Abstract
The accurate processing of neonatal and infant brain MRI data is crucial for developmental neuroscience but presents unique challenges that child and adult data do not. Tissue segmentation and image coregistration accuracy can be improved by optimizing template images and related segmentation procedures. Here, we describe the construction of the FinnBrain Neonate (FBN-125) template, a multi-contrast template with T1- and T2-weighted, as well as diffusion tensor imaging-derived fractional anisotropy and mean diffusivity images. The template is symmetric, aligned to the Talairach-like MNI-152 template, and has high spatial resolution (0.5 mm³). Additionally, we provide atlas labels, constructed from manual segmentations, for cortical grey matter, white matter, cerebrospinal fluid, brainstem, cerebellum as well as the bilateral hippocampi, amygdalae, caudate nuclei, putamina, globi pallidi, and thalami. This multi-contrast template and labelled atlases aim to advance developmental neuroscience by achieving reliable means for spatial normalization and measures of neonate brain structure via automated computational methods. We also provide standard volumetric and surface co-registration files to enable investigators to transform their statistical maps to the adult MNI space, improving the consistency and comparability of neonatal studies or the use of adult MNI space atlases in neonatal neuroimaging.
Similar content being viewed by others
Introduction
Neonatal and infant brain segmentation remains one of the biggest challenges for neuroscientists. Although multiple segmentation procedures have been developed, used, validated and published as openly available software1,2,3, it may be challenging to map and choose the best available tools that are likely to work across datasets. This is in stark contrast to operating with adult MRI data where already validated software is available. Neonatal MRI images have inconsistent tissue contrast that stems from the initial near absence of myelin-related contrast and its uneven pattern of development during the first year of life. In neonates, in areas with little to no myelin, the white matter is darker than the grey matter on T1-weighted images and lighter than the grey matter on T2-weighted images. Visually, the neonatal brain has roughly the reverse of the adult contrast in structural MR images. But, in areas showing early myelination, the two tissue classes can be almost indistinguishable. Important advancements in the field have been made by introducing high-quality templates and accurate anatomical labels to guide final segmentations that can then aid current and future segmentation algorithms4,5.
Currently, available neonatal and infant atlases are comprehensively introduced in recent review articles1,4,6. One of the reviews also aptly suggests that there may not be a one-size-fits-all atlas for neonates. Crucially, the neonatal period and early infancy are dynamic phases of brain development, and investigators likely benefit from having multiple available atlases4,5 and ultimately robust procedures across different stages of brain development (i.e. 4D templates and atlases across several ages). In addition to contributing to the available neonatal atlases, our work is especially motivated by a recent review pointing out that there is a lack of standard template spaces in neonatal/infant neuroimaging studies and that correspondence to adult MNI space would be helpful in supporting comparisons to adult studies, performing meta-analyses, and assuring reproducibility6. Finally, our review of available neonate atlases indicates that there is paucity of multimodal templates that include templates based on both structural and diffusion MRI for healthy infants (0–3 months of age) (Table 1 and Supplementary Note 1), and while our multimodal templates are not uncharted, we aimed to fill this gap with a set of templates that is based on a larger sample than used in prior studies. In summary, there are clear indications that age-appropriate templates and variability in available templates for specific age groups and MRI modalities to fit the needs of different studies are needed. The provision of standard registration files between neonatal and adult MNI space is a key need of the field6, and is one of the key novelties of the current article.
Our main objective was to create a new multi-contrast template for the neonate brain, comprised of T1- and T2-weighted data, as well as diffusion tensor imaging (DTI) data in terms of fractional anisotropy (FA) and mean diffusivity (MD) data. We also created accompanying neonatal brain atlases with the majority vote technique using manually defined labels of 21 subtemplates7. The atlases were created with: (1) gross tissue labels for grey matter, white matter, and cerebrospinal fluid (CSF); (2) symmetric labels for grey matter, white matter, cerebrospinal fluid (CSF), brainstem and cerebellum, as well as labels of the bilateral amygdala, hippocampus, caudate, putamen, globus pallidus and thalamus; and (3) corresponding asymmetric labels for the left and right hemisphere with FreeSurfer lookup table labels (the templates and labels themselves are symmetric). We also included standard surface transforms that are based on FreeSurfer processing and a volumetric comparison between our template created with joint label fusion and segmentations from the FreeSurfer-based synthetic T1 pipeline. Finally, we provide standard coregistration files to enable standard transforms to adult MNI coordinates for existing and future studies.
Methods
The study was conducted according to the Declaration of Helsinki and was reviewed and approved by the Ethics Committee of the Hospital District of Southwest Finland (ETMK:31/180/2011).
