Abstract
The role of peridroplet mitochondria (PDM) in diseased liver, such as during the progression of metabolic dysfunction-associated steatohepatitis (MASH), remains unknown. We isolated hepatic cytoplasmic mitochondria (CM) and PDM from a mouse model of diet-induced MASLD/MASH to characterize their functions from simple steatosis to advanced MASH, using chow-fed mice as controls. Our findings show an inverse relationship between hepatic CM and PDM levels from healthy to steatosis to advanced MASH. Proteomics analysis revealed these two mitochondrial populations are compositionally and functionally distinct. We found that hepatic PDM are more bioenergetically active than CM, with higher pyruvate oxidation capacity in both healthy and diseased liver. Higher respiration capacity of PDM was associated with elevated OXPHOS protein complexes and increased TCA cycle flux. In contrast, CM showed higher fatty acid oxidation capacity with MASH progression. Transmission electron microscopy revealed larger and elongated mitochondria during healthy and early steatosis, which appeared small and fragmented during MASH progression. These changes coincided with higher MFN2 protein levels in hepatic PDM and higher DRP1 protein levels in hepatic CM. These findings highlight the distinct roles of hepatic CM and PDM in MASLD progression towards MASH.

Similar content being viewed by others
Introduction
The hepatic manifestation of metabolic syndrome, in the absence of excessive alcohol, is metabolic dysfunction-associated steatotic liver disease (MASLD)1 or fatty liver disease (MAFLD)2, formerly known as non-alcoholic fatty liver disease (NAFLD)3,4,5. Hepatic triglyceride (TG) accumulation is the hallmark feature of MASLD, which encompasses a spectrum of pathologies ranging from simple steatosis to steatohepatitis (MASH) and fibrosis3,4,5. It is estimated that 20–30% of the population worldwide are affected by MASLD6. Most importantly, not all patients progress from steatosis to MASH and in fact, only 3–5% of population worldwide are affected by MASH7. MASH further progresses to liver cirrhosis and hepatocellular carcinoma (HCC), the leading cause of liver related morbidity or mortality8. Unfortunately, despite significant research efforts, we still do not completely understand what factors govern this progression. A large body of studies have shown that mitochondrial dysfunction9,10,11,12,13,14 and genetic factors15,16,17 influence the disease. Thus, it is important to understand, how mitochondria functions in MASLD/MASH livers to prevent and/or to reverse this condition.
Lipid droplets (LDs) are highly dynamic energy storage organelles consisting of a hydrophobic core of neutral lipids surrounded by a phospholipid monolayer containing a variety of specific proteins. LDs play an important role in MASLD development18 and in fact, several genome-wide association study (GWAS) loci have identified genetic variants that impact LD biology and MASH progression19,20,21,22,23. LDs interact with mitochondria that are essential for both de novo lipogenesis (DNL) as well as fatty acid oxidation (FAO). Early biochemical studies have documented physical contacts between LDs and mitochondria in various tissues. Conventionally, it is perceived that mitochondria binds to LDs to aid in FAO. For instance, upon starvation or nutrient deprived conditions, these interactions are enhanced to facilitate the trafficking of free fatty acids (FFA) towards mitochondria for FAO24,25 or to esterify the lipotoxic FFA to LDs increasing their size and/or number25,26. Conversely, LD-mitochondrial contacts in adipocytes have been shown to facilitate lipid storage by enhancing DNL27. These seemingly contradictory findings have prompted researchers to suggest the existence of multiple subpopulations of mitochondria, which can compartmentalize DNL, fatty acid esterification and FAO, within the same cell28. Indeed, investigators have identified and characterized two different mitochondrial subpopulations from brown adipose tissue (BAT)29, white adipose tissue (WAT)30 and liver31,32 from healthy animals.
The mitochondria surrounding the LDs are referred to as peridroplet mitochondria (PDM) or lipid droplet associated mitochondria (LDM), whereas those freely floating in the cytosol are termed cytoplasmic mitochondria (CM). In BAT, during thermoneutral conditions, high respiration capacities of PDM facilitated LD expansion by providing ATP for TG synthesis, while CM supported thermogenic FAO when exposed to cold temperature29. Conversely, PDM in WAT had reduced respiration and ATP synthesis capacities than CM, which varied depending on the size of LDs to which they were bound30. Similar to WAT, in healthy rat liver under fed conditions, CM exhibited higher respiration capacities, while PDM promoted FAO31. Conversely, similar to BAT, PDM isolated from overnight fasted mouse livers facilitated fatty acid esterification and LD expansion32. Nevertheless, the role of PDM function in diseased liver such as during MASH progression remains unknown. Thus, it is important to further understand and characterize these functions in the context of MASH because that knowledge can be used for therapeutic interventions.
To this end, using a mouse model of diet-induced MASLD/MASH, we have isolated and characterized hepatic CM and PDM from different stages of MASLD progression. We demonstrated an inverse relationship between hepatic CM and PDM levels from healthy to MASH phenotypes. We also report that their proteomes are compositionally and functionally distinct. We next found that, compared to CM, PDM are bioenergetically active with higher pyruvate oxidation capacities in both healthy and diseased liver. Additionally, we found that higher respiration capacities of PDM were associated with higher levels of OXPHOS protein complexes as well as higher TCA cycle flux. On the contrary, CM had higher FAO capacities with MASH progression. Lastly, we show that hepatic PDM are elongated with higher levels of MFN2 proteins during healthy and early steatosis conditions, which is lost with MASH progression, during which hepatic CM show higher levels of DRP1 proteins.
Results
Mitochondrial dynamics and LD size were affected during MASLD progression
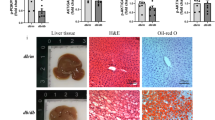
To characterize the various stages of MASLD, we used the choline-deficient L-amino acid-defined high fat diet (CDAHFD) mouse model as it is widely used to study hepatic steatosis, inflammation, and fibrosis in an accelerated manner33,34,35. Briefly, 8-weeks-old male C57BL/6J mice were fed a CDAHFD over a period of 3, 6, 9 or 15 weeks, while mice fed a standard chow diet for 15 weeks were used as healthy controls. In all our conditions, we fasted the mice for four hours before euthanasia. Subsequent histological examinations revealed that healthy controls displayed little to no evidence of TG accumulation, manifesting as LDs with the absence of fibrosis (Fig. 1A). In contrast, the livers of mice fed a CDAHFD at various timepoints showed increased TG accumulation characterized by the presence of both small and large LDs (Fig. 1A). Specifically, as originally demonstrated by Matsumoto et al.34, 3 weeks of CDAHFD induced steatosis, with LD increasing in size at 6 weeks of CDAHFD, which then reduced at 9 and 15 weeks of CDAHFD coinciding with fibrosis commencement. Also, we observed a time-dependent progressive increase in liver fibrosis in CDAHFD livers as indicated by the presence of perisinusoidal collagen deposition illuminated by trichrome staining (Fig. 1A) and their respective quantification (Fig. 1B). The severity of hepatic steatosis, ballooning, and inflammation was also evaluated in the liver histopathology specimens (Supplementary Fig. 1), which confirmed what Matsumoto et al., had previously observed34. Based on these observations, we classify the 3 and 6 weeks of CDAHFD as representing early and late steatosis, respectively, while designating the 9 and 15 weeks of CDAHFD as early and late fibrosis, respectively, for the purposes of this study.
A Representative H&E and trichrome images showing liver steatosis and fibrosis, B percent liver fibrosis area and C representative TEM images showing LD-mitochondrial contacts, CM and PDM, of livers harvested from healthy controls (i.e., 15 weeks Chow diet), early steatosis (i.e., 3 weeks CDAHFD), late steatosis (i.e., 6 weeks CDAHFD), early fibrosis (i.e., 9 weeks CDAHFD) and late fibrosis (i.e., 15 weeks CDAHFD) are shown. Percent fibrosis area is presented as mean ± SEM (n = 3 mice per group). P values were calculated by one-factor ANOVA with Holm-Sidak’s post hoc test. *P < 0.05; ***P < 0. 001.
