Abstract
Cyclophilin 38 (CYP38) plays a crucial role in the assembly and stability of photosystem II (PSII), but its molecular mechanism remains unclear. In this study, we identified thylakoid lumen protein 7.6 (TLP7.6) as an in vivo interactor of CYP38. Under normal growth conditions, the tlp7.6 single mutant exhibited no significant phenotypic differences compared to wild-type Col-0. However, the cyp38-2/tlp7.6-1 double mutant displayed severe developmental defects, including stunted growth, delayed flowering, yellowish leaf, short primary roots, abnormal chloroplast ultrastructure, and reduced biomass, which were more pronounced than those in either the tlp7.6 or cyp38-2 single mutant. Photosynthetic analysis revealed that PSII capacities in cyp38-2/tlp7.6-1 and cyp38-2 mutants were significantly reduced, consistent with their slow-growth phenotype. Blue native PAGE analysis demonstrated a substantial reduction in PSII supercomplexes and light-harvesting complex II (LHCII) in cyp38-2/tlp7.6-1, while PSII monomer (PSII-M) were significantly increased. Immunoblotting and two-dimensional gel electrophoresis further confirmed decreased levels of key components of PSII, PSI, and ATPase subunits in the double mutant. Altogether, these results highlight the role of TLP7.6 as an assistive factor in CYP38-mediated PSII assembly, and provide insights into thylakoid lumen protein function in photosynthesis.
Similar content being viewed by others
Introduction
Photosynthetic electron transfer occurs on chloroplast thylakoid membranes, which enclose a continuous aqueous lumenal space. This space was originally considered to be primarily responsible for promoting electron transport and proton gradient formation1,2,3,4. In light of current understanding, proteins located in the thylakoid lumen not only play roles in photosynthetic electron transfer, but also are involved in regulating thylakoid biogenesis, maintaining the stability of photosynthetic protein complexes, and contributing to redox regulation and stress response5. Although several lumen proteins have been extensively studied in detail, particularly the lumenal subunits of photosystem II (PSII) and NADPH complexes, the functions of many other lumenal proteins remain elusive. Decoding the precise functions of lumen proteins is crucial for a fully understanding of the mechanisms of photosynthesis, which is essential for improving photosynthetic efficiency and productivity.
The thylakoid lumen proteome consists of approximately 100 proteins, identified through proteomic analyses in Arabidopsis6,7,8. There proteins exhibit diverse functions, with a large subset involved in PSII assembly, repair, and stabilization6,7,8,9,10. Recent studies classified thylakoid lumen proteins into two parts: soluble free lumen (FL) proteins and membrane-associated lumen (MAL) proteins. It was experimentally revealed that more than 60 proteins are present in lumen, with most FL proteins are closely associated with PSII biogenesis and repair, while MAL proteins are related in oxygen-evolving complex (OEC) function11.
PSII biogenesis and repair are complex processes, which require over 20 protein subunits and multiple cofactors. During PSII repair, the damaged D1 protein undergoes rapid turnover, facilitated by lumenal proteins, such as Deg1, which cleaves the damaged D1 protein. After the damaged D1 was removed, the vacant place was incorporated in by a newly synthesized D1, with the assist of the precursor D1 (pD1) processing related protein, Low PSII Accumulation (LPA)12,13,14. Other proteins, such as TLP18.3, Psb27, and LPA19, further promote PSII assembly by stabilizing and facilitating the incorporation of PSII subunits14,15,16,17,18. Furthermore, during the assembly of PSII reaction centers, CYP38 and High Chlorophyll Fluorescence 136 (HCF136) assist the newly synthesized D1 protein in inserting into the thylakoid membrane and the core complex19,20,21,22.
CYP38, a thylakoid lumen-localized immunophilin, plays a pivotal role in PSII assembly and stabilization. Immunophilins are receptor proteins binding immunosuppressive drugs in animal, including cyclophilins (receptors for cyclosporin A), and FKBPs (receptors for FKBP506), and they mainly regulate protein folding in various cellular compartments23,24. In Arabidopsis, CYP38 contains an N-terminal binding domain and a C-terminal cyclophilin domain connected by a flexible acidic loop chain25. The cyp38 mutant exhibits defects in thylakoid grana structure, reduced PSII supercomplex accumulation, and increased sensitivity to high light stress26. Previous studies suggest that CYP38 facilitates the proper folding of PSII subunits, including D1 and CP43, and supports the assembly of the Mn4-Ca cluster required for water oxidation in PSII19,20. Additionally, the homologous protein TLP40 in spinach regulates PSII assembly through modulating protein phosphatase activity on the thylakoid membrane26,27.
Despite a growing understanding of CYP38, the regulatory mechanisms by which the CYP38 protein affects PSII assembly remains unclear, especially the identity of interacting proteins assisting its activity is still not illustrated. In this study, we identified and characterized a thylakoid lumen protein, thylakoid lumen protein 7.6 (TLP7.6), which interacts with CYP38. Using the CRISPR/Cas9 technology, we generated tlp7.6 knockout mutants and analyzed their phenotypes in combination with the cyp38-2 mutant. While tlp7.6 single mutants displayed no discernible defects under normal growth conditions, the cyp38-2/tlp7.6-1 double mutants exhibited severe growth abnormalities, and impaired chloroplast ultrastructure, much severe than the tlp7.6-1 or cyp38-2 single mutant. Our findings indicate that TLP7.6 functions as a co-factor that assists CYP38 in PSII supercomplex assembly and stabilization. By investigating the interaction between TLP7.6 and CYP38, we provide new insights into the molecular mechanisms regulating PSII biogenesis and highlight the functional diversity of thylakoid lumen proteins in chloroplasts.
