Abstract
The Neotropical armored catfish Harttia is a valuable model for studying sex chromosome evolution, featuring two independently evolved male-heterogametic systems. This study examined satellitomes—sets of satellite DNAs—from four Amazonian species: H. duriventris (X1X2Y), H. rondoni (XY), H. punctata (X1X2Y), and H. villasboas (X1X2Y). These species share homologous sex chromosomes, with their satellitomes showing a high number of homologous satellite DNAs (satDNAs), primarily located on centromeres or telomeres, and varying by species. Each species revealed a distinct satDNA profile, with independent amplification and homogenization events occurring, suggesting an important role of these repetitive sequences in sex chromosome differentiation in a short evolutionary time, especially in recently originated sex chromosomes. Whole chromosome painting and bioinformatics revealed that in Harttia species without heteromorphic sex chromosomes, a specific satDNA (HviSat08-4011) is amplified in the same linkage group associated with sex chromosomes, suggesting an ancestral system. Such sequence (HviSat08-4011) has partial homology with the ZP4 gene responsible for the formation of the egg envelope, in which its role is discussed. This study indicates that these homologous sex chromosomes have diverged rapidly, recently, and independently in their satDNA content, with transposable elements playing a minor role when compared their roles on autosomal chromosome evolution.
Similar content being viewed by others
Introduction
Multiple sex chromosome systems arise from rearrangements between XY or ZW chromosomes and autosomes, this being the most common way, observed in several animal and plant groups1,2,3,4,5, or even from fissions within the ancestral sex pairs without the involvement of additional autosomes6,7,8,9,10,11,12. In contrast, the differentiation of sex chromosomes in simple sex chromosome systems (i.e., XX female/XY male; ZZ male/ZW female), which is based on autosomal ancestry, is more extensively clarified13,14,15,16,17. An autosomal pair generally acquires a sex-determining locus, which may be a dosage-dependent or sex-determining allele. Subsequent modifications include the suppression of recombination either via the accumulation of various repetitive DNA classes or via chromosome rearrangements18,19,20,21,22,23. Undoubtedly, compared to the extensive research that has already been done on simple XY and ZW systems, multiple systems still have significant shortcomings in evolutionary research.
While some taxa, like birds and mammals, have retained the same mode of sex chromosome system (i.e., ZW and XY, respectively) in most of their extant species throughout their evolutionary history, fish and amphibians show regular turnovers, with a large number of species presenting homomorphic, simple and multiple sex chromosomes, including alternative modes of sex chromosome system even within closely related species5,24,25,26,27. Among vertebrates, multiple sex chromosome systems derived from the ZZ/ZW system are extremely rare, only known in a few birds, lizards, and fish species (revised in ref. 5). On the other hand, the X1X1X2X2/X1X2Y system is the most widespread XY-derived system, which has been found in several mammals and reptiles, frogs, and fishes3,5. Fishes possess the most abundant and diverse range of multiple sex chromosome systems among vertebrates. Approximately 5% of the teleost species examined had heteromorphic sex chromosomes, with a total of 81 instances of multiple systems2,4,5,28,29,30,31.
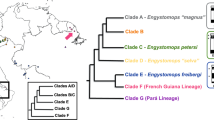
Harttia (Siluriformes, Loricariidae) is a Neotropical fish genus with significant karyotype diversity among its 28 recognized and several cryptic species, which are widespread in the waters of the Orinoco and Guyana shields, as well as most Brazilian rivers32,33. The genus Harttia records the highest number of species with multiple sex chromosomes among fishes, with six male-heterogametic occurrences, comprising two independently evolved sex chromosome systems: XX/XY1Y2 and X1X1X2X2/X1X2Y (Supplementary Table 1, Fig. 1). Whole chromosome painting (WCP) studies combined with the fluorescence in situ (FISH) mapping of repetitive sequences such as ribosomal DNA (rDNA) and microsatellites highlighted the putative origins and evolutionary relationships of the sex chromosomes, indicating that independent fission events occurred in the origin of the multiple systems10,11,34,35.
The map highlights the hydrographic basins of Brazil and the rivers of South America. Dashed lines indicate the inferred relationships of species based on chromosomal data10,11,34,87,90,124 that are not included in the previous70 phylogeny. Numbers on the map indicate the collection sites (detailed in the material and methods section Supplementary Table 11) of the same species presented in phylogeny. Idiograms illustrate the homologous sex chromosome systems according to their linkage groups, following previous whole-chromosome painting experiments10,11,34,35. Nodes marked in red indicate the divergence between clades recovered from TimeTree 5141. Blue dots indicate the species targeted for satellitome characterization, while the white dots indicate the ones used for comparative FISH experiments.
SatDNAs are fast-evolving repetitive sequences that show species-specific profiles36,37. Notwithstanding, these sequences might have a particular role in gene regulation, chromatin modification, or as chromosomal functional components36,38. The extensive mapping of repetitive DNA sequences in fish chromosomes has provided an unparalleled opportunity to understand the role of such sequences in the evolutionary process39. Although fish genomics has been studied since the late 1980s, the broad incorporation of next-generation sequencing methods in the study of non-model organisms (e.g. refs. 40,41,42,43), allowed the expansion of chromosomics44. More recently, this combination of techniques (cytogenetics and genomics) is being used to characterize and map full catalogs of satellite DNA sequences (satDNAs) in several non-model fish (e.g. refs. 41,45,46,47,48), revealing the essential role of those repetitive sequences in sex chromosomal differentiation49,50,51,52,53.
In standard XX/XY and ZZ/ZW systems, the association of satDNA sequences with the differentiation of the sex-specific (Y or W) chromosome has been clearly demonstrated (e.g. refs. 49,50,52,54,55,56,57,58,59). The few studies on the satDNA content of multiple sex chromosome systems also demonstrate an accumulation of these repetitive sequences in distinct regions of the sex chromosomes, as demonstrated for the cricket Eneoptera surinamensis (X1X2Y) that carries satDNAs in the Y60 and the anostomid Megaleporinus elongatus (Z1W1Z2W2) whose accumulation occurs in the W141. The sorrel plant (Rumex acetosa, XY1Y2) presents satDNAs accumulated at both Y1 and Y2 sex chromosomes with a different satDNA profile from those at the X chromosome, suggesting an older origin when compared to other dioecious plants with simple sex chromosomes61,62,63,64,65,66,67. The Oxycarenus hyalinipennis (X1X2Y) bug has a satDNA family distributed through the entire length of the Y chromosome and at least 26 satDNA families with a bias towards the male genome68. Although lacking Y-specific motifs, several satDNAs of Pyrrhulina semifasciata (X1X2Y) are accumulated in the proto-sex pairs in relative species69. Except for the few studies described above, no research on fish has been reported comparing species satellitomes with homologous multiple sex chromosomes. In the absence of chromosome-scale genome assemblies for the genus (and the whole family), we integrate genomic and chromosomal data to uncovered and compared the complete catalogs of satDNA families of four Harttia species named H. duriventris, H. punctata, and H. villasboas (X1X2Y) and H. rondoni (XY). This combined approach allowed us to investigate the evolutionary trajectory of satDNA families and expose their recent and accelerated patterns of evolution in these rearranged multiple sex chromosomes.
Results
Characteristics of Harttia satellitomes
The main features of the satellitomes are compiled in Table 1 and Supplementary Data 1, including the number of satellite sequences, the maximum and minimal values for the A + T proportion, and the range of repeat unit lengths. The maximum number of satellites was recovered in H. rondoni, with 25 satDNA families. This species also has the largest variation in the repeat unit length, with satDNAs varying from 4165 bp to 19 bp. Long satDNAs predominate among the four satellitomes, corresponding to 76.2% of the sequences in H. villasboas, 64% in H. rondoni, 61.2% in H. duriventris, and 52.6% in H. punctata. Putative ORFs were identified in four HviSatDNAs, five in HroSatDNAs, three in HduSatDNAs, and four in HpuSatDNAs (Supplementary Table 3). We have further investigated the satellitome of H. villasboas and studied it comparatively with the rest of the species.
Comparative fluorescence in situ hybridization
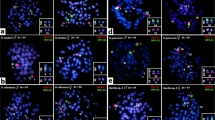
The FISH mapping of satellite sequences revealed distinct patterns for each Harttia species. Here we present the results in males, while hybridizations on females are presented in Supplementary Figs. 2 and 3. In the donor species H. villasboas (Fig. 2), concerning the sex chromosomes, only HviSat13-730 and HviSat18-1068 were present both at the secondary constriction of X2 and Y. Among autosomes, HviSat01-2531 was found in the pericentromeric regions of five chromosome pairs, while HviSat02-2707 was scattered in all pairs in a non-clustered pattern. Both HviSat04-2959 and HviSat05-177 were hybridized in the short arms of a small metacentric pair, and HviSat08-4011 was found in the long arms at the terminal region of a submetacentric pair. No visible signals were identified with the HviSat03-997 probe.