Next, to examine the LD-mitochondrial interactions from early steatosis to MASH, we analyzed liver sections obtained from healthy controls and CDAHFD fed mice by transmission electron microscopy (TEM). We observed that both healthy controls and early steatotic livers had larger and elongated mitochondria (Fig. 1C), while mitochondria were small or fragmented as the disease progresses from late steatosis to MASH phenotypes. We also noticed that LD size increased remarkably at 6 weeks of CDAHFD (Fig. 1C) corroborating our histological examinations (Fig. 1A). These results revealed that CDAHFD feeding differentially affects the overall mitochondria structure and/or shape from simple steatosis to MASH. However, the potential role of mitochondria structure on LD accumulation or MASH progression is unknown.
Inverse relationship between hepatic CM and PDM levels during MASLD progression
Conventionally, mitochondria isolation from homogenized animal tissues involves dual-speed centrifugation with the initial low-speed to remove debris and the second high-speed to pellet the mitochondria. In lipogenic tissues such as adipose, the initial low-speed centrifugation revealed an intact floating fat cake, facilitating the straightforward separation of PDM and CM29,30. However, one of the major obstacles in isolating liver PDM is that the low-speed centrifugation generates a thin floating fat layer (as opposed to a fat cake in adipose) that was very difficult to separate. To circumvent this problem, we developed a new separation method based on differences in sucrose gradients (described in the Methods and Fig. 2A) to isolate PDM from healthy, steatotic or fibrotic livers and to characterize their importance in hepatic LD accumulation and/or MASLD/MASH progression. Briefly, after low-speed centrifugation, we first separated the fat layer by overlaying with low sucrose buffer to bring the floating fat layer to the top. We next collected the fat layer to isolate PDM, while the supernatant was used to isolate CM (Fig. 2A). Our confocal analysis using neutral lipid dye (BODIPY) and mitochondrial dye (MTDR) clearly shows that LD-mitochondrial contacts were preserved in the floating fat layer, while such contacts were absent in the isolated hepatic CM and PDM (Supplementary Fig. 2A). Altogether, we manipulated the differences in sucrose gradients to isolate hepatic CM and PDM subpopulations at different levels of progression of MASLD.
A Schematic representation of hepatic CM and PDM isolation from healthy, steatotic and/or fibrotic mouse liver. Low-speed centrifugation of homogenized liver revealed a thin floating fat layer (as opposed to a fat cake in adipose tissues), which was then separated by overlaying with low sucrose buffer. The fat layer was then used to isolate PDM, while the supernatant was used for CM isolation. Comparisons of B, C CM or D, E PDM levels across different MASLD stages and percent fibrosis area are shown. Non-linear regression was used to fit the curve and the R squared values are shown. Data are presented as mean ± SEM (n = 4–5 livers per group for different MASLD stages and 3 for percent fibrosis area). P values were calculated by one-factor ANOVA with Holm-Sidak’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0. 001.
Given the existence of two different mitochondrial subpopulations in liver, we first aimed to test if differences in their levels drive the progression from early steatosis to advanced MASH. Overall, we observed the hepatic CM levels to be significantly greater than PDM irrespective of the conditions (Fig. 2B–E). We also observed that compared to healthy controls, CDAHFD significantly reduced hepatic CM levels (Fig. 2B). Specifically, correlation analyses with fibrosis quantification revealed that the hepatic CM levels (R2 = 0.73) initially decreased with early steatosis (i.e., 3 weeks) reaching its lowest at late steatosis (i.e., 6 weeks) and then appeared to increase as MASH progresses (Fig. 2C). In strong contrast, the hepatic PDM levels (R2 = 0.76) increased with early steatosis (i.e., 3 weeks) reaching its peak at late steatosis (i.e., 6 weeks), and then significantly decreased with disease progression i.e., advanced MASH (Fig. 2D, E and Supplementary Fig. 2B). This inverse relationship between hepatic CM and PDM levels from healthy to MASH phenotypes allowed us to speculate their functional differences as the disease progresses from steatosis to fibrosis. For instance, since the hepatic CM and PDM levels reached their respective lowest and peak at late steatosis (i.e., 6 weeks) and also, our histology and TEM examinations showed larger LDs at late steatosis (i.e., 6 weeks), we reasoned that hepatic PDM is highly active in DNL and/or LD expansion, whereas hepatic CM may promote FAO. Taken together, we speculated MASLD disease progression is under the tight control of hepatic mitochondrial subpopulations i.e., CM and PDM.
The proteomes of hepatic CM and PDM are compositionally and functionally distinct
To characterize the differences between hepatic CM and PDM across different stages of MASLD along with healthy controls, we subjected them to quantitative proteomics. We confirmed that both mitochondrial preparations from different conditions had similar protein levels (Supplementary Fig. 3). The principal component analysis (PCA) of the proteomic data revealed distinct clusters between hepatic CM and PDM under healthy and MASLD conditions (Fig. 3A), suggesting subpopulation- and disease-biased changes in proteomic profiles. Specifically, we found two significant observations. First, the fundamental differences in proteomic profiles were observed between two groups: healthy controls and early steatosis (i.e., 3 weeks) grouped together on one side, and advanced MASLD (i.e., late steatosis to advanced fibrosis) grouped on the other (explained by PC1). Next, within each group, distinct clustering of hepatic CM and PDM was observed in healthy controls and early steatosis (i.e., 3 weeks). However, this distinctive clustering was no longer apparent in advanced MASLD, with advanced MASH (i.e., 15 weeks) having completely overlapping clusters (Fig. 3A). Subsequent analyses, such as Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) as described36. Only enrichment terms that were statistically significant after FDR correction were included. Subcellular enrichment analyses by DAVID revealed ~57-61% of the proteins mapped to mitochondria followed by endoplasmic reticulum and peroxisomes (Fig. 3B). These findings suggest that mitochondrial protein enrichment in CM and PDM is comparable to and within the range of previously published methods29,32. Network enrichment analyses by DAVID revealed multiple metabolic pathways including electron transport, OXPHOS, TCA cycle, fatty acid, amino acid and steroid metabolism and urea cycle (Fig. 3C), comparable to prior studies29,32.
A PCA plot of proteomic data revealing distinct clusters between hepatic CM and PDM across healthy controls and different MASLD stages (n = 5 livers per group) are shown. DAVID B cellular compartment and C pathway enrichment analyses of the entire proteomic dataset are shown. Volcano plots showing significantly different proteomic changes and their respective DAVID enrichment analyses between hepatic CM and PDM isolated from D healthy controls, E early steatosis, F late steatosis, G early fibrosis and H late fibrosis are shown. I–L Distinct clusters of proteins identified by k-means clustering analyses and their respective DAVID enrichment analyses are shown. P values were calculated by FDR for DAVID enrichment analyses; D–H t test.
Next, we identified significantly different proteins and their respective network enrichments between hepatic CM and PDM in healthy and different MASLD stages (Fig. 3D–H). Specifically, we identified 139 CM and 19 PDM proteins that were significantly different in healthy controls (Fig. 3D), 90 CM and 28 PDM in early steatosis (Fig. 3E), 98 CM and 19 PDM in late steatosis (Fig. 3F), 36 CM and 41 PDM in early fibrosis (Fig. 3G) and 29 CM and 15 PDM in advanced MASH (Fig. 3H). It is worth noting that a greater number of proteins were significantly different between hepatic CM and PDM in healthy and steatotic livers and this difference reduced with fibrosis. Network enrichment analyses by DAVID revealed peroxisomal fatty acid oxidation, endoplasmic reticulum and peroxisomes in hepatic CM, while oxidoreductase, electron transport chain, ATP synthase, TCA cycle and amino acid biosynthesis were enriched in hepatic PDM irrespective of healthy or MASLD status (Fig. 3D–H). Interestingly, fatty acid oxidation was enriched in hepatic PDM from healthy controls (Fig. 3D), early steatosis (Fig. 3E) and early fibrosis (Fig. 3G). Lastly, we performed k-means clustering analysis37 on 169 proteins that were present in all conditions between hepatic CM and PDM and identified four distinct clusters (Fig. 3I–L). Cluster 1 (Fig. 3I) included proteins that were higher in hepatic PDM (i.e., Log2[PDM/CM] > 0) in early steatosis (i.e., 3 weeks) and early fibrosis (i.e., 9 weeks) and they notable mapped to mitochondrial FAO and TCA cycle. Cluster 2 (Fig. 3J) included proteins that were higher in hepatic PDM in early and late steatosis (i.e., 3 and 6 weeks), notably mapping to peroxisomal FAO. Clusters 3 and 4 (Fig. 3K, L) together included proteins that were higher in hepatic PDM in all conditions except late steatosis (i.e., 6 weeks), notably mapping to electron transport chain, OXPHOS, complex III, complex IV, ATP synthase and TCA cycle.