Results
Verification of the interaction between TLP7.6 and CYP38, and the evolutionary analysis of TLP7.6
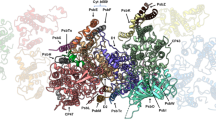
We previously demonstrated that TLP7.6 (At1g21500), a thylakoid lumen protein, interacts with CYP38 in yeast28. To confirm this interaction, we performed a Co-Immunoprecipitation (Co-IP) assay using transgenic plants expressing the 35Spro:CYP38-His in the cyp38-2 or Col-0 genetic background. The results (Fig. 1a) confirmed that CYP38 interacts with TLP7.6 in vivo. To further investigate the relationship between TLP7.6 and CYP38, we employed AlphaFold3 (https://github.com/google-deepmind/alphafold3) to predict their binding sites. The predicted interface reveals complementary electrostatic surfaces and hydrophobic regions between two proteins, which is essential for their stable interaction (Supplementary Fig. 1). The proximity of their functional domains suggests that TLP7.6 and CYP38 likely have a cooperative functional relationship. We next analyzed the evolutionary conservation of TLP7.6 across different species by searching for mature amino acid sequences (excluding the signal peptide) in the NCBI database and identifying conserved domains using the Pfam database. The evolutionary tree revealed that TLP7.6 is highly conserved in green terrestrial plants, and contains the BTB-BACK-Kelch (BBK) domain (Supplementary Fig. 2), which is known for its role in protein-protein interactions and recruitment of transcriptional corepressors29.
a Co-immunoprecipitation (Co-IP) assay shows the interaction between TLP7.6 and CYP38. Thylakoid membranes from Col-0, and transgenic cyp38-2 mutants expressing 35S-driven C-terminal His-tagged CYP38 were solubilized with 0.1% (w/v) n-dodecyl-β-D-maltoside (β-DM), incubated with Ni-NTA Agarose, and eluted proteins analyzed by immunoblotting with TLP7.6 and His antibodies. b GFP signals show that the TLP7.6-GFP fusion protein expressed from the 35S::TLP7.6-GFP construct localized to the chloroplast. Green fluorescence indicates GFP, red fluorescence shows chloroplast autofluorescence, and yellow fluorescence represents the merged signals. Scale bar: 25 µm. c Immunoblot analysis of chloroplast components. Thylakoid membrane, thylakoid lumen and stroma fractions from Col-0 were probed with antibodies against D1, ClpC, PC, and TLP7.6. d Tissue-specific expression of TLP7.6. qRT-PCR analysis of TLP7.6 transcript levels in roots, stems, young leaves, and old leaves of Col-0. Error bars: mean ± SE (n = 3).
TLP7.6 is localized in the chloroplast thylakoid lumen and is highly expressed in leaves
To determine the subcellular localization of TLP7.6, we constructed a plasmid expressing the TLP7.6-GFP fusion protein, and transiently expressed it in Nicotiana benthamiana leaves. Confocal microscopy revealed that GFP signal co-localized with chloroplast autofluorescence, confirming that TLP7.6 is localized in the chloroplasts (Fig. 1b). Further fractionation of chloroplasts revealed that TLP7.6 is exclusively detected in the thylakoid lumen fraction (Fig. 1c), with D1, ClpC, and PC as marker proteins for the thylakoid membrane, stroma, and lumen, respectively. Additionally, we assessed the expression pattern of TLP7.6 across different tissues using the quantitative real time PCR (qRT-PCR). The most highly expression observed in stems and leaves (Fig. 1d), this tissue-specific expression suggests that TLP7.6 may play a significant role in photosynthesis.
Mutants deficient of TLP7.6 were obtained using the CRISPR/Cas9 technique
To investigate the physiological function of TLP7.6, we generated tlp7.6 mutants using the CRISPR/Cas9 genome editing system. The gene sequence (At1g21500) was obtained from the Arabidopsis database (https://www.arabidopsis.org), and guide RNA sequences (gRNAs) were designed using the CRISPR-PLANT tool (http:// www.genome.arizona.edu/crispr/CRISPR search.html) (Supplementary Table 1).
After screening transgenic plants carrying the designed CRISPR/Cas9 construct, we obtained two individual transgene-free homozygous gene-edited lines, designated as tlp7.6-1 and tlp7.6-2, respectively (Fig. 2a). Sequence analysis revealed that tlp7.6-1 harbors a deletion mutation of the adenine (A) of the start codon (ATG), which introduces an NcoI restriction site (CCATGG), while tlp7.6-2 contains a frameshift mutation with an extra adenine (A) base at 73 bp downstream of the start codon (ATG) (Fig. 2b). Western blot analysis using a TLP7.6-specific antibody confirmed that both lines were null mutants (Fig. 2c). To perform the functional complementation experiment, we expressed the genomic DNA of TLP7.6 into the tlp7.6-1 mutant background (referred to herein as Com). As shown in Fig. 2c, the TLP7.6 protein levels in Com plants were restored to the wild-type level, confirming successful complementation. For all subsequent functional analyses, tlp7.6-1 and Com plants were used.
a Illustrated of the mutation sites in the tlp7.6 mutant lines. The blue line indicates the genomic target site of the sgRNA for tlp7.6. The tlp7.6-1 mutant exhibits a deletion of the start codon ATG, confirmed by NcoI digestion. The tlp7.6-2 mutant carries a frameshift mutation caused by the insertion of an additional adenine (A) at 73 bp after the initial ATG. b Sequencing chromatograms of the mutation sites of tlp7.6-1 and tlp7.6-2 mutants. The gray boxes indicate the base mutation. The target site was amplified by PCR with gene-specific primers listed in Supplementary Table 1. c Western blot to detect TLP7.6 protein levels in Col-0, tlp7.6-1, tlp7.6-2 and complemented line (Com) using specific TLP7.6 antibodies. Samples were collected from 21-day-old seedlings. Ponceau S staining serves as a loading control.