Both H. rondoni and H. duriventris share similar distribution patterns of HviSatDNAs. Only HviSat13-730 and HviSat18-1068 were mapped in the X and Y sex chromosomes of H. rondoni and X2 and Y of H. duriventris (Fig. 3a, b). HviSat08-4011 was hybridized to the terminal portion of the long arms of a submetacentric pair, in addition to a pericentromeric signal on the X1 chromosome of H. duriventris. Following this, HviSat04-2959 and HviSat05-177 were found on the short arms of a small metacentric pair. HviSat02-2707 was found scattered in all chromosomes and HviSat03-997 does not produce visible signals, while HviSat01-2531 was hybridized to pericentromeric regions of six and three pairs in H. rondoni and H. duriventris, respectively.
For H. dissidens and H. guianensis (Fig. 3c, d), HviSat01-2531 produced strong hybridization signals in five chromosome pairs in the first and was restricted to a single sm pair at an interstitial position in the last. The HviSat13-730 was restricted to the secondary NOR constriction in a single pair in both species. The HviSat08-4011 followed the pattern for most other species, being accumulated in the telomeric region of the long arms of a submetacentric pair in both species. While HviSat05-177 was found in a single pair of H. dissidens, four pairs were probed in H. guianensis. Exclusively in H. dissidens, a scattered pattern was observed for HviSat03-997, and a single pair was labeled with HviSat04-2959, since no visible signals were found in H. guianensis for both satDNAs. Both HviSat18-1068 and HviSat02-2707 do not produce visible FISH signals. Figure 4 highlights H. villasboas satellite sequences that were hybridized to sex chromosomes in distinct selected Harttia species were schematized according to their phylogenetic relationships by Covain et al.70.
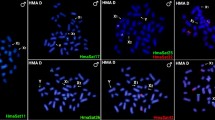
A highlight of H. villasboas satellite sequences that were hybridized to sex chromosomes in distinct selected Harttia species schematized according to their phylogenetic relationships70. H. intermontana was manually added to the tree based on geographical distribution and chromosomal characteristics, but its position must be confirmed since it was not included in Covain’s analysis. The combination of probes is indicated above each ideogram/FISH image. The colors of idiograms followed the homology of sex chromosomes obtained from whole chromosome painting with HPU-X1 (yellow) and HPU-X2 (gray) described by Deon et al.10,34 and11. See Supplementary Fig. 4 for WCP + HviSat08-4011 on species without heteromorphic sex chromosomes. Scale bar = 10 µm.
When hybridized with H. punctata chromosomes (Fig. 3e, f), the HviSatDNAs produced unexpected results. The sex chromosomes of this species accumulate several sequences, including HviSat08-4011 in a large pericentromeric block on X1, HviSat13-730 at the secondary constriction of X2, HviSat04-2959 also in a large pericentromeric block on X1, a small pericentromeric signal on X2 and Y, and HviSat05-177 at the centromere of X1 and X2, in addition to telomeric signals on X2 and in the Y, the last restricted to long-arms. HviSat04-2959 was also found in a small metacentric autosome pair and HviSat05-177 in the short arms of six autosomes. HviSat01-2531 was restricted to a large sm chromosome pair in an interstitial position on long arms, while HviSat02-2707 produced small signals in the pericentromeric region of a few chromosome pairs. The species Harttia sp. 3 (Fig. 3e, f) produced similar patterns to H. villasboas and H. rondoni. The at01-2531 was found in three chromosome pairs in the pericentromeric region, while HviSat03-997 was scattered in almost all chromosomes. HviSat18-1068 and HviSat13-730 were found in the secondary constriction of the first chromosome pair in the pericentromeric region. On the other hand, HviSat02-2707 was found slightly hybridized to some chromosome pairs, but only in one of the homologs of the first chromosome pair, all in the pericentromeric position. Both HviSat04-2959 and HviSat05-177 followed the pattern of a single chromosome pair hybridized with each probe, while HviSat08-4011 was found in the long arms of a submetacentric large pair.
Comparative satellitomics
19 of the 21 H. villasboas satDNAs have a homologous counterpart in one, two, or three other Harttia species (Table 2). Table 2 reflects that there are conserved satellites in all four species analyzed (e.g., the homologs of HviSat01-2531, HviSat04-2959 or HviSat05-177, among others). However, there are lineage-specific satellites: two satDNAs, HviSat17-49 and HviSat20-47, appear to be specific to H. villasboas, six satDNAs appear to be specific to H. rondoni, 1 to H. duriventris and 4 to H. punctata. In this context, we have found some satellites that are specific to the H. villasboas-H. rondoni lineage: HviSat08-4011, HviSat16-2480, HviSat19-589, and HviSat21-575 have homologues only in H. rondoni, the closest relative to H villasboas. But this table also reflects the saltational process of evolution of some satellites that form the satellitomes of these species since there are satDNAs shared by two or more species for which the pattern of sharing does not conform to the phylogeny of Fig. 1: among several others, one example is HviSat02-2702 for which we have found homologs in H. duriventris and H. punctata but not in H. rondoni. These data reflect differential amplifications of these satellites in different species at different evolutionary times.
Table 3 shows the genetic distances between the satDNAs of H. villasboas and the corresponding homologous satellites (i.e. the similarity between the homologous sequences of satDNAs, measured by the alignment of the sequences). In general, the greater the phylogenetic distance, the greater the genetic distance between homologous satellites. This is especially true in comparisons with H. punctata. But Table 3 also shows that there are highly accelerated or slowed rates of change for some satDNAs lineages, with genetic distances that are not consistent with phylogenetic proximity in some comparisons. For example, the homologous HviSat01-2531 in H. rondoni or the homologous HviSat04-2959 in H. duriventris show greater divergence from H. villasboas than homologs of phylogenetically more distant species. The table, in general, shows differential divergence patterns between different satellites and between different lineages. Taking as a reference the only node in which we have a clear dating to compare two species, Supplementary Table 9 shows CTR for each satDNA assuming a divergence time of 17.5 my between H. villasboas and H. punctata (Fig. 1), demonstrating very different rates of sequence change in different satDNAs.
Repeat landscapes of Harttia satDNAs
Supplementary Fig. 6 displays repeat landscape (RL) plots representing, for several Harttia satDNAs, abundance (y-axis) and divergence (x-axis) to the consensus sequence built for each satDNA repeat unit. satDNA degenerates through point mutation (increasing divergence) and homogenizes through amplification (decreasing divergence). Then, it is assumed that peaks at lower divergence values in the repeat landscape profiles are the product of recent amplifications, whereas those at higher divergence values are probably older variants degenerated by the accumulation of mutations. Following this argument, we have analyzed the RL plots of satDNAs found in the sex chromosomes of Harttia species. Thus, the satellite HviSat08-4011 and its counterpart HroSat14-4165 show a unique amplification peak at 0% divergence. A paired samples t-test found no significant differences between both satDNA repeat landscape profiles (t = −0.83, df = 40, p = 0.2). Then, although the graph shows sequence variants of these satellites with divergences of up to 40%, the amplification that resulted in the current status of this satellite in the H. villasboas-H. rondoni lineage must have been very recent after the lineage split that led to H. duriventris. In the case of the HviSat13-730 and HviSat18-1068 satellites and their counterparts, several peaks are observed that could reflect different, recent (the highest peaks are at 0% divergence) amplification waves of these satellites at different times in the distinct species. Paired samples t-test found significant differences for all pair comparisons between HviSat13-730 and HviSat18-1068 and their counterparts (Supplementary Table 8). The RLs of HviSat04-2959 and HviSat05-177, and their counterparts, show several peaks. Differences were significant in all pair comparisons between the repeat landscapes profiles, except for HviSat04-2959/HroSat09-2961 (Supplementary Table 8). The greatest peaks of the satellites HpuSat02-177 and HpuSat07-2750 stand out, which could be related to their incorporation and amplification in the sex chromosomes of H. punctata (these satellites are in autosomes in the other species) possibly after the separation of the lineages (17.5 mya; Fig. 1) since the divergence of 2% of these peaks would correspond to a divergence time of about 8 my (Table 3). Finally, the rest of the RL plots in Supplementary Fig. 6 represent examples of four autosomal satellites (HviSat01-2531, HviSat03-997, HviSat14-347, and HviSat19-589, and their counterparts). As shown, all these satellites show several peaks that could reflect different expansion waves, most of which are very old (there are divergence peaks of more than 20%), differentiated in time between the compared species (see also Supplementary Table 8).