Our goal in performing quantitative proteomics was to identify a unique hepatic CM- and/or PDM-specific protein that can be modulated for further analyses. However, our analyses could only identify hepatic CM- and/or PDM-biased protein profiles similar to prior studies29,32. Nevertheless, all our proteomic observations taken together with LD size differences as well as inverse levels of hepatic CM and PDM during MASLD/MASH progression suggest that hepatic CM predominantly plays an oxidative role, while PDM predominantly plays an anabolic role exhibiting higher respiration and TCA flux capacities, irrespective of heathy or MASLD status.
Hepatic PDM have enhanced lipogenic capacity irrespective of healthy or MASLD status
Given the significant differences in hepatic CM and PDM levels and their respective proteomes during MASLD progression, we next analyzed the oxidation capacities of freshly isolated hepatic CM and PDM using Seahorse Bioanalyzer. First, we measured the mitochondrial pyruvate oxidation capacity by using pyruvate and malate as fuel source and found that in all conditions from healthy controls, steatosis to fibrosis, hepatic PDM showed significantly higher State 3 and ATP linked respiration capacities compared to their respective CM counterparts (Fig. 4A–E). Furthermore, within groups, both hepatic CM and PDM showed significantly higher State 3 respiration capacities only during late fibrosis compared to healthy controls (Supplementary Fig. 4).
Respirometry traces of freshly isolated mitochondria driven by pyruvate and malate, and their respective State 3 and ATP-linked respiration from livers of A healthy controls, B early steatosis, C late steatosis, D early fibrosis and E late fibrosis are shown. F Quantification of CS activity between hepatic CM and PDM across healthy controls and different MASLD stages are shown. Comparisons of G CM or H PDM CS activity and percent fibrosis area are shown. Non-linear regression was used to fit the curve and the R squared values are shown. Data are presented as mean ± SEM (n = 4–5 livers per group for different MASLD stages and 3 for percent fibrosis area). P values were calculated by A–E two-factor ANOVA with Holm-Sidak’s post hoc test; F multiple t tests. *P < 0.05; **P < 0.01.
Since higher pyruvate oxidation by hepatic PDM directly affects TCA cycle flux, we next measured the activity of TCA cycle regulatory enzyme, citrate synthase (CS). We found three significant observations. First, hepatic PDM had significantly elevated CS activity compared to their respective CM counterparts irrespective of heathy or disease stages with the exception of 6 weeks (Fig. 4F). Next, compared to heathy controls, the CS activity of hepatic CM (R2 = 0.77) increased with early steatosis (i.e., 3 weeks), almost doubled, reaching its peak at late steatosis (i.e., 6 weeks), and then plateaued with MASLD progression i.e., advanced MASH (Fig. 4G). Lastly, compared to heathy controls, the CS activity of PDM (R2 = 0.87) gradually increased with early steatosis (i.e., 3 weeks) reaching its peak at early fibrosis (i.e., 9 weeks), and then plateaued (Fig. 4H). All these observations confirmed the proteomic data that hepatic PDM have higher levels of TCA cycle proteins (Fig. 3I–L). Taken together, we conclude that the overall enhanced pyruvate oxidation capacity and the corresponding increased CS activity in hepatic PDM supports LD biogenesis and/or expansion by providing citrate for DNL.
Hepatic PDM have higher respiration capacity irrespective of healthy or MASLD status
Next, we measured the respiration capacities of mitochondrial complexes II and IV in previously frozen hepatic CM and PDM using succinate and rotenone, or TMPD and ascorbate as fuel sources, respectively, as described38. In our healthy control, early steatosis, and fibrosis livers, hepatic PDM showed significantly elevated complex II and/or IV respiration capacities in comparison to CM (Figs. 5A, B, D, E). However, in late steatosis (i.e., 6 weeks), the hepatic PDM did not show any significant differences in complex II and/or IV respiration capacities compared to their respective CM counterparts (Fig. 5C). Within groups, the hepatic CM exhibited increased complex II and/or IV respiration capacities as the disease progressed compared to healthy controls, while the hepatic PDM showed no significant changes (Supplementary Fig. 5).
A–E Respirometry traces of previously frozen mitochondria driven by succinate and rotenone for Complex II, and TMPD and ascorbate for Complex IV respiration, and follow-up (F–J) immunoblot analyses and their respective quantifications of OXPHOS complex subunits I-V from livers of A, F healthy controls, B, G) early steatosis, C, H late steatosis, D, I early fibrosis and E, J late fibrosis are shown. VDAC was used as a loading control. Data are presented as mean ± SEM (n = 4 livers per group). P values were calculated by A–E two-factor ANOVA with Holm-Sidak’s post hoc test; F–J multiple t tests. *P < 0.05; **P < 0.01; ***P < 0. 001.
To test if the higher respiration capacities of hepatic PDM could result from increased levels of OXPHOS protein complexes, we next determined the relative levels of OXPHOS proteins by immunoblot analysis. Overall, as MASLD progressed from steatosis to fibrosis, we observed at least one mitochondrial OXPHOS protein to be elevated in hepatic PDM compared to CM (Fig. 5F–J). Specifically, in comparison to their respective CM counterparts, we found that hepatic PDM have elevated complex I protein levels among all conditions, thus explaining their elevated pyruvate oxidation capacities as seen earlier; elevated complex II protein levels in healthy controls and early steatosis livers; elevated complex III and/or IV protein levels in healthy controls, early steatosis, and fibrosis livers; and elevated complex V protein levels only in late fibrosis livers (Fig. 5F–J). Most importantly, hepatic PDM in late steatosis (i.e., 6 weeks) had no significant differences in complex II through V protein levels (Fig. 5H), thus explaining the lack of apparent differences observed in their complex II and/or IV respiration capacities (Fig. 5C). These observations also confirmed the proteomic data that hepatic PDM have higher levels of electron transport chain, complex III, complex IV, ATP synthase and OXPHOS proteins in all conditions except late steatosis (i.e., 6 weeks) (Fig. 3K, L). However, within groups, the majority of OXPHOS proteins in both hepatic CM and PDM were reduced as the disease progressed compared to healthy controls (Supplementary Fig. 6), consistent with previous findings in total mitochondria during MASH progression39. Taken together, we conclude that irrespective of the disease progression, hepatic PDM is bioenergetically active with higher respiration capacities along with elevated OXPHOS protein levels.