The absence of both CYP38 and TLP7.6 affects plant growth
To assess the functional relationship between TLP7.6 and CYP38, we analyzed the phenotypes of single and double mutants under standard growth conditions. While the tlp7.6-1 single mutant did not exhibit any visible defects compared to Col-0 (Fig. 3a). However, the cyp38-2/tlp7.6-1 double mutant displayed a significant growth retardation phenotype, characterized by yellowish leaves, delayed flowering, which was much more severe than the tlp7.6-1 or cyp38-2 single mutant (Fig. 3b, c). Additionally, the double mutant plants had reduced chlorophyll content (Fig. 3d), indicating impaired photosynthetic efficiency.
a Aboveground phenotype of 3-week-old plants. Scale bar: 1 cm. The CYP38 and TLP7.6 protein levels were confirmed by western blotting using specific antibodies against TLP7.6 or CYP38, with Ponceau S staining serves as a loading control. b Bolting phenotypes under normal light condition. Scale bar: 1 cm. c Bolting days of plants, with data shown for three biological replicates (n ≥ 30). d Total chlorophyll content. Statistical significance determined by student’s t-test (p < 0.05). Different letters indicate significant differences (n = 3).
Previously studies have demonstrated that the absence of CYP38 affects aboveground biomass and primary root development30. The cyp38-2 mutant exhibits an elevated sensitivity to high light stress, though exogenous sucrose supplementation partially rescues this phenotype20,30. To investigate the role of CYP38 and TLP7.6 in root development, we grew plants on half-strength Murashige and Skoog (MS) medium supplemented 1% sucrose. Under these conditions, both cyp38-2 and cyp38-2/tlp7.6-1 mutants displayed a phenotype with shorter root lengths compared to Col-0, indicating that the loss of either protein impacts root growth (Fig. 4a). Strikingly, in the absence of exogenous sucrose, the cyp38-2/tlp7.6-1 double mutant exhibited the most severe growth inhibition across all genotypes (Fig. 4b). These results suggest that TLP7.6 and CYP38 collectively enhance the plant’s capacity to synthesize and utilize sucrose for energy and grown. To evaluate the impact of these mutants on biomass accumulation, we measured the aboveground fresh weight of 7-day-old seedlings grown on sucrose-supplemented or sucrose-free media. The biomass of the cyp38-2/tlp7.6-1 double mutant was consistently lower than that of both Col-0 and single mutants, irrespective of the sucrose availability (Fig. 4c). This indicates that the combined loss of TLP7.6 and CYP38 compromises overall plant growth and productivity, reinforcing their synergistic role in supporting efficient photosynthesis and carbon utilization. The exacerbated phenotypes in the double mutant relative to single mutants provide compelling evidence for a functional interaction between TLP7.6 and CYP38. This partnership appears critical under growth-limiting conditions, such as sucrose deprivation, highlighting the significance of TLP7.6 and CYP38 in regulating plant development and stress response mechanisms.
a Primary root length of Col-0, cyp38-2, tlp7.6-1, cyp38-2/tlp7.6-1 and Com plants grown on 1/2 MS medium with 1% sucrose for 7 days. Scale bar: 1 cm. Quantitative analysis is shown in the lower panel. Data are presented as mean ± SE (n ≥ 3). b Primary root length of Col-0, cyp38-2, tlp7.6-1, cyp38-2/tlp7.6-1, Com plants grown on 1/2 MS medium without sucrose for 7 days, with quantitative analysis shown in lower panel. Scale bar: 1 cm. Data are presented as mean ± SE (n ≥ 3). c Aboveground biomass of 7-day-old seedlings in Col-0, cyp38-2, tlp7.6-1, cyp38-2/tlp7.6-1. Different letters above the bars indicate significant differences among group at p < 0.05 as determined by Student’s t-test. (n ≥ 25).
Chloroplast ultrastructure defects in cyp38-2/tlp7.6-1 double mutant
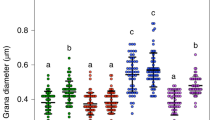
Given the observed growth retardation in the cyp38-2/tlp7.6-1 double mutant, we investigated the formation and development of the chloroplasts using transmission electron microscopy (TEM). In contrast to the well-organized grana stacks observed in Col-0 and tlp7.6-1, the cyp38-2 mutant exhibited thicker grana stacks (Fig. 5a-c, Supplementary Fig. 3), a phenotype consistent with previous characterization of CYP38-deficient plants26. This structural change may represent a compensatory response to altered light sensitivity in the absence of CYP38. Intriguingly, the cyp38-2/tlp7.6-1 double mutant displayed thinner grana stacks (Fig. 5d), exhibited more severe defects, including reduced starch accumulation and lower biomass (Fig. 4c). Starch quantification in 14-day-old seedlings confirmed that the cyp38-2/tlp7.6-1 double mutant had significantly lower starch levels than Col-0 (Supplementary Fig. 4). These paradoxical observation suggests that the increased grana thickness in cyp38-2 may serve as a protective adaptation, possibly stabilizing PSII under light stress. However, in the double mutant, the loss of TLP7.6 might impair thylakoid remodeling, preventing this compensatory mechanism, and then leading to growth defects. Additionally, chloroplast ultrastructure analysis revealed a significant increase in osmiophilic granules (plastoglobules) in both cyp38-2 and cyp38-2/tlp7.6-1, suggesting increased plastid remodeling or stress-related degradation processes (Supplementary Figs. 5 and 6). These findings indicate that TLP7.6 and CYP38 may be involved in early chloroplast development and thylakoid organization, potentially influencing PSII stability and photoprotection mechanisms.
a Chloroplast from a 14-day-old Col-0 plant. b Chloroplast from a 14-day-old cyp38-2 mutant plant. c Chloroplast from a 14-day-old tlp7.6-1 mutant plant. d Chloroplast from a 14-day-old cyp38-2/tlp7.6-1 mutant plant. Scale bar: 500 nm. CW Cell wall, ChM Chloroplast membrane, OS Osmiophilic granules, SG Starch granules, Thl thylakoid lamella.