The complex nature of HviSat08-4011
As indicated, homologous satellites to the satellite HviSat08-4011 and its counterpart HroSat14-4165 in H. rondoni were not isolated from the genomes of H. duriventris and H. punctata. However, FISH with probes derived from HviSat08-4011 detected large blocks of hybridization of this satellite on chromosome X1 of both species, in addition to an autosomal locus (only in H. duriventris), which contrasts with the unique autosomal location of this satellite in H. villasboas and H. rondoni (as well as in other species lacking sex-chromosomes) (Fig. 4). BLASTn and BLASTx searches of HviSat08-4011 reveal a complex satDNA composed of at least two differentiated parts (Supplementary Fig. 5A). The largest part (about 2300 bp) is a sequence with no homology to any other known sequence. However, about 1700 bp of HviSat08-4011 shows homology and high identity (~80%) with the zona pellucida sperm-binding protein 4-like (ZP4) gene of diverse species of the orders Siluriformes (such as Ictalurus, Silurus, Neoarius and Tachysurus) and Characiformes (such as Colossoma, Pygocentrus and Astyanax). As Supplementary Fig. 5A shows, the homologous parts are discontinuous, separated by intervening unknown sequences. However, the homologous parts align continuously with ~90% of the translated sequence of the gene in those other species, with small discrepancies at the boundaries.
The analogous region of the ZP4 gene is where the primers utilized in our initial FISH strategy were constructed (Supplementary Fig. 5A, F1R1). Two additional primer pairings have been put to the test. One is located in the ZP4 non-homologous region (Supplementary Fig. 5A, F2R2), while the other is made up of a reverse primer inside the ZP4 homologous region and a forward primer outside of it (Supplementary Fig. 5A, F3R3). In the first case, the results differ from those initially found (Supplementary Table 10 and Supplementary Fig. 5B): a) the two satellite parts (ZP4-homologous and non-ZP4-homologous) hybridize only on one autosome in H. rondoni and H. villasboas; b) the ZP4-homologous satellite part hybridizes on X1 and one autosome of H. duriventris but the satellite part not homologous to ZP4 hybridizes only on the autosome; c) the satellite part homologous to ZP4 hybridizes only on X1 of H. punctata and the satellite part not homologous to ZP4 does not hybridize on any chromosome. In the second case, with the F3R3-HviSat08-4011 primers, only H. villasboas and H. rondoni produced visible fragments in the agarose gel (Supplementary Table 10 and Supplementary Fig. 5B). Those last PCR product amplifications were then sequenced to check the composition of the amplified fragments, and they fully coincided with the expected region.
We have searched for HviSat08-4011 among the Illumina reads of H. duriventris and H. punctata using the script mapping_blat_gs.py (see Materials and Methods) but we obtained no results other than partial sequences with homology to the portion homologous to ZP4 (mostly) and some with homology to the portion not homologous to ZP4. We then mapped and quantified the raw reads of both species against both regions of HviSat08-4011 (repeat_masker_run_big.py) and obtained FISH-compatible results. Specifically, while this satDNA represents 0.05% and 0.01% of the genome of H. villasboas and H. rondoni, respectively (Supplementary Data 1), the part homologous to ZP4 accounts for 0.1% of the genome of H. punctata while the non-homologous part accounts for 0.03% of its genome, these percentages being 0.03% in both cases in H. duriventris. Supplementary Fig. 5C shows repeat landscape plots representing, for H. villasboas, H. rondoni, H. duriventris, and H. punctata, abundance (y-axis) and divergence (x-axis) with respect to a consensus sequence built for each part of the HviSat08-4011 repeat unit. The graphs show different evolutionary trajectories for each part (homologous to ZP4 and non-homologous to ZP4) in H. duriventris and H. punctata. Thus, we can observe in H. duriventris several amplification peaks with divergences between 15% and 6% for the ZP4 gene homologous part, not found for the non-homologous part, in addition to a recent peak (0% divergence) that in this case does coincide with the major peak found for the non-homologous part. In H. punctata there are several major peaks at a divergence between 4 and 10% for sequences homologous to the ZP4 gene, while the non-homologous to ZP4 gene part shows smaller amplification peaks at divergences higher than 16%. Differences between the profiles of the two parts in H. punctata, in addition to revealing amplification events that are different in evolutionary times as in the case of H. duriventris, are statistically significant (t = −2.423, df = 40, p = 0.01). Furthermore, paired samples t-tests between each species and H. punctata found significant differences for the landscape profiles of the ZP-4 gene homologous part (p < 0.05).
Relationship of Harttia satDNAs with transposable elements (TEs)
Supplementary Tables 4–7 show the relationships between transposable elements (TEs) and the various groups of Harttia homologous satellites. The most remarkable results are: a) out of the several satellites found in the sex chromosomes in this study, only the satellite HviSat04-2959, and its homologs in the rest of the species, is related to this type of elements; b) most of the satellites are complex ones composed of different parts in which sequences derived from SINE and/or LINE elements and tRNA genes (and satDNAs derived from tRNAs) are intermingled with intervening sequences; c) HviSat02-2707 and its homolog are connected to Penelope elements, whereas HduSat17-150, which has no homologs in other species, is related to Helitron elements; d) HviSat06-200, but not its shorter homologous HduSat04-31, is analogous to a mosaic zebrafish satDNA.
Discussion
Our results show that the complex history of chromosomal evolution in Harttia is also reflected in its satDNA content. We demonstrated the occurrence of distinct profiles of satDNAs on homologous sex chromosomes, suggesting that these repeats diverged fast, recently, and independently in each species. SatDNA sequences are abundant on sex chromosomes and may play an important role in gene regulation and dosage compensation, resulting in hybrid incompatibilities36,71. Moreover, our data demonstrated that in other Harttia without heteromorphic sex chromosomes, a satDNA (HviSat08-4011) is amplified in the same linkage group that creates the sex chromosomes seen in the four target species of this study, suggesting an ancestral sex chromosome system, which will be discussed below.
Evolution of satellite DNAs in Harttia genomes
A large number of satDNA families are shared among the four Harttia species investigated in this study. This finding supports the library hypothesis72, which has already been reported for other groups, including fishes69,73,74, mammals75,76, reptiles77, insects78,79, and plants80,81,82. The library hypothesis states that species possess a repertoire of satellite DNA (satDNA) families, which exhibit variations in both the number of copies and their arrangement within the genome72. As seen in Table 2, which is the result of differential amplifications of each satDNA in different species at different evolutionary times, thus supports this hypothesis. Roughly, genetic distances between homologous satellites agree with the phylogenetic relationships shown in Fig. 1. Therefore, the distances between H. villasboas and H. rondoni are consistently the shortest, indicating that these two species are the closest in phylogenetic proximity. The primary differences consistently occurred between H. punctata and any other species, indicating that this lineage split from the lineage that originated the clade consisting of the other three species a long time ago (Fig. 1). However, in agreement with the library hypothesis, differential amplifications lead to accelerated rates of change in different lineages as disparate genetic distances that do not fit with phylogenetic relationships are observed (Table 3). In this context, the repeat landscapes indicated peaks in both higher and lower divergence values, probably due to the presence of older variants degenerated by the accumulation of mutations in addition to recent amplification and homogenization events (Supplementary Fig. 6).
Satellite DNAs (satDNAs) exhibit variation in their location on Harttia chromosomes, ranging from being scattered in a few places throughout the chromosome to being mostly confined to either the pericentromeric or telomeric regions (Figs. 2 and 3), the preferential places of clusterization of satDNAs36,83. It has been proposed that satDNAs may play a significant role in the structure and operation of both centromeric and telomeric regions since such distribution lines up within all eukaryotes84,85. In terms of size, there is a prevalence of long satDNAs (>100 bp), which is consistent with what has been seen in the related species H. carvalhoi74.
Some satDNAs of Harttia also share the position with the 18S rDNA secondary constriction, following HviSat13-730 and HviSat18-1068 associated with the secondary NOR constriction of most species, with the last being especially highlighted in H. villasboas, H. rondoni, H. duriventris, and Harttia sp. 3 (Figs. 1–4). All four species that harbor the multiple X1X2Y sex chromosome system demonstrated some ribosomal DNA loci in both X and Y chromosomes. While H. punctata86 carries both 5S (X1 and Y) and 18S rDNA (X2), only the 18S rDNA is found in the sex chromosomes of H. duriventris and H. villasboas (X2 and Y)87. The sister species with the homologous simple XY system (H. rondoni87) has the 18S rDNA locus in both sex chromosomes. Ribosomal sequences were suggested to increase the instability of genomes due to their high transcriptional activity, which can facilitate double-strand breaks88,89, as suggested for other Harttia species90, and also other loricariids such as Ancistrus91 and Rineloricaria92,93. In plants, numerous satDNAs arise from the intergenic spacer of the ribosomal locus94,95, which was not observed in Harttia genomes. Since the rDNAs located in the sex chromosomes of Harttia were suggested to surround or be inserted in an evolutionary breakpoint region that is reused in the karyotypic diversification of the genus34, both HviSat13-730 and HviSat18-1068 may likely have contributed to the genomic instability that led to the fission process the originates the multiple sex chromosomes herein investigated. SatDNAs are also associated with the formation of chromocenters and micronuclei (reviewed in ref. 96), which can trigger DNA breakage and incompatibilities between populations, leading to reproductively isolated populations97.