Hepatic CM and PDM differentially regulates FAO depending on MASLD status
So far, our results show that, compared to CM, hepatic PDM is bioenergetically active, has increased pyruvate oxidation capacity, increased TCA cycle flux and higher OXPHOS capacity as well as protein levels. Taken together, these findings indicate that hepatic PDM serve as specialized organelles for lipogenesis in healthy liver and during MASLD progression. However, hepatic and/or mitochondrial lipid homeostasis is maintained by a delicate balance between lipogenesis vs. oxidation. Therefore, in order to completely understand the role of hepatic PDM in lipid homeostasis during MASH progression, we wanted to test if there were any differences in FAO capacity between freshly isolated hepatic CM and PDM from healthy to disease phenotypes. For this, we used palmitoyl-CoA, carnitine, and malate as fuel sources. To our surprise, in both healthy controls and early steatosis, we found that hepatic PDM had elevated FAO capacity as shown by significantly higher State 3 and/or ATP linked respiration capacities compared to their respective CM counterparts (Fig. 6A, B). This perhaps supports the recent finding indicating hepatic PDM are specialized for FAO in healthy rat livers under fed conditions31. We further reasoned that during a healthy state, since hepatic PDM are bioenergetically active, they exhibit a preference for utilizing both substrates (pyruvate and palmitoyl CoA) to maintain a delicate stoichiometric balance between DNL and FAO. Also, we speculated that the increased FAO capacity in healthy hepatic PDM is maintained during early steatosis for clearing the lipotoxic FFA. In strong contrast, during later stages of the disease, the FAO capacity is comparable between hepatic CM and PDM with no significant differences (Fig. 6C–E). On the other hand, within groups, both hepatic CM and PDM showed significantly higher State 3 respiration capacities under most disease conditions compared to healthy controls (Supplementary Fig. 7). This suggests a dynamic shift in FAO activity during MASLD progression, likely reflecting adaptations to the increasing lipid burden.
Respirometry traces of freshly isolated mitochondria driven by palmitoyl-CoA, carnitine and malate, and their respective State 3 and ATP-linked respiration from livers of A healthy controls, B early steatosis, C late steatosis, D early fibrosis and E late fibrosis are shown. F Ratio of PDM to CM State 3 respiration fueled by pyruvate and malate (PyrO) or palmitoyl-CoA, carnitine, and malate (FAO) across healthy controls and different MASLD stages are shown. G, I Immunoblot analyses and their respective (H, J) quantifications of MFN2 and DRP1 protein levels across healthy controls and different MASLD stages are shown. VDAC was used as a loading control. Note: These images were derived from the same blots used to probe the OXPHOS proteins shown in Fig. 5 and, therefore, utilized the same VDAC loading control. Please refer to the uncropped and unedited blot/gel images in Supplementary Figs. 8–18 for further details. Data are presented as mean ± SEM (n = 4–5 livers per group). P values were calculated by (A–E) two-factor ANOVA with Holm-Sidak’s post hoc test; F one sample t test against a hypothetical value of PDM/CM = 1; H, J multiple t tests. *P < 0.05; **P < 0.01; ***P < 0. 001.
Given that DNL (from pyruvate oxidation and subsequent CS activity) and FAO are two competing pathways, we next tested if the elevated OXPHOS capacity in hepatic PDM is supported by pyruvate oxidation or FAO. In essence, we sought to determine the predominant substrate preference of hepatic PDM. For this, we calculated the ratio of hepatic PDM to CM oxidation capacities from healthy to disease phenotypes. We found three significant observations. First, hepatic PDM had higher pyruvate oxidation capacity compared to FAO capacity irrespective of the disease status (Fig. 6F). This finding reiterates that hepatic PDM primarily serve as specialized organelles for lipogenesis in healthy liver and during MASLD progression. However, they also retain their FAO capacity when required. Next, we found that the pyruvate oxidation capacity was higher in hepatic PDM from healthy controls and early steatosis, then decreased in late steatosis (i.e., 6 weeks), and subsequently increased in fibrosis samples (Fig. 6F). This finding correlates with the rapid increase in the CS activity of hepatic CM in late steatosis (i.e., 6 weeks) (Fig. 4F, G) as well as our histology and TEM examinations showing larger LDs at late steatosis (i.e., 6 weeks) (Fig. 1A, C). Altogether, our data indicate that hepatic CM along with PDM contribute to the enlargement of LDs during late steatosis (i.e., 6 weeks). However, beyond this point, PDM appears to revert to their lipogenic function. Lastly, we found that the FAO capacity was higher in hepatic PDM only from healthy controls and early steatosis, then decreased in late steatosis (i.e., 6 weeks), and remain unchanged in fibrosis samples (Fig. 6F). We believe this observation to be crucial as it links to our previous findings regarding the inverse relationship between hepatic CM and PDM levels (Fig. 2B–E). During late steatosis (i.e., 6 weeks), the hepatic CM level reaches its lowest while the PDM is at its highest, coinciding with the maximal size of hepatic LDs. These observations also confirmed the proteomic data that hepatic PDM have higher levels of FAO proteins only in healthy controls and early steatosis, while CM have higher levels of peroxisomal FAO proteins irrespective of the heathy or MASLD status (Fig. 3D–H). Taken together, we conclude that the enhanced FAO capacity of healthy hepatic PDM is maintained during early steatosis but is compromised as MASH progresses. At this point, the hepatic CM assume the compensatory FAO role as an adaptive mechanism.
Hepatic PDM have higher MFN2 protein levels in healthy and early steatosis
So far, our findings have indicated that hepatic PDM function as specialized organelles for lipogenesis, while also demonstrating their FAO capacity during both healthy and early steatosis conditions. Also, our proteomics data have shown that these subpopulations have distinct proteome. Furthermore, as seen in our TEM examinations, the mitochondria were larger and elongated in healthy and early steatosis, while they were reduced in size and appear fragmented with MASH progression (Fig. 1C). It is well known that mitochondria undergo continuous cycles of fusion and fission events to maintain their turnover and integrity. Therefore, we reasoned that the hepatic mitochondrial subpopulations are maintained distinct through their unique dynamics profile. To test this, we next assessed the protein levels of mitochondrial fusion protein 2 (MFN2) in hepatic CM and PDM. We found that MFN2 protein levels were higher in hepatic PDM compared to CM in both healthy controls and early steatosis (Fig. 6G, H). As the disease progressed, we did not see any significant differences with MFN2 protein levels between hepatic CM and PDM (Fig. 6G, H). Interestingly, we observed a trend toward higher levels of dynamin-related protein 1 (DRP1) protein levels (mitochondrial fission protein) in hepatic PDM compared to CM in healthy controls (Fig. 6I, J). In stark contrast, as the disease progressed, DRP1 protein levels were significantly elevated in hepatic CM compared to PDM during both early and late fibrosis (Fig. 6I, J). These findings further support our TEM examinations that mitochondrial fusion is compromised and replaced by fission as the disease progress to fibrosis (Fig. 1C). Also, this is consistent with previous findings that both human MASH subjects and mouse models of MASLD/MASH have reduced hepatic MFN2 protein levels compared to their respective controls40. Furthermore, it has been demonstrated that both in mouse BAT29 and healthy rat livers31, PDM exhibited a more elongated structure and had higher levels of MFN2 protein. Taken together, we conclude that hepatic PDM has a unique fusion dynamics to maintain their integrity in healthy and early steatosis conditions that is lost with MASH progression corroborating our proteomic data (Fig. 3A). At this point, the hepatic CM exhibits a unique fission dynamics, possibly serving as an adaptive mechanism, as previously described41.
Discussion
Liver TG accumulation is regulated by communication among metabolic tissues and any imbalance between lipid storage and clearance in these tissues due to metabolic syndrome traits such as obesity and hyperlipidemia manifests into metabolic diseases such as diabetes and MASLD42,43. On the other hand, several conflicting studies have demonstrated that liver TG accumulation is a consequence of enhanced or failure in liver mitochondrial functions (reviewed in ref. 28). Thus, it is currently debatable whether liver mitochondria can aggravate or alleviate the MASLD disease progression. Moreover, despite extensive research, only one MASH drug was recently approved by the FDA44, which may target liver mitochondria45. To this end, we have now identified and characterized two independent hepatic mitochondrial subpopulations with unique proteomes and fission/fusion dynamics that compartmentalize metabolic functions during MASLD progression.