Chlorophyll Fluorescence Analysis of Col-0, tlp7.6-1, cyp38-2, cyp38-2/tlp7.6-1, Com mutants
Chlorophyll fluorescence and photosynthetic efficiency are commonly used to evaluate the physiological status of plants. To investigate whether the thylakoid lumen-localized TLP7.6 protein affects photosynthesis, we measured chlorophyll fluorescence to monitor the physiological state of the photosynthetic apparatus31. We analyzed chlorophyll fluorescence parameters of Col-0, tlp7.6-1, cyp38-2, cyp38-2/tlp7.6-1, and Com to assess whether tlp7.6-1 and cyp38-2/tlp7.6-1 mutations impairs their photosynthetic function. The maximum quantum efficiency of PSII, represented by the Fv/Fm values, remained approximately 0.8 in Col-0 plants. However, the Fv/Fm ratio in cyp38-2/tlp7.6-1 was lower than that of Col-0 and tlp7.6-1 (Fig. 6a, b). Notably, cyp38-2 and the cyp38-2/tlp7.6 double mutant exhibited similar Fv/Fm values (Fig. 6b) suggesting that TLP7.6 may not directly affect the photochemical efficiency of PSII. Instead, it may contribute to stabilizing PSII supercomplex assembly, indicating that the absence of both CYP38 and TLP7.6 has a more severe impact on PSII organization. This also implies that TLP7.6 could partially support CYP38 function. To further assess the functional impact on PSII, we examined the effective quantum yield of PSII, which declined with increasing photosynthetic photon flux density (PPFD). While Col-0 and the tlp7.6-1 single mutant exhibited significantly higher PSII values compared to cyp38-2 or cyp38-2/tlp7.6-1 mutants. However, no significant differences were observed between cyp38-2 and cyp38-2/tlp7.6-1 mutants (Fig. 6c). Consistently, the electron transport rate ETR(II) of PSII was reduced in both cyp38-2 and cyp38-2/tlp7.6-1 compared to Col-0, with no significant differences between these two mutants (Fig. 6c). These findings suggest that the disruption of CYP38 alone already severely impairs PSII photochemistry, and the additional loss of TLP7.6 does not further exacerbate these effects. To investigate photoprotection mechanisms, we analyzed non-photochemical quenching (NPQ), which represents the dissipation of excess light energy. A slight decrease in NPQ was observed in cyp38-2/tlp7.6-1, indicating that the double mutant plants may be less efficient in dissipating excess excitation energy under high-light conditions. This suggested that the cyp38-2/tlp7.6-1 plants are impaired in their ability to protect themselves from photooxidative damage (Fig. 6c). Furthermore, we examined the quantum yield of non-regulated energy dissipation Y(NO), which reflects the ability of plants to dissipate energy through non-regulated processes. Both cyp38-2 and cyp38-2/tlp7.6-1 mutants exhibited similar Y(NO) values, suggesting that the double mutant does not exhibit a significant difference in the ability to dissipate excess energy compared to the cyp38-2 mutant (Fig. 6c). Taken together, these results suggest that the cyp38-2/tlp7.6-1 double mutant exhibits a similar impact on photochemical reactions and protective mechanisms as the cyp38-2 single mutant. This reinforces the hypothesis that TLP7.6 primarily contributes to PSII structural stability rather than directly influencing its photochemical efficiency.
a Chl fluorescence images of Fv/Fm for Col-0 and mutant plants, with Fv/Fm values shown in the right panel. Scale bar: 1 cm. b Quantification of Fv/Fm. Each column represents the mean with the standard deviation of three replicates. Fv/Fm reflects the maximum photochemical yield of PSII. Fm represents maximum chlorophyll fluorescence, and Fv is variable fluorescence. c Chlorophyll fluorescence and photosynthetic efficiency. Y(PSII), quantum yield of PSII. ETR(II), electron transport rate of PSII. Y(NPQ), quantum yield of non-photochemical quenching in PSII. Y(NO), quantum yield of non-regulated energy dissipation. d Efficiency and dynamics of photosystem I in plants. Y(I), quantum yield of PSI. ETR(I), electron transport rate through PSI. Y(ND), quantum yield of non-photochemical energy dissipation due to donor side limitation in PSI. Y(NA), quantum yield of on-photochemical energy dissipation due to acceptor side limitation in PSI.
In addition, we analyzed the efficiency and dynamics of Photosystem I by measuring additional parameters such as Y(I), ETR(I), Y(ND), and Y(NA). These parameters provide insights into the functional status of PSI. Similar to the results for PSII, the double mutant cyp38-2/tlp7.6-1 exhibited similar results to cyp38-2, indicating that TLP7.6 does not significantly affect the function of CYP38 in PSI (Fig. 6d).