Transposable elements (TEs) are known to contribute to genome expansion mechanisms primarily through retrotransposition, but also by creating new tandem repeats98 by partial TE duplication followed by uneven crossing-over99, or by transposase-induced breaks and repair mechanisms100. In Harttia, these elements have an intricate relation with the formation of satDNAs, as demonstrated by complex sequences composed of distinct parts in which motifs derived from short-interspersed elements (SINE) and/or long interspersed elements (LINE) and tRNA genes. For example, HviSat02-2707 and its homologs in other species are related to Penelope elements, a class of repetitive elements widely distributed in fish genomes, seen in medaka, pufferfishes, and cichlids101. Penelope-like elements (PLE) are predominantly found in animal genomes, with occasional losses in some lineages102,103. On the other hand, the H. duriventris-specific satellite HduSat17-150 is related to Helitron, a group of TEs that conserves sequences at ends but with satDNA-like central tandem-repeated motifs104, including real satDNAs embedded in such central position in Bivalves105,106. This demonstrates that the evolutionary history of satDNA sequences is also related to TEs, reflecting the complex history of chromosomal rearrangements that occurred in the genus. However, such TEs do not seem to participate in sex chromosome differentiation, as discussed in the next section. Most satDNAs in this genus presented rates of divergence between homologous sequences that vary according to the phylogenetic relationships, but repeat landscapes demonstrated species-specific amplification and homogenization events, especially in the repeats found in sex chromosomes. Such patterns indicate a contingent evolution scenario for the satDNAs of Harttia, as also seen in the repetitive content of grasshoppers107.
The role of satDNAs in the evolution of Harttia´s sex chromosomes
Although all Harttia species from Clade III share homologous sex chromosomes, as indicated by chromosome painting studies11, each demonstrated a distinct composition of satDNAs, indicating independent evolution of these sequences in each lineage. Sex chromosome differentiation by independent accumulation of satDNAs in a recent evolutionary time may reflect the neo origin of such systems, as also seen in other fish with recently originated sex chromosomes, such as Megaleporinus elongatus and Nothobranchius spp41,43,47. In Harttia, the discrepancy in satDNA profiles can be observed, especially in sequences that do not follow a phylogenetic arrangement, for example, HviSat08-4011, which was probably amplified in H. duriventris and H. punctata after their split, besides HviSat13-730 and HviSat18-1068, which we will discuss below.
The sex chromosomes of H. villasboas harbor only two satDNAs: HviSat13-730 and HviSat18-1068. These satellites share a minor part of their repeat sequence (about 6%) with 84% similarity, suggesting an ancient common origin. However, both satDNAs and their homologous counterparts in each species, have signals of recent amplifications in H. villasboas, H. rondoni, and H. duriventris, in addition to independent events of previous amplifications (Supplementary Fig. 6). For HviSat18-1068 and its homologs HroSat11-534 and Hdu15-534, which are smaller in size, a duplication process in H. villasboas was responsible for their difference in size. This satellite is presented in the secondary constriction formed by the 18S rDNA, where events of unequal crossing-over were proposed as the main driver for differences in the accumulation of this ribosomal sequence between the homologs87. Since these satellites are not linked to TEs, unequal recombination may have caused the duplication, as described for other satDNAs in rice and monkeys108,109. However, rolling circle replication may also be responsible for the duplication of such repetitive sequences110. Regarding HviSat13-730 and its homologs HroSat04-920 and HduSat09-771, which showed positive FISH signals in all four species, we could not identify homolog sequences in H. punctata genome. In this species, the FISH signal is restricted to the X2 chromosome and lacking in the Y, matching the 18S rDNA accumulation pattern86, suggesting that the fission event that created the multiple sex chromosomes may also explain this variation11.
On the other hand, out of the several satellites found in H. punctata sex chromosomes (see Fig. 3e) only HviSat04-2959 and its homologs (HroSat09-2961, HduSat08-1754, and HpuSat07-2750), are related to TEs. This satellite has partial homology with LINE-Rex-Babar and Tc/Mariner elements (see Supplementary Tables 4–7), which are usually inactivated or degenerated in fish genomes but can accumulate mutations at neutral rates until losing their molecular identity111,112,113,114. Both elements have been identified in related Loricariidae species, but as dispersed sequences without any accumulation on a specific chromosome pair115. In catfishes, Tc1-Mariner elements can be found associated with rDNA loci variation, suggesting activity in the transposition system116. Both satellites HviSat04-2959 and HviSat05-177 and their homologous counterparts, located on sex chromosomes in H. punctata but on autosomes in the rest of the species, have been expanding over more than ~20 million years in the four species. However, there has been a significant increase in the number of these satellites in H. punctata, which aligns with the observation of extra loci on the X2 chromosome of this species. TEs are less likely to build up on young X chromosomes in fishes compared to autosomes117, and this might be the reason for the scarcity of TE-derived satellite sequences in the sex chromosomes of Harttia.
The most intriguing relation between a satDNA sequence and sex chromosomes in the genus was observed with HviSat08-4011, which has a partial homology with the ZP4 gene responsible for the formation of the egg envelope. Zona pellucida glycoproteins are generated in the ovary or liver by ZP genes118. The HviSat08-4011 is consistently mapped to autosomes and the linkage group is mapped by the whole chromosome painting probe HPU-X1 across the Harttia phylogeny (Supplementary Fig. 6), except in H. villasboas and H. rondoni, where only autosomal FISH signals were detected. Repeat landscapes show a symmetrical graphical pattern for both homologous and non-homologous ZP4 satDNAs in H. villasboas and H. rondoni, which would indicate a single autosomal satellite recently originated in both species. However, the graph in H. duriventris would be compatible with an older amplification of a satellite formed from partial sequences of the ZP4 gene in X1, possibly in addition to a more recent amplification of the composite satellite (composed of the two parts, the homologous and the non-homologous parts to ZP4 gene) in autosomes as revealed by FISH. This is more evident in H. punctata where there are several major peaks representing the most abundant set of sequences homologous to the ZP4 gene, at a divergence between 4 and 10%, compatible with several waves of amplification of a ZP4-derived satDNA on chromosome X1. The non-homologous to ZP4 gene part, not detected by FISH in H. punctata, shows relatively smaller amplification peaks at divergences higher than 16%. The ZP genes in vertebrates undergo frequent gene duplication and loss events, suggesting that they are reproductive genes that evolve rapidly119. This rapid evolution may enable them to adapt to various ecological environments120 and serve as barriers to fertilization121,122, making them potential targets for natural selection. A fertilization-related gene in the same linkage group as the ancestral of the three Harttia clades may imply an ancestral sex chromosome system. Indeed, the linkage group mentioned is exclusively recruited as sex chromosomes in Clade III, which includes the species under investigation. This suggests a turnover event in the emergence of multiple XY1Y2 chromosomes in Clade II11,34,35,74,93.
The species with the most satDNAs accumulated in sex chromosomes (H. punctata) is also the oldest divergent in the clade (17.5 My), when compared to a much more recent diversification of H. villasboas, H. rondoni, and H. duriventris clade at 5.5 My123. However, each species revealed a distinct satDNA profile, with independent amplification and homogenization events occurring, suggesting an important role of these repetitive sequences in sex chromosome differentiation in a short evolutionary time, especially in recently originated sex chromosomes. Besides, satDNAs may have contributed to the speciation process in Harttia, since these sequences play important roles in chromosome pairing and segregation, indicating that the distinct profiles on homologous sex chromosomes may have contributed to reproductive isolation.
Material and methods
Specimens, chromosomal obtainment, and DNA sequencing
We collected adult samples of nine Harttia species, namely H. dissidens, H. duriventris, H. guianensis, H. kronei, H. intermontana, H. punctata, H. rondoni, H. villasboas, and Harttia sp. 3 (a distinct karyomorph from Rio do Peixe, PA, Brazil, described in ref. 124 that is not assembled to any known species), in distinct localities inside the Brazilian territory according to Fig. 1 and Supplementary Table 11. We have complied with all relevant ethical regulations for animal use, as indicated by ARRIVE guidelines. For most experiments, H. villasboas was chosen as the main target for comparisons because it represents the species with a multiple sex chromosome system (X1X2Y) that is more closely related to H. rondoni, which harbors the simple XY that is considered to be the ancestral condition for the sex chromosomes in the genus. After anesthesia with clove oil (Eugenol diluted in 95% ethanol in a final concentration of 60 mg/L in water), as approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 7994170423), chromosomes were obtained from the anterior kidney following the classical air-drying technique125. The liver and muscle tissues were stored at 4 °C in 100% ethanol for molecular analysis. Total DNA was extracted from these tissues using a silica spin-column-based kit (Cellco Biotech, São Carlos – SP, Brazil). The purified DNA of species with described sex chromosomes (i.e. H. duriventris, H. rondoni, H. villasboas, and H. punctata) was sequenced on BGISEQ-500 platform (BGI Genomics, Shenzhen, China), generating 2 ×150 bp short read sequences. For H. villasboas and H. rondoni, males and females were sequenced, while for H. duriventris and H. punctata, only male samples were targeted.