Although, two different mitochondrial subpopulations namely, LD-associated PDM and unassociated CM, have been identified and characterized in BAT29, WAT30 and liver31,32 tissues from healthy animals, their function during MASLD disease progression from steatosis to fibrosis remains unknown. To address this fundamental question, we employed a mouse model of diet-induced MASLD progression from early steatosis to advanced MASH. Currently, there exists no ‘one size fits all’ mouse model that recapitulates all subsets of human MASLD/MASH. Over the years, we have seen numerous diet-induced mouse models of MASH, but their metabolic profile, histological patterns and pathological profiles differ from human MASH. Despite these differences, there are a myriad of mouse models that recapitulate many important histopathological features of MASLD including hepatic steatosis, inflammation, and fibrosis33. For the purpose of our study, we employed a routinely used MASLD/MASH model that develops different stages of MASLD in a shorter duration, i.e., CDAHFD model over a period of 3, 6, 9 or 15 weeks33,34,35. As a healthy control, we used chow-fed animals. It is true that CDAHFD model does not encompass human MASLD/MASH comorbidities such as obesity, peripheral insulin resistance and dyslipidemia, but it enables rapid and stable evaluations of all MASLD stages from steatosis and fibrosis.
Next, we used TEM to characterize LD size, mitochondria shape and structure and demonstrate LD-mitochondrial contacts during MASLD progression. Throughout MASLD progression, we observed a remarkable difference in LD size and mitochondria structure. As shown in our TEM images, the mitochondria were larger and elongated in both healthy controls and early steatosis, while they were round, smaller, or fragmented in advanced MASLD/MASH. Corroborating this, we found that MFN2 protein levels were higher in hepatic PDM in both healthy controls and early steatosis, but not as the disease progressed. Conversely, DRP1 protein levels were higher in hepatic CM in both early and late fibrosis. Simultaneously, the LD increased in size with steatosis followed by a decline with MASH phenotypes. These observations made us hypothesize that liver LD size is regulated by the shape of the mitochondria and/or their contacts with LD. Our prediction was supported by previous findings in which LD-mitochondrial contacts were shown to regulate LD size29,30,31,32. Also, the differences in mitochondrial dynamics during MASLD progression are supported by previous findings demonstrating that loss of MFN2 drives steatosis to fibrosis phenotype40,46 possibly via inhibition of TGF-β1/Smad signaling pathway47. Furthermore, MFN2 is known to regulate LD-mitochondrial contacts in BAT48,49 and that PDM from both healthy BAT and rat livers had higher MFN2 levels29,31. Conversely, previous studies have shown that DRP1 loss can have dual effects: it alleviates diet-induced hepatic steatosis when disrupted early50 but exacerbates MASH when disrupted late41. Taken together, these observations suggest that specific changes in mitochondrial dynamics among hepatic CM and PDM may play a pivotal role in regulating LD size and driving the progression from steatosis to fibrosis.
Next, to characterize the individual roles of hepatic mitochondrial subpopulations in MASLD progression and LD homeostasis, we developed an approach to isolate them. The barrier in separating hepatic PDM was that after the initial low-speed centrifugation of liver homogenates, we observed a thin floating fat layer (as opposed to an intact floating fat cake in adipose tissues29,30). Therefore, we took advantage of the buoyancy of LDs and used differences in sucrose gradients to separate the LD-bound PDM from CM. Our method not only isolated hepatic PDM from steatotic livers but also from healthy livers where hepatic LD accumulation was minimal. Furthermore, our quantitative proteomic analyses revealed that the proteomes of the two mitochondrial subpopulations are compositionally and functionally distinct.
After separating the hepatic CM and PDM, we first quantified them at different stages of MASLD in comparison to healthy controls and uncovered an inverse relationship between their levels. Compared to healthy controls, we found that hepatic CM levels decreased with steatosis and concomitantly, there was an increase in PDM levels. In strong contrast, the hepatic CM levels appeared to increase with fibrosis, while PDM levels decreased. Thus, our study highlights their importance in determining the disease severity from steatosis to fibrosis. Furthermore, as seen in our H&E, trichrome staining and TEM imaging, the larger LD size that we observed during late steatosis (i.e., 6 weeks) was directly proportional to the highest PDM and lowest CM levels. Thus, we speculated that hepatic PDM facilitates LD expansion rather than oxidation during MASLD progression. Our speculations were further supported by studies that demonstrated PDM’s role in LD expansion in BAT and liver tissues29,32. Corroborating this, we indeed observed that hepatic PDM was bioenergetically active with higher pyruvate oxidation and respiration capacities along with higher OXPHOS protein levels in both healthy and diseased livers. Moreover, several studies have demonstrated that citrate-induced hepatic DNL causes steatosis in the liver51,52,53,54,55,56. Corroborating this, we found higher CS activities in hepatic PDM compared to CM, which significantly increased from healthy to MASLD progression, highlighting PDM’s role in lipid anabolism. Added to this, the significant increase in hepatic PDM levels from healthy to steatosis that coincides with increased LD size highlights the importance of PDM in LD expansion.
Since we had demonstrated an anabolic role for hepatic PDM, we next wanted to investigate which mitochondrial subpopulations play a catabolic role during MASLD progression. To address this, we performed mitochondrial bioenergetics using palmitoyl-CoA and carnitine (to measure CPT1-mediated FAO) as a fuel source. In the absence of steatosis (i.e., healthy controls) and at early steatosis (i.e., 3 weeks), the FAO capacity of hepatic PDM was higher in comparison to their respective CM. However, the PDM’s FAO capacity did not change from late steatosis to advanced MASH. Hence, we speculated that hepatic PDM’s compromised FAO capacity drives MASLD progression from steatosis to fibrosis. In other words, the LD accumulation along the spectrum of MASLD progression is largely dependent on the dynamic functioning of hepatic PDM. This was supported by prior research, which reported that hepatic PDM performs FAO in healthy conditions and that their compromised FAO capacity could cause LD accumulation31. In strong contrast, we observed an increase in CM’s FAO capacity as the disease progressed from late steatosis to advanced MASH, thus playing a compensatory catabolic role.
Overall, considering the demonstrated differences in their levels, proteome, dynamics, pyruvate oxidation, respiration, CS activities and FAO capacities between hepatic CM and PDM and the resulting LD size changes from healthy to steatosis and/or fibrosis, we conclude that i) hepatic PDM are specialized organelles for lipogenesis in both healthy and diseased livers, ii) hepatic PDM can also perform FAO in healthy conditions to regulate or maintain the LD size, and in early steatosis to prevent lipotoxicity due to nutrient abundance, iii) hepatic CM assumes a compensatory catabolic role in response to PDM’s compromised FAO capacity with MASH progression, and lastly iv) hepatic CM and PDM have unique fusion dynamics to maintain their proteomic integrity that is lost with MASH progression.
Although this study is the first to highlight the critical role of hepatic mitochondrial subpopulations in MASLD/MASH progression, we have few limitations. Our findings reveal distinct functional roles for hepatic CM and PDM at different stages of MASLD, suggesting that selectively targeting specific mitochondrial subpopulations at specific disease stages could be a promising strategy to mitigate the progression from steatosis to fibrosis. However, the proteomics data did not identify any subpopulation-specific molecular targets. Additionally, developing a small molecule inhibitor capable of targeting one mitochondrial subpopulation without affecting the other remains a significant challenge so far, but we believe this will become feasible in the near future. Future research should systematically explore the potential of targeting individual hepatic mitochondrial subpopulations and their roles in lipid accumulation, inflammation, and fibrosis.
Methods
Animals
All mice were purchased from the Jackson Laboratory and maintained at the University of Cincinnati according to approved Institutional Animal Care and Use Committee (IACUC) protocols. We have complied with all relevant ethical regulations for animal use. All animal experiments performed in this study were approved by the IACUC of the University of Cincinnati. Animals were maintained under standard housing condition with 12 h light/dark cycle and health was monitored daily by vivarium personnel. For steatosis to fibrosis models, 8-week-old male C57BL/6J mice were fed a choline-deficient L-amino acid-defined high fat diet (CDAHFD) (Research Diets-A06071302) for 3, 6, 9 or 15 weeks.