PSII SC is compromised in the cyp38-2/tlp7.6-1 double mutants
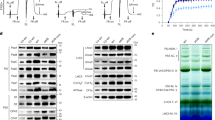
To investigate the functional interplay between TLP7.6 and CYP38 in photosystem assembly, BN-PAGE analysis was performed on thylakoid membranes isolated from Col-0, tlp7.6-1, cyp38-2, and cyp38-2/tlp7.6-1 plants under normal growth conditions (Fig. 7a). The results indicated a reduction in the PSII SC in tlp7.6-1, cyp38-2 and cyp38-2/tlp7.6-1 compared to Col-0. However, this decrease was not pronounced in the double mutant relative to the cyp38-2 single mutant. In addition, the assembly of LHCII was less pronounced in the cyp38-2/tlp7.6-1 compared to both Col-0 and cyp38-2, further supporting the idea that TLP7.6 is involved in the stabilization or proper assembly of photosystem complexes. Interestingly, we observed a significant increase in the PSII monomer (PSII-M) and cytb6f bands in the double mutant, which may indicate an attempt by the plant to compensate for the instability of the PSII supercomplex. Previous studies have shown that CYP38 deficiency leads to high light sensitivity, while the plants exhibit normal growth under low light conditions20. To further explore TLP7.6 functions, we performed BN-PAGE analysis under low light intensity (15 μmolm-2s-1), where no significant differences in PSII and PSI complex assembly were observed between the mutants and Col-0 (Fig. 7b). This suggests that under low light, the PSII supercomplex and other photosystem complexes, such as PSI, may remain stable despite the absence of TLP7.6, which aligns with the general finding that low light minimizes the demands for photosystem assembly and turnover. Silver staining of BN-PAGE gels further support for these observations. In particular, the cyp38-2/tlp7.6-1 double mutant displayed a marked reduction in PSII SC, reinforcing the idea of PSII instability in the absence of both CYP38 and TLP7.6 (Fig. 7c-e). Finally, we conducted 2D BN/PAGE analysis, which revealed a reduction in key core proteins of PSII, including D1, LHCII, and CP47 in cyp38-2/tlp7.6-1(Fig. 7f). This reduction of key PSII components further supports the hypothesis that TLP7.6 acts as an auxiliary factor in the proper assembly or stability of PSII, especially in conjunction with CYP38.
a Thylakoid membranes from plants grown under normal light conditions (60 μmolm-²s-¹) for 3 weeks. b Thylakoid membranes from plants grown under low light conditions (15 μmolm-²s-¹) for 3 weeks. a-b Thylakoid membranes (1 μg of chlorophyll) were solubilized with 1% w/v β-DM and separated by 5–13.5% acrylamide gradient gel. PSI-M, PSI monomer; PSII-D, PSII dimer; LHCII, PSII light-harvesting complex; LHCII-T, PSII light-harvesting complex trimer. c–e Native gel strips from thylakoid membrane complexes of Col-0, cyp38-2 and cyp38-2/tlp7.6-1 were subjected to 12% SDS-PAGE and stained with silver. f Thylakoid membranes solubilized by 1% w/v β-DM were separated by BN-PAGE (1 μg chlorophyll each lane). The individual lanes were placed on 12% SDS-PAGE, followed by immunoblotting with antibodies against D1, LHCII and CP47.
Given the role of CYP38 in plant high light sensitivity, we further examined the response of photosystem core proteins in mutants after short exposure to high light (650 μmolm-2s-1). The results showed that the cyp38-2/tlp7.6-1 double mutant was more sensitive to high light, showing a significant reduction in core proteins such as PsaD compared to the cyp38-2 mutant, but slightly changes under low light and normal light (Supplementary Fig. 7). These findings suggest that TLP7.6 is crucial for mitigating high-light induced damage to the photosynthetic apparatus, and that the double mutant exacerbates the instability of photosystem proteins under high light stress conditions.
Our results indicate that the cyp38-2/tlp7.6-1 double mutant exhibits compromised photosystem stability, particularly under high light stress. The significant reduction in PSII SC and core proteins in the double mutant compared to the single mutants supports the hypothesis that TLP7.6 functions as an auxiliary factor to CYP38, helping to maintain the integrity and stability of photosystem complexes under varying light conditions.
Discussion
Stunted growth and development in the double mutants
Plant growth and development are highly rely on photosynthesis to provide the energy and substance for both vegetative and reproductive stages. While the biochemical reactions involved in photosynthesis have been well-characterized, the mechanisms governing the assembly and macromolecular complexes remain unclear. Recent study has focused on identifying co-factors that assist in the assembly and functionality of these complexes. CYP38 is known to play a key role in assembly of PSII, but its mechanism is not well understood. To address this, we have identified several proteins that interact with CYP38 through a thylakoid lumen mini-library screening, revealing nine soluble thylakoid lumen proteins and six membrane proteins whose lumen-exposed domains interact with CYP3828. Further pull-down and Co-IP assays confirmed direct interactions between CYP38 and several of these proteins, both in vitro and in vivo28.
Among the identified interacting proteins, TLP7.6 (At1g21500) does not appear to have a significant impact on plant growth under normal growth conditions, but does show minor reductions in PSII SC and LHCII assembly, as indicated by BN-PAGE results. This suggests that TLP7.6 may contribute to the stability and function of PSII. Additionally, cyp38-2/tlp7.6-1 exhibits severely impaired growth and developmental delays (Figs. 4–6). Therefore, TLP7.6 may be one of several auxiliary factors that supports the proper assembly and stability of PSII.
Potential mechanisms of CYP38 and TLP7.6 interaction
CYP38 contains an N-terminal α-helical domain and a C-terminal cyclophilin domain, which are closely packed to form a strong intramolecular interaction25. The N-terminal domain has a low affinity for interacting with other proteins, as evidenced by yeast two hybrid (Y2H) screenings, whereas the C-terminal cyclophilin domain is crucial for its interaction with target proteins, including PSII subunits29. Based on previous studies, it is proposed that CYP38 function as a chaperone protein, binding to and stabilizing PSII subunits during their assembly. It is hypothesized that CYP38 exists in a packed state in the thylakoid lumen, becoming activated by light-induced proton flux, which promotes its interaction with PSII subunits. After assisting with the assembly of PSII, CYP38 likely returns to its inactive state32.
