer’s protocol (Qiagen, Germany). To knock down genes, dsRNA was generated following the protocols of flyrnai.org. The following oligonucleotide sequences were used to generate T7-promoter–containing amplicons: slimb-R 5’-TAATACGACTCACTATAGGGAGCTCATCGAACGCAAGGTG-3’ and slimb-S 5’- TAATACGACTCACTATAGGGTGCGCACGAATTCACAGCTA-3’. For RNAi in cultured cells, we followed a previously described protocol26 using two rounds of incubation with 20 µg of dsRNA at days 1 and 3 to enhance knockdown efficiency. On day 4, the described cDNAs were transfected using the Effectene reagent.
Immunoblotting and immunoprecipitation
Drosophila S2 cells were lysed using modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, and 0.25% sodium deoxycholate) with a protease inhibitor cocktail (Roche) and PhosSTOP. For immunoblotting of the fly heads, the heads were collected by freezing at the indicated times in LD and were lysed using lysis buffer (10 mM HEPES at pH 7.5, 5 mM Tris-HCl at pH 7.5, 50 mM KCl, 10% glycerol, 2.5 mM EDTA, 5 mM dithiothreitol, and 0.2% Triton X-100) with a protease inhibitor cocktail (Roche). After centrifugation at 15,700 × g for 10 min, proteins in the supernatant were separated using SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Merck Millipore, Billerica, MA, USA). The antibodies used in this study are listed in S1 Table.
Cycloheximide protein stability assay
Drosophila S2 cells expressing the indicated genes were treated with 1 μg/mL or 10 μg/mL cycloheximide (Sigma-Aldrich), and cells were harvested at the indicated time points. Cell lysates were obtained using modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, and 0.25% sodium deoxycholate) with a protease inhibitor cocktail (Roche) and PhosSTOP and were subsequently subjected to immunoblotting.
Immunohistochemistry
The fly heads were cut open, fixed in 2% formaldehyde, and washed with PAXD buffer (1 × PBS, 5% BSA, 0.03% sodium deoxycholate, and 0.03% Triton X-100)27. The fixed heads were dissected, and the isolated brains were permeabilized in 1% PBT for 20 min and then blocked in PAXD containing 5% horse serum for 1 h. The following primary antibodies were directly added to the mixtures at a 1:200 dilution: anti-PDF antibody (C7) (DSHB, Iowa City, IA, USA) and anti-PER antibody (Rb1)23. The brains were washed with PAXD and incubated overnight with secondary antibodies in a blocking solution at 4 °C. The following secondary antibodies were used at a 1:200 dilution: goat anti-rabbit Alexa-488 (Thermo Fisher Scientific, Waltham, MA, USA) and goat anti-mouse Alexa-555 (Thermo Fisher Scientific). The immunostained brain samples were washed with PAXD, incubated in 0.1 M phosphate buffer containing 50% glycerol for 30 min, and mounted using a mounting medium. Confocal images were obtained using the LSM 800 confocal microscope (Carl Zeiss, Germany) and were processed using the Zen software (ZEN Digital Imaging for Light Microscopy, Carl Zeiss). For signal quantification, the pixel intensity of each cell was determined using the ImageJ software (NIH, Bethesda, MD, USA).
Real-time RT-PCR
Total RNA was isolated from flies using TRIzol reagent (Invitrogen, USA), and 100 ng of total RNA was used for reverse transcription with the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Quantitative PCR was performed for 40 cycles using the TOPreal qPCR 2× PreMIX (SYBR Green with high ROX) on a LightCycler® 480 Real-time PCR system. The following primer sequences were used: per_F 5′-AACATGCTGCTCGTCATCTG, per_R 5′-GAACTTGGGGCTCTTCTGTG, tim_F 5’-CAAGAGCGTGGTGGAGTACA, tim_R 5’-TCTCAGCAGCAGCAGACAGT, rp49_F 5’-AGATCGTGAAGAAGGCACCAAG, rp49-R 5’-CACCAGGAACTTCTTGAATCCGG, slimb_F 5’-CGTCAATGTGGTGGACTTTG, and slimb_R 5’- CGCACGAATTCACAGCTAGA.