Histology
All animals were subjected to 4 h fasting prior to sacrifice. Livers were extracted and a 5 mm cubes were cut and fixed in ice cold 4% paraformaldehyde for 72 h and processed for paraffin sections. 5 µm thin paraffin sections were used for H&E’s and trichrome staining. H&E’s were stained using the VENTANA HE 600 system (Roche) and the Trichrome was stained using the BenchMark Special Stains system (Roche) and the VENTANA HE 600 system (Roche). ImageJ software was then used to quantify the collagen deposition in Trichrome images. Briefly, using the ‘Colour Deconvolution 2’ ImageJ plugin and selecting the Masson Trichrome setting, the color threshold was adjusted, and the percent area of collagen deposition was quantified in each image. H&E images were scored by a professional liver pathologist for the severity of hepatocellular steatosis, ballooning and inflammation according to the NASH-CRN criteria57.
Transmission electron microscopy (TEM)
For transmission electron microscopy (TEM), livers were cut into 1 mm cubes and fixed in 2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 (Electron Microscopy Sciences #15960-01). Post fixing, tissues were further fixed in 1% osmium tetroxide in 0.15 M sodium cacodylate buffer, processed through a series of alcohols, infiltrated, and embedded in the LX-112 resin. After polymerization at 60 °C for three days, ultrathin sections (120 nm) were cut using a Leica EM UC7 ultramicrotome and counterstained in 2% aqueous uranyl acetate and Reynold’s lead citrate. Images were taken with a transmission electron microscope (Hitachi H-7650) equipped with a digital camera (Biosprint 16).
Isolation of hepatic CM and PDM
It is widely understood that mitochondrial isolation procedures commonly use a four hour to overnight fasting period to remove differences in hormonal and/or nutrient levels that can affect homogeneous mitochondrial preparations58,59. However, we did not follow overnight or 16 h fasting because it exhibited fasting-induced hepatic steatosis32,60. Our study focused on investigating the hepatic mitochondrial subpopulations in diet-induced MASLD/MASH, thus we fasted for four hours, as previously reported61,62,63. Livers were harvested and washed in ice cold 1X PBS to remove blood and then transferred to homogenization buffer with BSA (250 mM Sucrose, 5 mM HEPES, 2 mM EGTA, 2% BSA, pH 7.2) and minced finely. The minced liver pieces were transferred into glass-Teflon homogenizer and homogenized with 15 up and down strokes. The homogenate was collected into 15 mL tubes and centrifuged at 900 × g for 10 min at 4 °C. Post centrifugation, the floating fat layer was carefully layered with ice-cold layering buffer with BSA (70 mM Sucrose, 210 mM Mannitol, 5 mM HEPES, 1 mM EGTA, 2% BSA, pH 7.2) allowing the fat layer to separate, which was then transferred into a new 15 mL tube. After collecting the fat layer, the supernatant was collected into a new 15 mL tube. Both the fat layer and the supernatant tubes were centrifuged twice at 900 × g for 10 min at 4 °C to remove any debris, followed by high-speed centrifugation at 9000 × g for 10 min at 4 °C to pellet down PDM and CM, respectively. Both the pellets were washed by resuspending them in homogenization buffer with BSA and centrifuged at 10,000 × g for 10 min at 4 °C. The washing step was repeated by resuspending the pellets in homogenization buffer without BSA and centrifuged at 10,000 × g for 10 min at 4°C. Final resuspension of mitochondrial pellets was done in 1X MAS buffer (70 mM Sucrose, 220 mM Mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1 mM EGTA, pH 7.2) and protein estimation was determined by BCA (Thermo Scientific).
Mass spectrometry and quantitative proteomic analyses
Fifty samples each containing 2.25 µg of protein were constituted in 25.5 µL of 2X Laemmli sample buffer. The samples were vortexed, heated 100 °C for 10 min and loaded on a 1.5 mm, 4–12% gradient gels using MOPS buffer with prestained molecular weight marker lanes in between. They were electrophoresed until the samples ran 1.5 cm into the gel. The gel was fixed in 50% ethanol/10% acetic acid overnight at RT, then washed in 30% ethanol for 10 min followed by two 10 min washes in MilliQ water (MilliQ Gradient system). The lanes were cut out of the gel based on the boundaries from the prestained protein marker lanes, and further cut into small (~2 mm) squares. The samples were subjected to in-gel digestion and recovery of the peptides as described64. Briefly, this included reduction of disulfide bonds with dithiothreitol, alkylation of cysteine with iodoacetamide, digestion with trypsin, extracted of the peptide and drying of the samples in a SpeedVac concentrator and storage at −80 °C until ready for tandem isotope tagging. The fifty samples plus the five bridge pools were Tandem Mass Tag (TMT)-labeled (Thermo Scientific) following the vendor protocol for 0.2 mg of TMT10plex isobaric labels. TMT-labeled peptides were combined in equal portions (2.25 µg) from each sample. Each of the TMT sets were dried in a SpeedVac and were reconstituted in 0.1% Formic acid (FA) for mass spectrometry.
Data were collected in duplicate on an Orbitrap Eclipse mass spectrometer (Thermo Scientific) coupled to a Dionex Ultimate 300 RSLCnano system (Thermo Scientific). Three microgram of the total TMT labeled peptides combined from each sample was injected onto a 5 mm nanoviper μ-Precolumn (i.d.300 μm, C18 PepMap 100, 5.0 μm, 100 Å) from Thermo Scientific at 5 µl/min in formic acid/H2O 0.1/99.9 (v/v) for 5 min to desalt and concentrate the samples. For the chromatographic separation of peptides, the trap-column was switched to align with the EASY-Spray column PepMap RSLC C18 with a 150 mm column (i.d. 75 μm, C18, 3.0 μm, 100 Å). The peptides were eluted using a variable mobile phase (MP) gradient from 98% phase A (Formic acid/H2O 0.1/99.9, v/v) to 4% phase B (Formic Acid/Acetonitrile 0.1/99.9, v/v) for 3 min, from 4% phase B to 16% phase B for 83 min, from 16% phase B to 25% phase B for 45 min, from 25% phase B to 85% phase B for 5 min and then keeping the same mobile phase composition for 5 more minutes at 300 nL/min. MS1 data were collected in the orbitrap (120,000 resolution; maximum injection time 50 ms; AGC was standard which was selected via automated fashion per scan type). Charge states between 2 and 6 were required for MS2 analysis, precursor selection mass range 400 m/z to 1400 m/z, and a 20 s dynamic exclusion window was used. Cycle time was set at 3.0 s. MS2 scans were performed in the quadrupole with CID fragmentation (isolation window 0.7 m/z; NCE 35%; maximum injection time was auto for maximize sensitivity while maintaining maximum scan rate; AGC was set to standard, which was selected via automated fashion per scan type; isobaric tag loss exclusion TMT; SPS 10 precursors). MS3 scans were collected in the orbitrap with HCD fragmentation (50,000 resolution; isolation window 2.0 m/z; NCE 65%; AGC target 200%, maximum injection time 1.5 ms). The data was recorded using Xcalibur 4.5.445.18 software (Thermo Scientific).
Peptide identification and TMT-based reporter ion quantitation was achieved using Proteome Discoverer 3.0 (Thermo Scientific) searched against a combined contaminant and Uniprot mus musculus database (UP000000589) with the Sequest HT search algorithm. The samples were grouped into five categories: BRIDGE POOL; week 3 and week 15 chow CM; week 3 and week 15 chow PDM; week 6, week 9, and week 15 PDM; week 6, week 9 and week 15 CM. In the processing workflow, the mass recalibration node (spectrum files RC) along with the standard spectrum selector, Reporter ions quantifier, Sequest HT and Percolator nodes were used. The precursor mass tolerance was 10ppm and the fragment mass tolerance was set to 0.06 Da as these were run using a SPS MS3 instrument method. The search parameters included 2 missed trypsin cleavages and a variable peptide modification of oxidized methionine. Static modifications were TMT 10plex (+229.163) on lysine, end terminus TMT 10plex and carbamidomethyl on cysteines. Default variable N-terminal protein modifications of Acetyl, Met loss and acetyl+met loss were also used. The TMT quantitation method was a 10plex TMT + 131 C (TMT11plex 10plex YG375131 + 131 C WK307858) and adjustments were made for isotope impurities in the reporter ions. FDR tolerances in the Percolator node were set to 0.05 for medium confidence and 0.01 for high confidence peptides. The consensus was the standard workflow using normalization to total peptides and scaling to a bridge control average. The bridge control was made from an equal amount of all the samples. Peptide abundance calculation was based on the summed abundance of the TMT reported ions for the collective peptides for each protein compared across each sample group. P values were calculated using a t-test. In order to minimize missing abundance values, the S/N was set to 5 and the SPS mass match percentage to 50%. The maximum allowed fold change is 100-fold, which is thus represented as 100 for increased ratio or 0.01 for the maximum decreased ratio. The exported data was filtered for medium (95%) FDR confidence, 1 peptide per protein, Sequest score >0 and removal of contaminants such as trypsin and keratin.