Satellite DNA library, sequence analysis and primer design
The raw reads obtained after the shotgun sequencing were trimmed with Trimmomatic126 to select pair-end reads with Q > 20 for all nucleotides. Then, the catalogs of satellite DNAs (satellitome) of each analyzed species (i.e. H. duriventris, H. rondoni, H. punctata, and H. villasboas) were characterized by running several iterations of the TAREAN tool127, each followed by filtering out the reads with similarity to the identified satDNAs using DeconSeq128, until no satDNA was found. Subsequently, we performed a homology search with RepeatMasker129 to group the sequences into variants, families, and superfamilies, as suggested by Ruiz-Ruano et al.130. Abundance and divergence values of each satDNA were estimated by masking male and female genomic libraries against the catalogs of satDNAs with RepeatMasker129 with the publicly available script (https://github.com/fjruizruano/satminer/blob/master/repeat_masker_run_big.py), using 10,000,000 reads (2 × 5,000,000). Additionally, repeat landscapes were generated to estimate the average divergence of satDNAs considering the genetic distances between sequences based on the Kimura-2-parameter model using the script calcDivergenceFromAlign.pl of the RepeatMasker suite (i.e. the similarity between the homologous sequences of satDNAs, measured by the alignment of the sequences).
Based on the abundance, satellites were named using the first letter of the genus and the two first letters of the specific epithet (HviSat for H. villasboas, HroSat for H. rondoni, Hdu for H. duriventris, and Hpu for H. punctata). To detect sex-specific accumulated satDNAs, we selected those with an F/M ratio (quotient of satDNA female and male abundances) higher than 1.2.
We searched for homology between Harttia satellitomes with the rm_homology script130 that makes all-to-all alignments with RepeatMasker v4.0.5129. For correspondent satellites between species, we did pairwise alignments using ClustalX131, which were manually revised. Sequence divergence between satDNA families of each pairwise comparison was calculated following Kimura’s two-parameter method (K2P)132 using MEGA11133. A consensus turnover rate (CTR) was calculated using the CTR = K/2 T equation, where T = divergence time (see nodes in the phylogenetic tree of Fig. 1) between species and K = K2P distance107.
In the absence of a chromosome-scale genome assembly for the genus, the whole satellitome was BLAST-searched134 against NCBI nucleotide collection to check for the presence of conserved satDNAs. Putative open reading frames were prospected in the satellitomes using “orfipy”135 and Geneious 7.1.3, with a minimum size of 100 bp. Identified ORFs were translated to protein sequences using SMS136 and searched against the NCBI database using BLASTp137, following filtering for query cover and percentage identity >70%.
In addition, we searched Repbase138 for homologies with transposable elements with RepeatMasker129 with “no_low” and “no_is” options.
We searched for some satDNAs like HviSat08-4011 among the Illumina reads of species like H. duriventris and H. punctata, from which we could not isolate its homologous counterpart with TAREAN, using the publicly available script mapping_blat_gs.py (https://github.com/fjruizruano/ngs-protocols/blob/master/mapping_blat_gs.py). Raw reads of both species were mapped and quantified against every two parts described in HviSat08-4011 using the script repeat_masker_run_big.py, as described above.
Primers were designed for the eight most abundant of the total 21 satDNAs identified in H. villasboas using the abovementioned process. We selected H. villasboas for the construction of probes since this species has multiple X1X2Y and is more closely related to the XY-carrier species H. rondoni (when compared to H. duriventris and H. punctata), which allows us to infer the role of the rearrangements that occurred in the clade. The consensus sequences for each of those satDNAs were manually investigated to design PCR primers (Supplementary Table 2). Then, satDNAs were amplified by PCR using a starting denaturation step of 95 °C for 5 min, 30–35 cycles with 95 °C during 20 s, with 39.1–54.2 °C as annealing temperature during 40 s, 72 °C during 30 s, and a final extension step of 10 min (Supplementary Table 2). The resulting PCR products were checked on a 1% agarose gel to confirm the typical ladder pattern characteristic of satDNAs (Supplementary Fig. 1). Sequences were deposited on Genbank (Access numbers: OR827473-OR827555).
Fluorescence in situ hybridization
According to the manufacturer’s instructions, the selected PCR-amplified satellites of H. villasboas were directly labeled with Atto550-dUTP or Atto488-dUTP by Nick-Translation (Jena Biosciences, Jena, Germany). Probes were composed of 100 ng of each labeled satellite DNA plus 50% formamide, 2× SSC, 10% SDS, 10% dextran sulfate, and Denhardt’s buffer at pH 7.0 in a total volume of 20 µl. FISH experiments were conducted firstly in H. villasboas slides with every single probe, then in double-FISH for sister species H. rondoni, H. duriventris, and H. punctata, in addition to H. dissidens, H. guianensis, and Harttia sp. 3. The sex chromosomes were identified using their morphological characteristics, as established in prior studies refs. 87,124. The following morphological traits were employed to discern the sex chromosomes: of both H. villasboas and H. duriventris, both X2 and Y possess a prominent secondary constriction owed to the 18S rDNA loci. In H. rondoni, the same applies to both the X and Y chromosomes. In H. punctata, the 18S rDNA loci is confined to the X2 chromosome, while the Y chromosome corresponds to the largest metacentric chromosome of its karyotype. Finally, the X1 in H. villasboas, H. duriventris, and H. punctata represents the largest acrocentric chromosome of their karyotypes. Following the results of satDNAs probed on sex chromosomes, we performed whole chromosome painting (WCP), together with a FISH mapping of these satellites, using probes derived from microdissection of the sex chromosomes of H. punctata (HPU-X1 and HPU-X2), as previously described in refs. 11,34. For the investigation of sex chromosome evolution, we included two species (H. kronei and H. intermontana) that carry the ancestral condition of the linkage groups that compose the multiple X1X2Y (i.e., both HPU-X1 and HPU-X2 located in synteny on the same chromosome)34. In these last two species, we mapped the HviSatDNAs that were found accumulated in the sex chromosomes of the four main targeted species and detected the ancestral linkage groups through WCP. Chromosomes were denatured in 70% Formamide/2 × SSC at 72 °C for 3 min, probes at 86 °C for 8 min, then cooled at 4 °C for 2 min before being applied to slides. The hybridization was carried out for 16 h in a dark, moist chamber at 37 °C. Following a 5-min post-hybridization wash with 1×SSC at 65 °C, next with a 4×SSC/Tween solution at room temperature. Finally, chromosomes were counterstained with DAPI mounted in Vectashield (Vector Laboratories, Burlingame, USA). For the WCP experiment, chromosomes were denatured as abovementioned, but the probes were denatured at 85 °C for 8 min, cooled at 4 °C for 2 min, and pre-hybridized with the blocking sequence (unlabeled C0t-1 DNA, following139) at 37 °C for 1 h. Hybridization occurred for 48 h overnight at 37 °C in a dark, moist chamber. The final wash was performed using the same procedure as described above.
Statistics and reproducibility
To confirm the 2n number, karyotype structure, and FISH results, at least 30 metaphase spreads per individual were examined. Images were captured with CoolSNAP on an Axioplan II microscope (Carl Zeiss Jena GmbH, Germany) and processed with ISIS (MetaSystems Hard & Software GmbH, Altlussheim, Germany). The map was generated using QGIS 3.32 (Lima) with a Natural Earth package and riverine information from HydroRIVERS140. Statistical analysis (t-test) was performed in Microsoft Excel (Office 365, Microsoft).
Data availability
Sequencing data that support the findings of this study have been deposited in Genbank, with their accession codes OR827473-OR827555 (http://www.ncbi.nlm.nih.gov/nuccore/). All other data is available from the corresponding author on reasonable request.
References
White, M. The origin and evolution of multiple sex chromosome mechanisms. J. Genet. 40, 303–306 (1940).
Kitano, J. & Peichel, C. L. Turnover of sex chromosomes and speciation in fishes. Environ. Biol. Fishes 94, 549–558 (2012).
Pokorná, M., Altmanová, M. & Kratochvíl, L. Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrates. Chromosome Res. 22, 35–44 (2014).
Pennell, M. W. et al. Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles. PLoS Genet. 11, 1–17 (2015).
Sember, A. et al. Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: state of the art and future challenges. Philos. Trans. R. Soc. B 376, 20200098 (2021).