Since TLP7.6 interacts with the C-terminal of CYP38, we hypothesize that the combination of TLP7.6 and CYP38 assists in the correct assembly of different forms of the PSII complex. This interaction likely facilitates the stabilization and maintenance of PSII SC. However, our BN-PAGE data show that in the absence of both proteins, the levels of PSII SC are drastically reduced, supporting the hypothesis that TLP7.6 functions as an auxiliary protein in the PSII assembly. It is also possible that TLP7.6’s role is partly redundant with other lumenal proteins that collaborate CYP38 to assemble PSII. The double mutant shows chloroplasts with abnormal architecture, as evidenced by transmission electron microscopy (Fig. 5). These physiological and structural impairments likely contribute to the observed growth retardation in the double mutants, as plants are unable to efficiently store energy or protect themselves from light-induced stress.
To further investigate the role of TLP7.6 in photosynthesis, we performed Y2H assay to access whether TLP7.6 interacts with core subunits of PSII. As we know, the core subunits of PSII, D1 (PsbA), D2 (PsbD), CP43 (PsbC) and CP47 (PsbB), are membrane proteins. Using yeast vectors with the lumenal regions of these proteins, we found that TLP7.6 do not interact with these core PSII subunits, suggesting that TLP7.6 interacts specifically with CYP38 rather than directly with the PSII core components (Supplementary Fig. 8).
In conclusion, our study highlights the critical role of both CYP38 and TLP7.6 in photosynthesis and plant growth. While TLP7.6 is not essential for normal plant development under standard conditions, its absence in the cyp38-2 mutant severely disrupts PSII SC assembly and function, impairing photosynthesis and resulting in slowed growth, suggesting that TLP7.6 may play the less important role in plant growth and development process, but may play a role in stabilizing PSII supercomplex assembly rather than directly affecting its photochemical efficiency. These findings provide valuable insights into the cooperative functions of luminal proteins in thylakoid and underscore the complexity of the molecular machinery involved in photosystem assembly. Our findings lay the foundation for further studies on CYP38 and enhance our understanding of the functions of thylakoid lumen proteins. Future research will focus on how this auxiliary protein assists CYP38, as well as exploring other potential auxiliary proteins involved in PSII assembly. Understanding the full spectrum of proteins that interact with CYP38 and their respective roles in photosynthesis will be critical for uncovering the detailed mechanisms behind PSII complex formation and its regulation in response to environmental conditions.
Methods
Plant Materials and Growth Conditions
Seeds of the wild-type and cyp38-2 mutant, both in Columbia-0 (Col-0) ecotype, were obtained from Syngenta (Research Triangle Park, NC). The tlp7.6-1 and tlp7.6-2 single mutants were generated using the CRISPR/Cas9 gene editing system. The cyp38-2 mutant line was then crossed with tlp7.6-1 to produce the cyp38-2/tlp7.6-1 double mutant. For soil-grown plants, seeds were vernalized for 3 days and then grown at 23 °C under continuous light at 60 μmolm-2s-1. For plate-grown plants, seeds were surface-sterilized with 70% ethanol for 5 min, followed by 50% bleach supplemented with 0.05% Tween 20 for 10 min, and then washed four times with sterilized water. The sterilized seeds were plated on 0.8% agar plates containing 1/2 MS medium with or without 1% sucrose under long-day conditions (16-h of 60 μmolm-2s-1 and 8-h dark cycle).
Identification of Mutants and Complementation with Transgenic Plants
Two TLP7.6 single-mutant plants were produced using the CRISPR/Cas9 system and were named as tlp7.6-1 and tlp7.6-2. Homozygous transgenic lines were confirmed using the NcoI restriction enzyme. The Col-0 digested by NcoI results in two bands, while tlp7.6-1 mutant failed to digest by NcoI due to absence of an A base in the genome (all the primer sequences used in this study are presented in Supplementary Table 2). qRT-PCR was utilized to examine the expression levels of tlp7.6.
For reverse transcription-PCR analysis, total RNA was isolated from 3-week-old leaves using a total RNA extraction kit (RNAprep Pure Tissue Kit, TIANGEN). qRT-PCR reactions were performed with the SuperScript III First-Strand Synthesis System (TransGen Biotech) using gene-specific primers. For complementing the tlp7.6-1 mutant, a 908-bp genomic DNA fragment containing the At1g21500 gene, along with 554-bp upstream and 545-bp downstream sequences, was amplified by PCR. The fragment was cloned behind the native promoter in the binary vector pCambia3300. The construct was further introduced into the tlp7.6-1 mutant using the floral dip method.
Co-immunoprecipitation assay
The Co-IP assay was conducted with slight modifications33. Briefly, intact chloroplasts from Col-0 or 35Spro:CYP38-His/cyp38-2 mutant were carefully collected and suspended in SH buffer (0.33 M sorbitol, 50 mM HEPES, pH 8.0). The suspension was centrifuged at 2600×g for 10 min, and then resuspended with lysis buffer (10 mM HEPES, pH 8.0) for 10 min on ice. After centrifugation at 10,000 rpm for 10 min, the pellet was resuspended in SH buffer with 0.1% (w/v) n-dodecyl-β-D-maltoside (β-DM). The thylakoid lumen was recovered by centrifugation at 12,000 rpm for 1 h at 4 °C and incubated with the Ni-NTA agarose separately at 4 °C overnight under gentle rotation. After washing with pre-cold PBS (phosphate-buffered saline, PH 7.4) buffer 5 times, the beads were boiled in 2×SDS loading buffer at 98 °C for 10 min, these samples were then fractionated on 12% SDS-PAGE and analyzed by immunoblotting with specific antibodies.