MRI acquisition
The participants underwent an MRI scan solely for research purposes and without clinical indications. The scanning was performed at the Medical Imaging Centre of the Hospital District of Southwest Finland by an experienced radiographer, without anaesthesia, during natural sleep using the “feed and swaddle” procedure8. We used a Siemens Magnetom Verio 3 T scanner (Siemens Medical Solutions, Erlangen, Germany). The 60-minute protocol included a PD-T2-TSE (dual-echo turbo spin echo) sequence with a repetition time (TR) of 12,070 ms and effective echo times (TE) of 13 and 102 ms (PD-weighted and T2-weighted images, respectively), and a sagittal 3D T1-weighted MPRAGE sequence with a TR of 1900 ms, a TE of 3.26 ms and an inversion time (TI) of 900 ms. The total number of slices was 128, the resolution was 1.0 mm3 isotropic for both the T1- and T2-weighted images, and the images covered the whole brain. Sequence parameters were optimised so that the 'whisper' gradient mode could be used in the PD-T2-TSE and 3D T1-sequences to reduce acoustic noise during the scan. Single shell diffusion-weighted data was acquired with a standard twice-refocused spin echo-echo planar imaging (SE-EPI) sequence (field of view (FOV) 208 mm; 64 slices; TR 9300 ms; TE 87 ms), with 2 mm3 isotropic resolution and a b-value of 1000 s/mm. There were, in total, 96 unique diffusion encoding directions in a three-part DTI sequence. Each part consisted of uniformly distributed 31, 32 or 33 directions and three b0 images (images without diffusion encoding) that were taken in the beginning, in the middle, and at the end of each scan9,10.
All the brain images were assessed by a paediatric neuroradiologist for any incidental findings9. Developmental status has thereafter been normal for all the participants, including those with incidental findings. The incidental findings were deemed not to affect the brain anatomy/volume estimates of the participants in the current study. It is important to note that the encountered incidental findings have been found to be common and clinically insignificant in previous studies; see our recent article for more details11. More detailed information about the scanning visits and tips for investigators can be found in our review12.
Template creation
Creation of population-specific FBN-125 structural templates
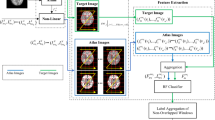
The images that were not suitable for data analysis (excessive number of artefacts) were excluded, leaving 125/180 successful structural MRI for template creation (69.4% success rate), which is comparably low and was due to technical issues with scanning that went unnoticed during data collection12. The MRIs that passed this quality control were used to construct a population-specific dual-contrast template (Fig. 1A). The procedures are based on prior work by ref. 13 and have been successfully applied to create neonatal templates14 (https://nist.mni.mcgill.ca/infant-atlases-0-4-5-years/). The T1 template was first created from T1-weighted images and linearly registered to the MNI-152 template13. The average scaling from the native MRIs to the MNI-152 template was then computed, and the inverse was used to scale the MNI-152 template to the average size of our neonate population, which served as an initial target for construction of the population-specific template. The T2-weighted images were linearly registered to the T1 and subsequently to the neonate template space with the transforms estimated from T1 scans. The template construction procedure is described in a prior article by ref. 13 and is based on the work of ref. 15; the method employs the principles of average model construction using elastic body deformations from ref. 16. It is an iterative procedure that, given a set of MRI volumes, builds a template that minimises the mean squared intensity difference between the template and each subject’s MRI and minimises the magnitude of all deformations used to map the template to each subject’s MRI.
A Iterative construction of the infant template as described in Fonov et al. (2011). B Labelling the infant template. The data were registered to the infant template and then clustered based on the amount of distortion required to do that into 21 clusters representing the morphological variability in the population. The template was then warped to the centralmost subject of each cluster, providing 21 subtemplates for manual segmentation. After manual segmentation, the labels were then unwarped back to the base infant template and merged via voxel-wise majority vote to create the consensus labels. This figure is modified from Acosta et al., Cerebral Cortex, 2020 | reprinted with permission.
Creation of 21 subtemplates for manual segmentation
The non-linear transformations derived in the construction of the template were then used to cluster the subjects into 21 clusters, from which we used the centre-most subject as the basis to construct 21 targets for manual segmentation (Fig. 1B). As the basis for clustering, the Jacobian was computed for the non-linear transform mapping each subject to the template. The values in the Jacobian were extracted as a vector for each voxel within the template brain mask and clustered using an equal combination of cosine similarity and Euclidean distance with Ward’s clustering method17. We chose the number of clusters to be 21, which provided a good balance between reliable analysis procedures and the amount of work needed for manual labelling.