Coupled bioenergetics in hepatic CM and PDM
Seahorse XF Pro Analyzer (Agilent) was used to measure coupled mitochondrial respiration. Equal amounts (5 µg) of freshly isolated hepatic CM and PDM were used for both Pyruvate/Malate mediated respiration and fatty acid oxidation (FAO) assay. For Pyruvate/Malate mediated respiration, 5 mM pyruvate, 0.5 mM malate and 4 mM ADP in assay buffer (1X MAS with 0.2% BSA) were used. For FAO, 80 µM Palmitoyl Co-A, 0.5 mM malate, 0.5 mM Carnitine and 4 mM ADP in assay buffer were used. Then, oxygen consumption rates (OCR) were measured before and after the sequential injections of 2.5 µM oligomycin, 4 µM FCCP, and 1 µM of rotenone/antimycin A. Measures were normalized by total protein.
Frozen bioenergetics in hepatic CM and PDM
Seahorse XF Pro Analyzer (Agilent) was used to measure frozen mitochondrial respirometry as described38. Equal amounts (1 µg) of previously frozen hepatic CM and PDM were taken in assay buffer (1X MAS with 10 µg/mL of Cytochrome C). Then, OCR were measured before and after the sequential injections of Port A: 5 mM Succinate and 2 µM Rotenone (Complex II substrate and Complex I inhibitor respectively), Port B: 4 µM Antimycin (Complex III inhibitor), Port C: 0.5 mM TMPD and 1 mM Ascorbate (Complex IV substrate) and Port D: 50 mM Sodium azide (Complex IV inhibitor). Measures were normalized by total protein.
Citrate synthase assay in hepatic CM and PDM
Citrate synthase assays were performed with equal amounts (5 µg) of hepatic CM and PDM as described65. Reaction mixture of 100 mM Tris buffer with 0.1% triton X 100, 100 μM DTNB, and 0.3 mM Acetyl Co-A was incubated with mitochondria for 2 min. Reaction started by adding 0.5 mM oxaloacetic acid (OAA) to the mitochondria reaction mixture and absorption increase was recorded for 3 min at 412 nm.
Immunoblotting analyses
Equal amounts (10 µg) of hepatic CM and PDM were resolved in 4%-12% Bis-Tris gels (Invitrogen). Proteins were then transferred to polyvinylidene difluoride (PVDF) membrane (Thermo Scientific) and probed by using rabbit monoclonal VDAC (Abcam #ab154856), rabbit monoclonal MFN2 (Cell Signaling #9482), total OXPHOS rodent western blotting antibody cocktail (Abcam #ab110413), and their respective fluorescent labeled secondary antibodies. As depicted in the Supplementary Fig. 8, each blot was divided into two sections along the 70 kDa marker. The upper section (>70 kDa) was used to probe either the MFN2 (Fig. 6G) or DRP1 (Fig. 6I) proteins, while the lower section (<70 kDa) was used to probe both VDAC (Figs. 5F–J and 6G, I) and OXPHOS proteins (Fig. 5F–J). Consequently, both OXPHOS and MFN2 or DRP1 proteins were normalized using the same VDAC probe and are presented in Figs. 5 and 6. Blots were visualized in Infrared Imager LiCor Odyssey CLx and quantified using ImageJ.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Statistics and reproducibility
Graphs and statistical analyses were performed using Prism v10.2.3 (GraphPad Software). Error bars plotted on graphs are presented as the mean ± standard error of the mean (SEM) unless reported otherwise. The sample sizes (n ≥ 3) in each figure are indicated in the respective figure legends. The critical significance value (α) was set at 0.05, and if the P values were less than α, we reported that by rejecting the null hypothesis, the observed differences were statistically significant.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE66 partner repository with the dataset identifier PXD052526. Uncropped and unedited blot/gel images are shown in Supplementary Figs. 8–18. The source data can be obtained in Supplementary Data 1. All other data are available from the corresponding author (or other sources, as applicable) on reasonable request.
References
Rinella, M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 78, 1966–1986 (2023).
Eslam, M., Sanyal, A. J., George, J. & International Consensus, P. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 158, 1999–2014.e1991 (2020).
Adams, L. A. et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology 129, 113–121 (2005).
Browning, J. D. et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 40, 1387–1395 (2004).
Kopec, K. L. & Burns, D. Nonalcoholic fatty liver disease: a review of the spectrum of disease, diagnosis, and therapy. Nutr. Clin. Pr. 26, 565–576 (2011).
Vernon, G., Baranova, A. & Younossi, Z. M. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharm. Ther. 34, 274–285 (2011).
Povsic, M., Wong, O. Y., Perry, R. & Bottomley, J. A Structured Literature Review of the Epidemiology and Disease Burden of Non-Alcoholic Steatohepatitis (NASH). Adv. Ther. 36, 1574–1594 (2019).
Ratziu, V., Bellentani, S., Cortez-Pinto, H., Day, C. & Marchesini, G. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J. Hepatol. 53, 372–384 (2010).
Pessayre, D. & Fromenty, B. NASH: a mitochondrial disease. J. Hepatol. 42, 928–940 (2005).
Caldwell, S. H. et al. Mitochondrial abnormalities in non-alcoholic steatohepatitis. J. Hepatol. 31, 430–434 (1999).
Sanyal, A. J. et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120, 1183–1192 (2001).
Ibdah, J. A. et al. Mice heterozygous for a defect in mitochondrial trifunctional protein develop hepatic steatosis and insulin resistance. Gastroenterology 128, 1381–1390 (2005).
Sobaniec-Lotowska, M. E. & Lebensztejn, D. M. Ultrastructure of hepatocyte mitochondria in nonalcoholic steatohepatitis in pediatric patients: usefulness of electron microscopy in the diagnosis of the disease. Am. J. Gastroenterol. 98, 1664–1665 (2003).
Begriche, K., Igoudjil, A., Pessayre, D. & Fromenty, B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion 6, 1–28 (2006).
Struben, V. M., Hespenheide, E. E. & Caldwell, S. H. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am. J. Med. 108, 9–13 (2000).
Willner, I. R. et al. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am. J. Gastroenterol. 96, 2957–2961 (2001).
Wagenknecht, L. E. et al. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity 17, 1240–1246 (2009).
Mashek, D. G. Hepatic lipid droplets: A balancing act between energy storage and metabolic dysfunction in NAFLD. Mol. Metab. 50, 101115 (2021).
Romeo, S. et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet 40, 1461–1465 (2008).
Su, W. et al. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA 111, 11437–11442 (2014).
Faulkner, C. S., White, C. M., Shah, V. H. & Jophlin, L. L. A single nucleotide polymorphism of PLIN2 is associated with nonalcoholic steatohepatitis and causes phenotypic changes in hepatocyte lipid droplets: A pilot study. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158637 (2020).
Mancina, R. M. et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 150, 1219–1230.e1216 (2016).
Kozlitina, J. et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet 46, 352–356 (2014).
Kramer, D. A., Quiroga, A. D., Lian, J., Fahlman, R. P. & Lehner, R. Fasting and refeeding induces changes in the mouse hepatic lipid droplet proteome. J. Proteom. 181, 213–224 (2018).