For TLP7.6 antibody preparation and purification, the GST-TLP7.6 recombinant protein was expressed and purified from E.coli BL21 transformed with the construct of pGEX-TLP7.6, in which mature DNA (without signal peptide) fragment was cloned into expression vector. The antibody was generated by immunizing a rabbit with TLP7.6 recombinant protein as antigen, and then purified from serum. Other antibodies used in this study are listed in Supplementary Table 3.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence parameters were measured on the leaves of three-week-old seedlings grown under continuous illumination (60 μmolm-2s-1) using a Dual Pulse-Amplitude Modulation100 (Dual-PAM) chlorophyll fluorometry (Walz Effeltrich, Germany). The chlorophyll fluorescence parameters were calculated as follows: Fv/Fm was defined as (Fm -Fo)/Fm, and Y(PSII) was defined as (F’m-Fs)/F’m. NPQ was calculated as (Fm-F’m)/F’m. The terms Y(PSII), ETR(II), Y(NPQ), and Y(NO) are key parameters used in the study of chlorophyll fluorescence and photosynthetic efficiency in plants. The terms Y(I), ETR(I), Y(ND), and Y(NA) are parameters used for evaluating the efficiency and dynamics of Photosystem I (PSI).
Subcellular localization
The full-length coding sequence of TLP7.6 was cloned into the binary expression vector pMDC32-GFP, creating the pMDC32-TLP7.6-GFP construct. This construct was introduced into Agrobacterium tumefaciens strain GV3101, which was further used to transiently transform Nicotiana benthamiana leaves via infiltration buffer. GFP fluorescence was observed using an Olympus FV-1000 confocal laser-scanning microscopy. GFP was excited at 488 nm and detected using a 510 nm emission filter. Chlorophyll autofluorescence was excited at 543 nm and collected with a 633 nm long-pass emission filter.
Chloroplast ultrastructure
Leaves from 21-day-old Col-0 and mutant seedlings were analyzed using transmission electron microscopy. Briefly, leaf samples were fixed in 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 24 h at 40 °C, followed by overnight fixation in 1% OsO4 at 4 °C. The tissues were then dehydrated in a gradient of ethanol solutions and embedded in resin. Ultrathin (80–100 nm) sections were prepared using a diamond knife on a Reichert OM2 ultramicrotome, stained with 2% (v/v) uranyl acetate (pH 5.0), and subsequently with 10 mM lead citrate (pH 12), and then examined with a transmission electron microscope (Hitachi, Tokyo, Japan).
BN-PAGE and 2D SDS-PAGE
For BN-PAGE, thylakoid membranes of all lines were resuspended in 25BTH20G buffer (50 mM BisTris, pH 7.0, and 40% glycerol) to a final concentration of 1 mg/mL chlorophyll. The membrances were solubilized with 1% (w/v final concentration) β-DM on ice for 10 min. After centrifugation at 13,000 rpm for 10 min, the supernatant was mixed with BN sample buffer (100 mM BisTris-HCl, pH 7.0, 5% Serva Blue G, 0.5 M 6-amino-n-caproic acid, 30% (w/v) sucrose), and separated by electrophoresis on a 5–13.5% polyacrylamide gradient gel. After electrophoresis, the gel strips were cut and incubated in 2×SDS sample buffer for 30 min. Proteins were separated by SDS-PAGE, following electrophoresis, the gels were subjected to immunoblot analysis34,35,36.
Silver staining
To perform silver staining, the gel was fixed in the destaining buffer and gentle agitation overnight, washed three times for 20 min, and then incubated in formaldehyde fixing solution with gentle agitating for 30 min. The gel was rinsed twice with nonionized water slowly for 15 min. The gel was rinsed by incubate 0.1% silver nitrate solution for 30 min in the dark. After staining, the gel was rinsed twice with non-ionic water for 30 s each. The gel was developed in a thiosulfate solution, shaken gently until the protein bands become visible, and then the development was stopped by immersing the gel in 10 mL citric acid solution, shaking for over 1 min until the reaction was completed.
Yeast two hybrid
The sequence encoding the mature TLP7.6 protein was cloned into pGBKT7 vector to construct the bait vector. The lumenal regions of D1 (PsbA), D2 (PsbD), CP43 (PsbC) and CP47 (PsbB) were separately inserted into pGADT7 vector as prey, which were maintained in our laboratory. These vectors were then separately transformed into yeast strain AH109 and co-cultured on the SD medium lacking Leu and Trp. Positive clones were identified by the ability to grow on SD/-Trp-Leu-His or SD/-Trp-Leu-His-Ade medium for evaluation of the transcriptional activation activity37.
Starch content
Starch content was determined by first extracting soluble sugars with 80% ethanol. The starch was then hydrolyzed with acid to break it down into glucose. The glucose was quantified using the anthrone colorimetric method, and starch content was calculated based on glucose concentration (Starch content assay kit, Solarbio).
Statistics and reproducibility
All statistical analyses were performed using GraphPad Prism software 8.0 (GraphPad Software, USA). Comparisons between two groups were conducted using the Student’s t-test, with a significance threshold of p < 0.05. All analysis presented in this study were based on a minimum samples size of three replications. Root length and thylakoid stack heights were measured using ImageJ software (NIH, Maryland, USA). Each experiment was repeated at least three times to ensure reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data used in this study is available as described in the methods section and include in the article and its supplementary materials. The uncropped blot images for figures are available in the Supplementary Information file. Sequencing data details are included in Supplementary Data 1. The numerical source data for graphs are available in Supplementary Data 2. All other relevant data are available from the corresponding author upon reasonable request.