Cryosection
Fly heads were embedded in optimal cutting temperature (OCT) compound and snap-frozen using dry ice. Sections were cut at a thickness of 10 µm using a cryostat set at -25 °C and transferred to Superfrost Plus slides for drying. The slides were then fixed with 4% formaldehyde in PBS for 20 min and permeabilized in 0.3% PBT for 20 min. Immunostaining was performed using an anti-PER antibody at a 1:200 dilution, followed by a secondary goat anti-rabbit Alexa-405 antibody at a 1:500 dilution. Confocal images were acquired using an LSM 710 confocal microscope (Carl Zeiss, Germany).
On-bead trypsin digestion
The bead pellet from immunoprecipitation was suspended with a 5% SDS buffer with 50 mM ammonium bicarbonate. To reduce disulfide bonds, dithiothreitol was added to a final concentration of 20 mM, and the samples were incubated at 95 °C and 1000 rpm for 10 min. Subsequently, a final concentration of 40 mM iodoacetamide was added, and the samples were incubated in the dark at 25 °C for 30 min. The samples were acidified by adding a 10-fold dilution of 12% phosphoric acid. The acidified samples were then added to 500 µL of Suspension Trap digestion (S-Trap) binding buffer (90% methanol and 100 mM Triethylammonium bicarbonate buffer [TEAB] [pH 7.55]). The S-Trap spin column (ProtiFi, Long Island, New York, USA) was then used to perform centrifugation at 4000 × g for 30 s. After washing the spin column with 150 µL of S-trap binding buffer and centrifuging at 4000 × g for 30 s, the washing step was repeated twice. Finally, the spin column was transferred to a new 1.5 mL sample tube, and a trypsin/Lys-C Mix with a protein to Trypsin/Lys-C mixture ratio of 100:1 (Promega, Madison, WI, USA) dissolved in 50 mM TEAB was added to the S-trap spin column. The column containing Trypsin/Lys-C was incubated at 37 °C for 16 h without shaking28. Peptide elution was performed thrice, first by adding 40 µL of 50 mM TEAB, centrifuging at 1000 × g for 1 min, and then by adding 40 µL of 0.2% formic acid, followed by centrifugation at 1000 × g for 1 min. In the final elution step, 40 µL of 0.2% formic acid and 50% acetonitrile were added, and the sample was centrifuged at 4000 × g for 1 min to elute the peptides. The eluted peptides were dried using an evaporator combined with a cold trap and stored at -80 °C until use.
LC-MS analysis and database search
The peptide mixture for each sample set was reconstituted in 0.1% formic acid, and peptide separation was performed using the Ultimate3000 RSLC system coupled with a Q Exactive HFx mass spectrometer (Thermo Fisher Scientific). The liquid chromatography gradient and data-dependent acquisition-MS options followed previously published methods29. The resulting acquired MS spectra were searched using Sequest HT on Proteome discoverer (version 2.3, Thermo Scientific, USA) against the SwissProt Drosophila melanogaster proteome sequence database (Taxon ID 7227) using variable modification at lysine residues with di-glycine modification. Label-free quantities of each di-glycine attached peptide of the target protein were extracted and used for further analysis.
Statistics and Reproducibility
At least three independent biological replicates were analyzed, and the results are presented as the mean ± SEM. Statistical analyses were conducted using GraphPad Prism 10 (GraphPad, San Diego, CA, USA). A Student’s t-test was used for comparisons between two groups, while comparisons among three or more groups were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test or the Mann–Whitney test. A P-value of <0.05 was considered statistically significant.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Uncropped gel images are presented in Supplementary Fig. 3, and the source data for all figures in this study are included in Supplementary Data.
References
Patke, A., Young, M. W. & Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 (2020).
Allada, R. & Chung, B. Y. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624 (2010).
Hardin, P. E. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 74, 141–173 (2011).
Kloss, B. et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94, 97–107 (1998).
Price, J. L. et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95 (1998).
Grima, B. et al. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178–182 (2002).