Then, within each of the 21 clusters, the sum-squared distance from each subject to every other subject was computed, and the subject with the minimum sum-squared distance was taken as the centralmost subject of the cluster and used as the basis to create a subtemplate. The dual-contrast template constructed in the previous step was then warped to overlay the MRIs of these 21 subjects. These 21 subtemplates were then provided for manual segmentation without those doing that segmentation being made aware that these were, in fact, 21 versions or warped copies of the template. The demographics of the neonates whose brain images were determined to be one of the 21 cluster centroids are provided in Table 2.
Creation of FBN-125 DTI templates
Good-quality b0 images were chosen manually, coregistered, averaged, and moved in front of each 4D series. Brain masks were created based on the b0 volumes with the Brain Extraction Tool18 (BET) from FSL19 (FMRIB Software Library v 5.0.9). DTIPrep software20 was used to inspect the quality of the data. Low-quality diffusion images identified by DTIprep were discarded. The remaining images were then visually inspected following the automated quality control of DTIprep, and more directions were excluded as needed. We have found that after the quality control steps, datasets that have more than 20 diffusion encoding directions will yield reliable tensor estimates9,10. Here all infants with at least 20 diffusion encoding directions were selected, and we used all available participant’s data thereafter (N = 122). Eddy current and motion correction steps were conducted with FSL21, and the b-vector matrix was rotated accordingly. A diffusion tensor model was fitted to each voxel included in the brain mask using the DTIFIT tool in FDT (FMRIB’s Diffusion Toolbox) of FSL using ordinary least squares (OLS) fit. Our DTI preprocessing steps have been provided in detail in our previous publications that also report good test-retest repeatability in between segments of the multi-part DTI sequences9,10. The DTI template creation was carried out by rigidly registering the b0 images to the nonuniformity-corrected T1-weighted data and combining the transformations from b0-to-T1 and the T1-to- The FinnBrain Neonatal (FBN-125) template space for FA and MD maps7,22. The registrations were carried out with ‘antsRegistration’. The FA and MD template images were then created by averaging the images with FSL’s fslmaths, part of FMRIB Software Library v6.019.
Manual segmentation
The manual segmentation procedures and tools
Manual neonate brain segmentation is extremely labour-intensive and requires considerable knowledge of the developmental characteristics of the various tissues. Full manual segmentation of the brain, including cortical and subcortical grey matter, white matter, and the CSF slice-by-slice, is very time-consuming. In our experience, working at 1 mm3 resolution, this task takes around 1 month of full-time work. A higher 0.5 \(\times\) 0.5 \(\times\) 0.5 resolution of our subtemplates would have made this process even more labour-intensive, and we thus employed a model where we start the work from initial estimates for the gross tissue segmentation as outlined below. We were able to divide the work among research assistants whom we quickly and successfully trained to perform the manual segmentations.
Another key thing that affected the workflow was the good initial tissue contrast in the created 21 subtemplates (due to averaging). Namely, the brain structures and their boundaries against neighbouring structures are relatively easy to detect. Manual segmentation is always prone to inter-rater and even intra-rater discrepancies, which may affect the statistical power of studies and lead to inaccurate estimates of outcome metrics. Here, the use of 21 subtemplates to delineate the final segmentation on the FBN-125 template alleviated the final effect of minor errors and variability that stem from using multiple raters and additionally allowed us to quantify the quality of segmentations.
We used teams of junior raters, supervised by senior investigators, to accomplish the work. For the subcortical grey matter nuclei, hippocampus and amygdala, we started with one template jointly segmented for all subcortical structures by the primary rater NH and senior rater JJT (externally reviewed by JDL). The final subcortical segmentations of the 21 subtemplates were performed by three research assistants, supported by author NH on a regular basis, and all working under the supervision of JJT. The final labels on the 21 subtemplates were critically reviewed and corrected by JJT for consistency and externally reviewed by JDL. The final labels are thus a consensus between two senior raters. The manual segmentation of the amygdala, hippocampus, and subcortical grey matter nuclei was done with Display software, part of the MINC Tool Kit (https://bic-mni.github.io/).
For the cortical and gross anatomy segmentations, we made prior estimates of the structures that we manually corrected. The segmentations were performed by three research assistants. To aid the work, JJT prepared a detailed manual and video material showing model edits on each step. JJT also performed weekly quality controls to check all the segmentations as well as a final check on all the images. As before, the images were externally reviewed by JDL. The gross anatomical segmentations were done with FSL tools and manual segmentation with FSLeyes23.