References
Nelson, N. & Ben-Shem, A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982 (2004).
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 (2004).
Kieselbach, T., Hagman, A., Andersson, B. & Schröder, W. P. The thylakoid lumen of chloroplasts Isolation and characterization. J. Biol. Chem. 273, 6710–6716 (1998).
Kieselbach, T. & Schröder, W. P. The proteome of the chloroplast lumen of higher plants. Photosynth Res 78, 249–264 (2003).
Järvi, S., Gollan, P. J. & Aro, E. M. Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front Plant Sci. 4, 434 (2013).
Schubert, M. et al. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–8365 (2002).
Peltier, J. B. et al. Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12, 319–341 (2000).
Peltier, J. B. et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236 (2002).
Goulas, E. et al. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 47, 720–734 (2006).
Fristedt, R. et al. PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol. 167, 481–492 (2015).
Gollan, P. J. et al. Characterization of the free and membrane-associated fractions of the thylakoid lumen proteome in Arabidopsis thaliana. Int J. Mol. Sci. 22, 8126 (2021).
Kapri-Pardes, E., Naveh, L. & Adam, Z. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19, 1039–1047 (2007).
Sun, X. et al. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19, 1347–1361 (2007).
Wei, L. et al. LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J. Biol. Chem. 285, 21391–21398 (2010).
Järvi, S., Suorsa, M. & Aro, E. M. Photosystem II repair in plant chloroplasts-Regulation assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys. Acta 1847, 900–909 (2015).
Sirpiö, S. et al. AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55, 639–651 (2008).
Chen, H. et al. A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61, 567–575 (2006).
Sirpiö, S. et al. TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem J. 406, 415–425 (2007).
Wu, H. Y., Liu, M. S., Lin, T. P. & Cheng, Y. S. Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen. Plant Physiol. 157, 1015–1025 (2011).
Fu, A. G. et al. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. Usa. 104, 15947–15952 (2007).
Komenda, J. et al. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 22390–22399 (2008).
Plöchinger, M., Schwenkert, S., Sydow von, L., Schröder, W. P. & Meurer, J. Functional update of the auxiliary proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in maintenance and assembly of PSII. Front Plant Sci. 7, 423 (2016).
Handschumacher, R. E., Harding, M. W., Rice, J., Drugge, R. J. & Speicher, D. W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226, 544–547 (1984).
Romano, M. F. et al. Rapamycin inhibits doxorubicin-induced NF-κB/Rel nuclear activity and enhances the apoptosis of melanoma cells. Eur. J. Cancer 40, 2829–2836 (2004).
Vasudevan, D., Fu, A., Luan, S. & Swaminathan, K. Crystal structure of Arabidopsis cyclophilin38 reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism. Plant Cell 24, 2666–2674 (2012).
Vojta, L. et al. Complex lumenal immunophilin AtCYP38 influences thylakoid remodelling in Arabidopsis thaliana. J. Plant Physiol. 243, 153048 (2019).
Rokka, A., Aro, E. M., Herrmann, R. G., Andersson, B. & Vener, A. V. Dephosphorylation of photosystem II reaction center proteins in plant photosynthetic membranes as an immediate response to abrupt elevation of temperature. Plant Physiol. 123, 1525–1536 (2000).
Hao, Y. et al. Identification of interacting proteins of Arabidopsis cyclophilin38 (AtCYP38) via multiple screening approaches reveals its possible broad functions in chloroplasts. J. Plant Physiol. 264, 153487 (2021).
Perez-Torrado, R., Yamada, D. & Defossez, P. A. Born to bind: the BTB protein-protein interaction domain. Bioessays 28, 1194–1202 (2006).
Duan, L., Pérez-Ruiz, J. M., Cejudo, F. J. & Dinneny, J. R. Characterization of CYCLOPHILLIN38 shows that a photosynthesis-derived systemic signal controls lateral root emergence. Plant Physiol. 185, 503–518 (2021).
Baker, N. R. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev. Plant Biol. 59, 89–113 (2008).
Shi, L. et al. Conserved Residues in the C-Terminal Domain Affect the Structure and Function of CYP38 in Arabidopsis. Front Plant Sci. 12, 630644 (2021).
Armbruster, U. et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25, 2661–2678 (2013).
Asakura, Y. et al. Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16, 201–214 (2004).
Heinemeyer, J., Eubel, H., Wehmhöner, D., Jänsch, L. & Braun, H. P. Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65, 1683–1692 (2004).
Lima, A. et al. A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. Usa. 103, 12631–12636 (2006).
Fields, S. & Song, O. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China to Y.H (NO.31300204) and to A.F. (NO.32070269), Research fund from Shaanxi Provincial Science and Technology Department to Y.H (NO.2023-JC-YB-153) and the Shaanxi Fundamental Science Research Project for Chemistry & Biology to Y.H (NO.22JHQ062).
Author information
Authors and Affiliations
Contributions
Y.H. and A.F. designed research, P.R., R.L., K.C., C.Z., Z.L., T.W., C.M., B.L., X.W., F.S., T.Z., Y.X., X.H., and H.L. performed research and analyzed data. Y.H., A.F., W.Y. and P.R. wrote and revised the paper. All authors contributed to the article and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Xin Hou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Xiaoling Xu and David Favero.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ren, P., Li, R., Chen, K. et al. Characterization of Arabidopsis thaliana thylakoid lumen 7.6 protein functions in photosystem II assembly. Commun Biol 8, 490 (2025). https://doi.org/10.1038/s42003-025-07907-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07907-1