Rambold, A. S., Cohen, S. & Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015).
Nguyen, T. B. et al. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell 42, 9–21.e25 (2017).
Freyre, C. A. C., Rauher, P. C., Ejsing, C. S. & Klemm, R. W. MIGA2 Links Mitochondria, the ER, and Lipid Droplets and Promotes De Novo Lipogenesis in Adipocytes. Mol. Cell 76, 811–825.e814 (2019).
Shum, M., Ngo, J., Shirihai, O. S. & Liesa, M. Mitochondrial oxidative function in NAFLD: Friend or foe? Mol. Metab. 50, 101134 (2021).
Benador, I. Y. et al. Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab. 27, 869–885.e866 (2018).
Brownstein, A. J. et al. Mitochondria isolated from lipid droplets of white adipose tissue reveal functional differences based on lipid droplet size. Life Sci. Alliance 7, e202301934 (2024).
Talari, N. K. et al. Lipid-droplet associated mitochondria promote fatty-acid oxidation through a distinct bioenergetic pattern in male Wistar rats. Nat. Commun. 14, 766 (2023).
Najt, C. P. et al. Organelle interactions compartmentalize hepatic fatty acid trafficking and metabolism. Cell Rep. 42, 112435 (2023).
Gallage, S. et al. A researcher’s guide to preclinical mouse NASH models. Nat. Metab. 4, 1632–1649 (2022).
Matsumoto, M. et al. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J. Exp. Pathol. 94, 93–103 (2013).
Cheng, D. et al. MGAT2 inhibitor decreases liver fibrosis and inflammation in murine NASH models and reduces body weight in human adults with obesity. Cell Metab. 34, 1732–1748.e1735 (2022).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Hartigan, J. A. & Wong, M. A. Algorithm AS 136: A K-Means Clustering Algorithm. Appl. Stat. 28, 100–108 (1979).
Acin-Perez, R. et al. A novel approach to measure mitochondrial respiration in frozen biological samples. EMBO J. 39, e104073 (2020).
Sugasawa, T. et al. One Week of CDAHFD Induces Steatohepatitis and Mitochondrial Dysfunction with Oxidative Stress in Liver. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22115851 (2021).
Hernandez-Alvarez, M. I. et al. Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease. Cell 177, 881–895.e817 (2019).
Steffen, J. et al. The mitochondrial fission protein Drp1 in liver is required to mitigate NASH and prevents the activation of the mitochondrial ISR. Mol. Metab. 64, 101566 (2022).
Green, C. J. & Hodson, L. The influence of dietary fat on liver fat accumulation. Nutrients 6, 5018–5033 (2014).
Scheja, L. & Heeren, J. Metabolic interplay between white, beige, brown adipocytes and the liver. J. Hepatol. 64, 1176–1186 (2016).
FDA approves first MASH drug. Nat. Biotechnol. 42, 540 (2024).
Karim, G. & Bansal, M. B. Resmetirom: An Orally Administered, Smallmolecule, Liver-directed, beta-selective THR Agonist for the Treatment of Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis. touchREV Endocrinol. 19, 60–70 (2023).
Sebastian, D. et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl. Acad. Sci. USA 109, 5523–5528 (2012).
Zhu, H. et al. Specific Overexpression of Mitofusin-2 in Hepatic Stellate Cells Ameliorates Liver Fibrosis in Mice Model. Hum. Gene Ther. 31, 103–109 (2020).
Boutant, M. et al. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36, 1543–1558 (2017).
Mahdaviani, K. et al. Mfn2 deletion in brown adipose tissue protects from insulin resistance and impairs thermogenesis. EMBO Rep. 18, 1123–1138 (2017).
Galloway, C. A., Lee, H., Brookes, P. S. & Yoon, Y. Decreasing mitochondrial fission alleviates hepatic steatosis in a murine model of nonalcoholic fatty liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G632–G641 (2014).
Ferre, P. & Foufelle, F. Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP-1c. Diab. Obes. Metab. 12, 83–92 (2010).
Belew, G. D. & Jones, J. G. De novo lipogenesis in non-alcoholic fatty liver disease: Quantification with stable isotope tracers. Eur. J. Clin. Invest 52, e13733 (2022).
Knebel, B. et al. Fatty Liver Due to Increased de novo Lipogenesis: Alterations in the Hepatic Peroxisomal Proteome. Front. Cell Dev. Biol. 7, 248 (2019).
Lambert, J. E., Ramos-Roman, M. A., Browning, J. D. & Parks, E. J. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology 146, 726–735 (2014).
Schwarz, J. M., Clearfield, M. & Mulligan, K. Conversion of Sugar to Fat: Is Hepatic de Novo Lipogenesis Leading to Metabolic Syndrome and Associated Chronic Diseases? J. Am. Osteopath Assoc. 117, 520–527 (2017).
Talari, N. K. et al. Hepatokine ITIH3 protects against hepatic steatosis by downregulating mitochondrial bioenergetics and de novo lipogenesis. iScience 27, 109709 (2024).
Kleiner, D. E. et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41, 1313–1321 (2005).
Frezza, C., Cipolat, S. & Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2, 287–295 (2007).
Jensen, T. L., Kiersgaard, M. K., Sorensen, D. B. & Mikkelsen, L. F. Fasting of mice: a review. Lab. Anim. 47, 225–240 (2013).
Heijboer, A. C. et al. Sixteen hours of fasting differentially affects hepatic and muscle insulin sensitivity in mice. J. Lipid Res. 46, 582–588 (2005).
Hui, S. T. et al. The Genetic Architecture of Diet-Induced Hepatic Fibrosis in Mice. Hepatology 68, 2182–2196 (2018).
Hui, S. T. et al. The genetic architecture of NAFLD among inbred strains of mice. Elife 4, e05607 (2015).
Norheim, F. et al. Genetic and hormonal control of hepatic steatosis in female and male mice. J. Lipid Res. 58, 178–187 (2017).
Eismann, T. et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G266–G274 (2009).
Spinazzi, M., Casarin, A., Pertegato, V., Salviati, L. & Angelini, C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7, 1235–1246 (2012).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022).
Acknowledgements
We thank all members of the K lab for their assistance and helpful discussion. We thank the Integrated Pathology Research Facility at Cincinnati Children’s Hospital Medical Center and the University of Cincinnati Proteomics Laboratory and Live Microscopy Core for their assistance. The graphical abstract was created with BioRender.com. This work was supported by R00 DK120875 (K.C.K), R01 HL167670 (K.C.K) and P30 DK078392 (K.C.K., Integrated Pathology Research facility and Live Microscopy Core) of the Digestive Diseases Research Core Center in Cincinnati. The proteomics data for this study were all collected on an orbitrap mass spectrometer funded in part by an NIH High-End Instrumentation grant S10OD026717 (K.D.G). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Contributions
K.C.K. conceived the study. K.C.K., U.M., and N.K.T. designed, performed experiments, or analyzed the data. A.P.R., and B.K.H. assisted in animal experiments. P.B.S. performed liver histological scoring. M.W., and K.D.G. performed quantitative proteomics. K.C.K., U.M., and N.K.T. drafted the manuscript, and all authors read or revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
K.C.K., and U.M. have a patent pending for the method described herein. All other authors declare no conflict of interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Christopher Hine and Ophelia Bu.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Talari, N.K., Mattam, U., Rahman, A.P. et al. Functional compartmentalization of hepatic mitochondrial subpopulations during MASH progression. Commun Biol 8, 258 (2025). https://doi.org/10.1038/s42003-025-07713-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07713-9
This article is cited by
-
Si-Ni-San improves the deposition of lipid droplets in MAFLD through modulating the FXR-GPAT4 axis
Chinese Medicine (2026)
-
Interactions between lipid droplets and mitochondria in metabolic diseases
Lipids in Health and Disease (2025)
-
Morphological alterations of peridroplet mitochondria in human liver biopsy
Scientific Reports (2025)