Ming, R., Bendahmane, A. & Renner, S. S. Sex Chromosomes in Land Plants. Annu. Rev. Plant Biol. 62, 485–514 (2011).
Blanco, D. R. et al. The role of the Robertsonian rearrangements in the origin of the XX/XY1Y2 sex chromosome system and in the chromosomal differentiation in Harttia species (Siluriformes, Loricariidae). Rev. Fish. Biol. Fish. 23, 127–134 (2013).
Šíchová, J. et al. Fissions, fusions, and translocations shaped the karyotype and multiple sex chromosome constitution of the northeast-Asian wood white butterfly, Leptidea amurensis. Biol. J. Linn. Soc. 118, 457–471 (2016).
Giovannotti, M. et al. New insights into sex chromosome evolution in anole lizards (Reptilia, Dactyloidae). Chromosoma 126, 245–260 (2017).
Deon, G. A. et al. Chromosomal Rearrangements and Origin of the Multiple XX/XY1Y2 Sex Chromosome System in Harttia Species (Siluriformes: Loricariidae). Front. Genet. 13, 877522 (2022).
Sassi, F. M. C. et al. Homeology of sex chromosomes in Amazonian Harttia armored catfishes supports the X-fission hypothesis for the X1X2Y sex chromosome system origin. Sci. Rep. 13, 15756 (2023).
Dutrillaux, B., Dutrillaux, A.-M., Salazar, K. & Boucher, S. Multiple Chromosome Fissions, Including That of the X Chromosome, in Aulacocyclus tricuspis Kaup (Coleoptera, Passalidae) from New Caledonia: Characterization of a Rare but Recurrent Pathway of Chromosome Evolution in Animals. Genes 14, 1487 (2023).
Muller, H. J. A Factor for the Fourth Chromosome of Drosophila. Science 39, 906–906 (1914).
Ohno, S. Conservation of the Original Z-Chromosome by Diverse Avian Species and Homology of the Z-linked Genes. In Sex Chromosomes and Sex-linked Genes (ed. Ohno, S.) 73–81 (Springer, Berlin, Heidelberg, 1966). https://doi.org/10.1007/978-3-662-35113-0_6.
Bull, J. J. Evolution of Sex Determining Mechanisms. 1 (The Benjamin/Cummings Publishing Company, Inc., Menlo Park, California, USA, 1983).
Charlesworth, B. The Evolution of Sex Chromosomes. Science 251, 1030–1033 (1991).
Graves, J. A. M. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet 17, 33–46 (2016).
Fisher, R. A. The evolution of dominance. Biol. Rev. Camb. Philos. Soc. 6, 345–368 (1931).
Nei, M. Linkage modification and sex difference in recombination. Genetics 63, 681–699 (1969).
Rice, W. R. Sex Chromosomes and the Evolution of Sexual Dimorphism. Evolution 38, 735–742 (1984).
Charlesworth, B., Jordan, C. Y. & Charlesworth, D. The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 68, 1339–1350 (2014).
Evans, B. J. et al. Rapid Sex Chromosome Turnover in African Clawed Frogs (Xenopus) and the Origins of New Sex Chromosomes. Mol. Biol. Evol. 41, msae234 (2024).
Kratochvíl, L. et al. Expanding the classical paradigm: what we have learnt from vertebrates about sex chromosome evolution. Philos. Trans. R. Soc. B Biol. Sci. 376, 20200097 (2021).
Nanda, I., Schartl, M., Feichtinger, W., Epplen, J. T. & Schmid, M. Early stages of sex chromosome differentiation in fish as analysed by simple repetitive DNA sequences. Chromosoma 101, 301–310 (1992).
Rovatsos, M., Vukić, J., Lymberakis, P. & Kratochvíl, L. Evolutionary stability of sex chromosomes in snakes. Proc. R. Soc. B Biol. Sci. 282, 20151992 (2015).
Stöck, M. et al. Sex chromosomes in meiotic, hemiclonal, clonal and polyploid hybrid vertebrates: along the ‘extended speciation continuum’. Philos. Trans. R. Soc. B Biol. Sci. 376, 20200103 (2021).
Pla, S., Benvenuto, C., Capellini, I. & Piferrer, F. Switches, stability and reversals in the evolutionary history of sexual systems in fish. Nat. Commun. 13, 3029 (2022).
Arai, R. Fish Karyotypes: A Check List (Springer Science & Business Media, 2011).
Marajó, L. et al. Chromosomal rearrangements and the first indication of an ♀X1X1X2X2/♂X1X2Y sex chromosome system in Rineloricaria fishes (Teleostei: Siluriformes). J. Fish. Biol. 102, 443–454 (2023).
Nirchio, M. et al. Occurrence of Sex Chromosomes in Fish of the Genus Ancistrus with a New Description of Multiple Sex Chromosomes in the Ecuadorian Endemic Ancistrus clementinae (Loricariidae). Genes 14, 306 (2023).
de Araújo, L. et al. Cytogenetic and Molecular Characterization of Eigenmannia aff. desantanai (Gymnotiformes: Sternopygidae): A First Report of System of Sex Chromosomes ZW1W2/ZZ in Gymnotiformes. Zebrafish 20, 77–85 (2023).
Oyakawa, O. T., Fichberg, I. & Py-Daniel, L. R. Three new species of Harttia (Loricariidae: Loricariinae) from Serra do Cachimbo, Rio Xingu basin, Pará, Northern Brazil. Zootaxa 4387, 75–90 (2018).
Fricke, R., Eschmeyer, W. N. & van der Laan, R. Eschmeyer’s catalog of fishes: Genera, Species, References. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (2024).
Deon, G. A. et al. Evolutionary breakpoint regions and chromosomal remodeling in Harttia (Siluriformes: Loricariidae) species diversification. Genet. Mol. Biol. 45, e20210170 (2022).
Sassi, F. M. C. et al. Turnover of multiple sex chromosomes in Harttia catfish (Siluriformes, Loricariidae): a glimpse from whole chromosome painting. Front. Genet. 14, 1226222 (2023).
Garrido-Ramos, M. A. Satellite DNA: An evolving topic. Genes 8, 230 (2017).
Thakur, J., Packiaraj, J. & Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 22, 4309 (2021).
Kuhn, G. C. S. Satellite DNA transcripts have diverse biological roles in Drosophila. Heredity 115, 1–2 (2015).
Cioffi, M. de B. & Bertollo, L. A. C. Chromosomal distribution and evolution of repetitive DNAs in fish. in Repetitive DNA (ed. Garrido-Ramos, M. A.) 197–221 (Karger, Basel, 2012). https://doi.org/10.1159/000337950.
Gaffaroglu, M. et al. Present and Future Salmonid Cytogenetics. Genes 11, 1462 (2020).
Crepaldi, C. & Parise-Maltempi, P. P. Heteromorphic Sex Chromosomes and Their DNA Content in Fish: An Insight through Satellite DNA Accumulation in Megaleporinus elongatus. Cytogenetic Genome Res. 160, 38–46 (2020).
Favarato, R. M. et al. Comparative cytogenetics of Serrasalmidae (Teleostei: Characiformes): The relationship between chromosomal evolution and molecular phylogenies. PLOS ONE 16, e0258003 (2021).
Voleníková, A. et al. Fast satellite DNA evolution in Nothobranchius annual killifishes. Chromosome Res. 31, 33 (2023).
Deakin, J. E. et al. Chromosomics: Bridging the Gap between Genomes and Chromosomes. Genes 10, 627 (2019).
Serrano-Freitas, É. A. et al. Satellite DNA content of B chromosomes in the characid fish Characidium gomesi supports their origin from sex chromosomes. Mol. Genet Genomics 295, 195–207 (2020).
Goes, C. A. G. et al. Revealing the Satellite DNA History in Psalidodon and Astyanax Characid Fish by Comparative Satellitomics. Front. Genet. 13, 884072 (2022).
Štundlová, J. et al. Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci. Chromosome Res 30, 309–333 (2022).
Rocha-Reis, D. A. et al. In Silico characterization of satellitomes and cross-amplification of putative satDNAs in two species of the Hypostomus ancistroides complex (Siluriformes, Loricariidae). Cytogenetic Genome Res. 1, https://doi.org/10.1159/000539429 (2024).
Utsunomia, R. et al. Satellitome landscape analysis of Megaleporinus macrocephalus (Teleostei, Anostomidae) reveals intense accumulation of satellite sequences on the heteromorphic sex chromosome. Sci. Rep. 9, 5856 (2019).
Ferretti, A. B. S. M., Milani, D., Palacios-Gimenez, O. M., Ruiz-Ruano, F. J. & Cabral-de-Mello, D. C. High dynamism for neo-sex chromosomes: satellite DNAs reveal complex evolution in a grasshopper. Heredity 125, 124–137 (2020).