Ko, H. W., Jiang, J. & Edery, I. Role for Slimb in the degradation of Drosophila period protein phosphorylated by doubletime. Nature 420, 673–678 (2002).
Chiu, J. C., Vanselow, J. T., Kramer, A. & Edery, I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 22, 1758–1772 (2008).
Shirogane, T., Jin, J., Ang, X. L. & Harper, J. W. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–26872 (2005).
Ohsaki, K. et al. The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J. Biochem. 144, 609–618 (2008).
He, Q. et al. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 22, 4421–4430 (2003).
Siepka, S. M. et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023 (2007).
D’Alessandro, M. et al. Stability of wake-sleep cycles requires robust degradation of the PERIOD protein. Curr. Biol. 27, 3454–3467.e3458 (2017).
Stoleru, D., Peng, Y., Agosto, J. & Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868 (2004).
Grima, B., Chelot, E., Xia, R. & Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873 (2004).
Rieger, D., Shafer, O. T., Tomioka, K. & Helfrich-Forster, C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26, 2531–2543 (2006).
Yao, Z., Bennett, A. J., Clem, J. L. & Shafer, O. T. The drosophila clock neuron network features diverse coupling modes and requires network-wide coherence for robust circadian rhythms. Cell Rep. 17, 2873–2881 (2016).
Delventhal, R. et al. Dissection of central clock function in Drosophila through cell-specific CRISPR-mediated clock gene disruption. Elife 8. https://doi.org/10.7554/eLife.48308 (2019).
Schlichting, M., Diaz, M. M., Xin, J. & Rosbash, M. Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. Elife 8. https://doi.org/10.7554/eLife.48301 (2019).
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C. & Taghert, P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999).
Bae, K., Lee, C., Hardin, P. E. & Edery, I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 20, 1746–1753 (2000).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Kim, E. Y. et al. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490–502 (2012).
Schmid, B., Helfrich-Forster, C. & Yoshii, T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J. Biol. Rhythms 26, 464–467 (2011).
Park, J. E. et al. The function of Drosophila USP14 in endoplasmic reticulum stress and retinal degeneration in a model for autosomal dominant retinitis pigmentosa. Biology (Basel) 9. https://doi.org/10.3390/biology9100332 (2020)
Kang, M. J. et al. 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. J. Cell Biol. 216, 115–129 (2017).
Gunawardhana, K. L. & Hardin, P. E. VRILLE controls PDF neuropeptide accumulation and arborization rhythms in small ventrolateral neurons to drive rhythmic behavior in drosophila. Curr. Biol. 27, 3442–3453.e3444 (2017).
HaileMariam, M. et al. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 17, 2917–2924 (2018).
Ahn, H. S. et al. Differential urinary proteome analysis for predicting prognosis in type 2 diabetes patients with and without renal dysfunction. Int. J. Mol. Sci. 21. https://doi.org/10.3390/ijms21124236 (2020).
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea, NRF- 2022R1A2C1003431 (to M.-J.K.) and NRF-2019R1A5A2026045, RS-2023-00208490 (to E.Y.K.), and from the Asan Institute for Life Sciences (Seoul, Republic of Korea), 2024IL0005 and 2024IP0038. All Drosophila stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University (NIH P40OD018537).
Author information
Authors and Affiliations
Contributions
S.W.K., J.-E.P., E.Y.K, and M.-J.K. conceptualized and designed the project, S.W.K., J.-E.P., S.O, M.U., H.S., S.B., N.P., S.J.L., T.X.T.R., G.K., J.Y., K.K., E.Y.K. and M.-J.K. performed experimental work and analyzed data; E.Y.K. and M.-J.K. prepared the manuscript; All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Choogon Lee and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Min Zhuang and Christina Karlsson Rosenthal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kang, S.W., Park, JE., Ok, S. et al. Drosophila ubiquitin-specific peptidase 14 stabilizes the PERIOD protein by regulating a ubiquitin ligase SLIMB. Commun Biol 8, 191 (2025). https://doi.org/10.1038/s42003-025-07632-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07632-9
This article is cited by
-
Role of circadian rhythms in heart failure: insights from myocardial energy metabolism
Journal of Translational Medicine (2025)