Shatskikh, A. S., Kotov, A. A., Adashev, V. E., Bazylev, S. S. & Olenina, L. V. Functional Significance of Satellite DNAs: Insights From Drosophila. Front. Cell Dev. Biol. 8, 312 (2020).
Cabral-de-Mello, D. C., Zrzavá, M., Kubíčková, S., Rendón, P. & Marec, F. The Role of Satellite DNAs in Genome Architecture and Sex Chromosome Evolution in Crambidae Moths. Front. Genet. 12, 661417 (2021).
de Oliveira, M. P. B. et al. Following the Pathway of W Chromosome Differentiation in Triportheus (Teleostei: Characiformes). Biology 12, 1114 (2023).
Yunis, J. J. & Yasmineh, W. G. Heterochromatin, Satellite DNA, and Cell Function. Science 174, 1200–1209 (1971).
Bonaccorsi, S. & Lohe, A. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics 129, 177–189 (1991).
Harvey, S. C. et al. Analysis of repetitive DNA sequences in the sex chromosomes of Oreochromis niloticus. Cytogenetic Genome Res. 101, 314–319 (2003).
Krzywinski, J., Sangaré, D. & Besansky, N. J. Satellite DNA From the Y Chromosome of the Malaria Vector Anopheles gambiae. Genetics 169, 185–196 (2005).
Schemberger, M. O. et al. Construction and Characterization of a Repetitive DNA Library in Parodontidae (Actinopterygii: Characiformes): A Genomic and Evolutionary Approach to the Degeneration of the W Sex Chromosome. Zebrafish 11, 518–527 (2014).
Kretschmer, R. et al. Satellitome analysis illuminates the evolution of ZW sex chromosomes of Triportheidae fishes (Teleostei: Characiformes). Chromosoma 131, 29–45 (2022).
Palacios-Gimenez, O. M. et al. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 7, 6422 (2017).
Navajas-Pérez, R. et al. The Evolution of Reproductive Systems and Sex-Determining Mechanisms Within Rumex (Polygonaceae) Inferred from Nuclear and Chloroplastidial Sequence Data. Mol. Biol. Evol. 22, 1929–1939 (2005).
Navajas-Pérez, R. et al. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene 368, 61–71 (2006).
Navajas-Pérez, R., Quesada del Bosque, M. E. & Garrido-Ramos, M. A. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Mol. Genet Genomics 282, 395–406 (2009).
Navajas-Pérez, R., Schwarzacher, T., Rejón, M. R. & Garrido-Ramos, M. A. Molecular cytogenetic characterization of Rumex papillaris, a dioecious plant with an XX/XY1Y2 sex chromosome system. Genetica 135, 87–93 (2009).
Hobza, R. et al. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 115, 376–382 (2006).
Steflova, P. et al. Contrasting Patterns of Transposable Element and Satellite Distribution on Sex Chromosomes (XY1Y2) in the Dioecious Plant Rumex acetosa. Genome Biol. Evol. 5, 769–782 (2013).
Jesionek, W. et al. Fundamentally different repetitive element composition of sex chromosomes in Rumex acetosa. Ann. Bot. 127, 33–47 (2021).
Cabral-de-Mello, D. C. et al. The spread of s atellite DNAs in euchromatin and insights into the multiple sex chromosome evolution in Hemiptera revealed by repeatome analysis of the bug Oxycarenus hyalinipennis. Insect Mol. Biol. 32, 725–737 (2023).
De Moraes, R. L. R. et al. Chromosomal Rearrangements and Satellite DNAs: Extensive Chromosome Reshuffling and the Evolution of Neo-Sex Chromosomes in the Genus Pyrrhulina (Teleostei; Characiformes). IJMS 24, 13654 (2023).
Covain, R. et al. Molecular phylogeny of the highly diversified catfish subfamily Loricariinae (Siluriformes, Loricariidae) reveals incongruences with morphological classification. Mol. Phylogenetics Evol. 94, 492–517 (2016).
Ramos, L. & Antunes, A. Decoding sex: Elucidating sex determination and how high-quality genome assemblies are untangling the evolutionary dynamics of sex chromosomes. Genomics 114, 110277 (2022).
Fry, K. & Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12, 1069–1084 (1977).
Utsunomia, R. et al. A glimpse into the satellite DNA library in Characidae fish (Teleostei, Characiformes). Front. Genet. 8, 103 (2017).
Deon, G. A. et al. The role of satellite DNAs in the chromosomal rearrangements and the evolution of the rare XY1Y2 sex system in Harttia (Siluriformes: Loricariidae). J. Heredity esae028, https://doi.org/10.1093/jhered/esae028 (2024).
Ahmad, S. F. et al. Dark Matter of Primate Genomes: Satellite DNA Repeats and Their Evolutionary Dynamics. Cells 9, 2714 (2020).
Gutiérrez, J. et al. Satellitome Analysis on Talpa aquitania Genome and Inferences about the satDNAs Evolution on Some Talpidae. Genes 14, 117 (2023).
Lisachov, A., Rumyantsev, A., Prokopov, D., Ferguson-Smith, M. & Trifonov, V. Conservation of Major Satellite DNAs in Snake Heterochromatin. Animals 13, 334 (2023).
Mestrović, N., Plohl, M., Mravinac, B. & Ugarković, D. Evolution of satellite DNAs from the genus Palorus–experimental evidence for the ‘library’ hypothesis. Mol. Biol. Evolution 15, 1062–1068 (1998).
Palacios-Gimenez, O. M. et al. Comparative analysis of morabine grasshopper genomes reveals highly abundant transposable elements and rapidly proliferating satellite DNA repeats. BMC Biol. 18, 199 (2020).
Quesada del Bosque, M. E. et al. satellite DNA evolutionary analysis in the North American endemic dioecious plant Rumex hastatulus (Polygonaceae). Genome 54, 253–260 (2011).
Quesada del Bosque, M. E., López-Flores, I., Suárez-Santiago, V. N. & Garrido-Ramos, M. A. Differential spreading of HinfI satellite DNA variants during radiation in Centaureinae. Ann. Bot. 112, 1793–1802 (2013).
del Bosque, M. E. Q., López-Flores, I., Suárez-Santiago, V. N. & Garrido-Ramos, M. A. Satellite-DNA diversification and the evolution of major lineages in Cardueae (Carduoideae Asteraceae). J. Plant Res. 127, 575–583 (2014).
Šatović-Vukšić, E. & Plohl, M. Satellite DNAs—From Localized to Highly Dispersed Genome Components. Genes 14, 742 (2023).
Casacuberta, E. Drosophila: Retrotransposons Making up Telomeres. Viruses 9, 192 (2017).
Talbert, P. B. & Henikoff, S. What makes a centromere? Exp. Cell Res. 389, 111895 (2020).
Blanco, D. R. et al. Origin of the X1X1X2X2/X1X2Y sex chromosome system of Harttia punctata (Siluriformes, Loricariidae) inferred from chromosome painting and FISH with ribosomal DNA markers. Genetica 142, 119–126 (2014).
Sassi, F. M. C. et al. Multiple sex chromosomes and evolutionary relationships in amazonian catfishes: The outstanding model of the genus Harttia (siluriformes: Loricariidae). Genes 11, 1179 (2020).
Berthelot, C., Muffato, M., Abecassis, J. & Crollius, H. R. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep. 10, 1913–1924 (2015).
Warmerdam, D. O. & Wolthuis, R. M. F. Keeping ribosomal DNA intact: a repeating challenge. Chromosome Res. 27, 57–72 (2019).
Deon, G. A. et al. Highly Rearranged Karyotypes and Multiple Sex Chromosome Systems in Armored Catfishes from the Genus Harttia (Teleostei, Siluriformes). Genes 11, 1366 (2020).
Barros, A. V. et al. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role? Gene 608, 20–27 (2017).
Glugoski, L., Giuliano-Caetano, L., Moreira-Filho, O., Vicari, M. R. & Nogaroto, V. Co-located hAT transposable element and 5S rDNA in an interstitial telomeric sequence suggest the formation of Robertsonian fusion in armored catfish. Gene 650, 49–54 (2018).
Glugoski, L., Deon, G. A., Nogaroto, V., Moreira-Filho, O. & Vicari, M. R. Robertsonian Fusion Site in Rineloricaria pentamaculata (Siluriformes: Loricariidae): Involvement of 5S Ribosomal DNA and Satellite Sequences. Cytogenet Genome Res. 162, 657–664 (2022).
Lim, K. Y. et al. Dynamic Changes in the Distribution of a Satellite Homologous to Intergenic 26-18S rDNA Spacer in the Evolution of Nicotiana. Genetics 166, 1935–1946 (2004).
Jo, S.-H. et al. Evolution of ribosomal DNA-derived satellite repeat in tomato genome. BMC Plant Biol. 9, 42 (2009).
Gerbi, S. A. Bundling up DNA. eLife 7, e37234 (2018).
Jagannathan, M., Cummings, R. & Yamashita, Y. M. A conserved function for pericentromeric satellite DNA. eLife 7, e34122 (2018).
Ahmed, M. & Liang, P. Transposable Elements Are a Significant Contributor to Tandem Repeats in the Human Genome. Int. J. Genomics 2012, 947089 (2012).
Wong, L. H. & Choo, K. H. A. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 20, 611–616 (2004).
Kapitonov, V. V. & Jurka, J. The Long Terminal Repeat of an Endogenous Retrovirus Induces Alternative Splicing and Encodes an Additional Carboxy-Terminal Sequence in the Human Leptin Receptor. J. Mol. Evol. 48, 248–251 (1999).
Volff, J.-N., Hornung, U. & Schartl, M. Fish retroposons related to the Penelope element of Drosophila virilis define a new group of retrotransposable elements. Mol. Gen. Genomics 265, 711–720 (2001).
Eickbush, T. H. & Malik, H. S. Origins and Evolution of Retrotransposons. in Mobile DNA II 1111–1144 (John Wiley & Sons, Ltd, 2007). https://doi.org/10.1128/9781555817954.ch49.
Arkhipova, I. R. Distribution and Phylogeny of Penelope-Like Elements in Eukaryotes. Syst. Biol. 55, 875–885 (2006).
Thomas, J. & Pritham, E. J. Helitrons, the Eukaryotic Rolling-circle Transposable Elements. in Mobile DNA III 891–924 (John Wiley & Sons, Ltd, 2015). https://doi.org/10.1128/9781555819217.ch40.
Satović, E., Vojvoda Zeljko, T., Luchetti, A., Mantovani, B. & Plohl, M. Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genomics 17, 997 (2016).
Vojvoda Zeljko, T., Pavlek, M., Meštrović, N. & Plohl, M. Satellite DNA-like repeats are dispersed throughout the genome of the Pacific oyster Crassostrea gigas carried by Helentron non-autonomous mobile elements. Sci. Rep. 10, 15107 (2020).
Camacho, J. P. M. et al. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biol. 20, 36 (2022).
Cardone, M. F. et al. Evolution of Beta Satellite DNA Sequences: Evidence for Duplication-Mediated Repeat Amplification and Spreading. Mol. Biol. Evol. 21, 1792–1799 (2004).
Ma, J. & Jackson, S. A. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res 16, 251–259 (2006).
Plohl, M., Meštrović, N. & Mravinac, B. Centromere identity from the DNA point of view. Chromosoma, https://doi.org/10.1007/s00412-014-0462-0 (2014).
Izsvák, Z., Ivics, Z. & Hackett, P. B. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio). Molec. Gen. Genet. 247, 312–322 (1995).
Munoz-Lopez, M. & Garcia-Perez, J. L. DNA Transposons: Nature and Applications in Genomics. Curr. Genomics 11, 115–128 (2010).
Primo, C. C., Glugoski, L., Vicari, M. R. & Nogaroto, V. Chromosome Mapping and Molecular Characterization of the Tc1/Mariner Element in Rineloricaria (Siluriformes: Loricariidae). Braz. Arch. Biol. Technol. 61, e18170623 (2018).
Schemberger, M. O. et al. DNA transposon invasion and microsatellite accumulation guide W chromosome differentiation in a Neotropical fish genome. Chromosoma 128, 547–560 (2019).
Ayres-Alves, T. et al. Karyotypic Evolution and Chromosomal Organization of Repetitive DNA Sequences in Species of Panaque, Panaqolus, and Scobinancistrus (Siluriformes and Loricariidae) from the Amazon Basin. Zebrafish 14, 251–260 (2017).
Gouveia, J. G. et al. Repetitive DNA in the Catfish Genome: rDNA, Microsatellites, and Tc1-Mariner Transposon Sequences in Imparfinis Species (Siluriformes, Heptapteridae). J. Heredity 108, 650–657 (2017).
Chalopin, D., Volff, J.-N., Galiana, D., Anderson, J. L. & Schartl, M. Transposable elements and early evolution of sex chromosomes in fish. Chromosome Res. 23, 545–560 (2015).
Bausek, N., Waclawek, M., Schneider, W. J. & Wohlrab, F. The Major Chicken Egg Envelope Protein ZP1 Is Different from ZPB and Is Synthesized in the Liver. J. Biol. Chem. 275, 28866–28872 (2000).
Feng, J.-M., Tian, H.-F., Hu, Q.-M., Meng, Y. & Xiao, H.-B. Evolution and multiple origins of zona pellucida genes in vertebrates. Biol. Open 7, bio036137 (2018).
Cao, L. et al. Neofunctionalization of zona pellucida proteins enhances freeze-prevention in the eggs of Antarctic notothenioids. Nat. Commun. 7, 12987 (2016).
Bianchi, E. & Wright, G. J. Cross-species fertilization: the hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140101 (2015).
Claw, K. G. & Swanson, W. J. Evolution of the Egg: New Findings and Challenges. Annu. Rev. Genomics Hum. Genet. 13, 109–125 (2012).
Cassemiro, F. A. S. et al. Landscape dynamics and diversification of the megadiverse South American freshwater fish fauna. Proc. Natl. Acad. Sci. 120, e2211974120 (2023).
Sassi, F. M. C. et al. Adding New Pieces to the Puzzle of Karyotype Evolution in Harttia (Siluriformes, Loricariidae): Investigation of Amazonian Species. Biology 10, 922 (2021).
Bertollo, L. A. C., Cioffi, M. de B. & Moreira-Filho, O. Direct chromosome preparation from Freshwater Teleost Fishes. in Fish cytogenetic techniques (Chondrichthyans and Teleosts) (eds. Ozouf-Costaz, C., Pisano, E., Foresti, F. & Almeida Toledo, L. F.) 21–26 (CRC Press, Enfield USA, 2015). https://doi.org/10.1201/b18534-4.
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Novák, P. et al. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 45, e111 (2017).
Schmieder, R. & Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLOS ONE 6, e17288 (2011).
Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0. http://www.repeatmasker.org (2015).
Ruiz-Ruano, F. J., López-León, M. D., Cabrero, J. & Camacho, J. P. M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 6, 28333 (2016).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Singh, U. & Wurtele, E. S. orfipy: a fast and flexible tool for extracting ORFs. Bioinformatics 37, 3019–3020 (2021).
Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102–1104 (2000).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Zwick, M. S. et al. A rapid procedure for the isolation of C 0 t-1 DNA from plants. Genome 40, 138–142 (1997).
Lehner, B. & Grill, G. Global river hydrography and network routing: baseline data and new approaches to study the world’s large river systems. Hydrological Process. 27, 2171–2186 (2013).
Kumar, S., Stecher, G., Suleski, M. & Hedges, S. B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 34, 1812–1819 (2017).
Acknowledgements
The authors are grateful to Flávio Henrique Silva and Eduardo Pereira de Souza for the help with sequencing, to Foyez Shamz for the valuable insights, and to Jhon Alex Dziechciarz Vidal for the assistance in the ORFs identification. We are also grateful to Eliana Feldberg, Osvaldo Takeshi Oyakawa and Lúcia Helena Rapp Py-Daniel for the identification of the samples, and to Luiz Henrique da Silva, Antônio Aparecido Donizetti da Silva, Ezequiel Aguiar de Oliveira and Patrik Viana for assistance in field work. We are also indebted to Juan Pedro Martinez Camacho for his advice on statistical analysis. F.M.C.S. was supported by FAPESP (Proc. numbers 2020/02681-9; 2022/04261-2 and 2023/08116-0). M.B.C. was founded by FAPESP (2023/00955-2) and CNPq (302928/2021-9). T.L. was founded by the German Research Foundation Projekt-Nr. 512648189 and the Open Access Publication Fund of the Thueringer Universitaets- und Landesbibliothek Jena. This study was partially funded by CAPES Finance code 001 and INCT – Peixes (proc. 405706/2022-7).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
F.M.C.S., M.A.G.R., G.A.D., and M.B.C. conceived and designed research. F.M.C.S., M.A.G.R., R.Z.S., R.U. and G.A.D. conducted experiments. F.M.C.S., G.A.D., R.Z.S., R.U., T.E., F.P.F., T.L., and M.B.C. analyzed the data. M.A.G.R., R.Z.S., and R.U. contributed with new methods. F.M.C.S., M.A.G.R., R.Z.S., R.U., T.E., G.A.D., F.P.F., T.L., and M.B.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Martin Knytl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: John Mulley, David Favero and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sassi, F.d.M.C., Garrido-Ramos, M.A., Utsunomia, R. et al. Independent evolution of satellite DNA sequences in homologous sex chromosomes of Neotropical armored catfish (Harttia). Commun Biol 8, 524 (2025). https://doi.org/10.1038/s42003-025-07891-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07891-6