A detailed description of the manual segmentation is provided in the supplementary information (Supplementary Notes 2, 3). Briefly, the segmentation steps included bilateral segmentation of the hippocampus and the amygdala24, the caudate nucleus25, the putamen (mainly in the coronal plane) and the globus pallidus was segmented after the putamen starting from the posterior border and moving in circular tracings in all planes, and the thalamus segmentation that was guided by prior work26. To decrease the time needed for manual segmentations of the cortical grey matter, white matter, and CSF, initial estimates for the 21 subtemplates were created with the FSL-VBM pipeline using the UNC neonate template grey matter probability mask to guide the segmentation27. These initial estimates were then manually corrected to yield binary labels for cortex, white matter, internal and external CSF (the labels were later combined), brainstem, cerebellum, and a 'deep grey' segmentation that intentionally covered the subcortical grey matter and the myelinated portions of white matter surrounding the nuclei. The previously created subcortical areas were subtracted from this label, and the remaining voxels were added to the binary white matter mask.
The creation of atlas labels from manual segmentations
After manual segmentation, the labels were unwarped back to the FBN-125 space and merged via voxel-wise majority vote to create the consensus labels, and the labels were assumed to be symmetric and complete through visual inspection.
Finally, we used the symmetric labels to create several atlases: (1) gross tissue labels for grey matter, white matter, and CSF; (2) symmetric labels for grey matter, white matter, CSF, brainstem and cerebellum, as well as labels of the bilateral amygdala, hippocampus, caudate, putamen, globus pallidus and thalamus; and (3) corresponding asymmetric labels for left and right hemispheres with FreeSurfer lookup table labels. For the creation of the asymmetric labels, we defined a right hemispheric binary mask to aid the separation of the hemispheres. The labels were created from symmetric labels with ‘fslmaths’ from the FMRIB Software Library v6.019. The FreeSurfer labels were obtained from: https://surfer.nmr.mgh.harvard.edu/fswiki/LabelsClutsAnnotationFiles.
We calculated the generalised conformity index (GCI) for all structures to quantify the agreement across the atlas labels. Here, the GCI quantified the spatial overlap among the manually defined atlas labels. GCI is a generalisation of the Jaccard score so that for two raters, the GCI equals the Jaccard score: GCI = (A1 ∩ )/Vol(A1∪). We quantified the GCI across the 21 manual segmentations by including segmentation j, its volume Vol(Aj), and Σ pairs (i > j) the summation over all combinations of unique pairs of labels, and defined GCI as:
We first binarized each manually created label and then added all unique pairs of these binarized labels so that all voxels with a value >0 as their union and all voxels with a value of 2 as their intersection. We then used ANTs ‘LabelGeometryMeasures’ to calculate the size of both the union and intersection and used those values in the formula for GCI28,29. Since the FBN-125 template is symmetric, we reported an average of bilateral labels.
We also ensured that the template remained close to the average neonate brain size throughout the creation process, as the template generation procedure that we used has a special 'de-drifting' step at each iteration, so that it is guaranteed to generate an unbiased template. The intracranial volume, total brain volume, total white matter volume, total grey volume, and total cortical volume were calculated from the template and shown to be consistent with previous literature8,30.
Benchmarking transfer of statistical maps of functional MRI activations from neonatal to adult MNI space
We created standard coregistration files from the FBN-125 neonate template to the adult MNI space and made them freely available with the templates and atlases. For these transforms, we estimated a transform from the adult MNI space template to the FBN-125 neonate template to prevent the effects of the minor differences in cortical anatomy on the final transforms. We used ‘antsRegistrationSyNQuick.sh’ and ‘antsApplyTransforms’ available from ANTs software for all coregistrations31,32.
The templates used for spatial normalisation in neonatal studies vary from using an MNI template33 or standard Talairach space34 for adults and for infants a study-specific template35 or off-the-shelf atlas33,36, such as the UNC-infant template33,37. We chose the UNC-0-1–2-year neonate template as the model template for coregistrations as it was identified as the most used off-the-shelf atlas used for infants6.
To test the utility of transferring statistical maps obtained in neonatal functional MRI (fMRI) from neonatal template space to adult MNI space, we used results from our recent fMRI study38. We first estimated transforms from UNC neonate template to FBN-125 template space. We then transformed statistical maps to adult MNI space by concatenating the warps from UNC–to FBN-125–to MNI template space.
Surface-based approach
We created the surface files separately from the volumetric image processing that was based on multi-atlas segmentation using manually defined labels. Our aim was to use existing tools to create standard transforms to volumetric MNI space and the Human Connectome Project surface spaces. We first ran the recon-all-clinical.sh pipeline39 to the T2-weighted FBN-125 template. This yielded a synthetic T1-weighted image of our template that has the white matter contrast normalised to 110. The synthetic T1-weighted image was used as an input to recon-all the pipeline of FreeSurfer 6.3 (the older version was used to assure compatibility with the next step). Finally, the outputs were used as input to ciftify that creates multiple standard space surface files and transforms40.
Results
High-resolution multimodal neonatal brain templates for structural and diffusion MRI and accompanying atlases
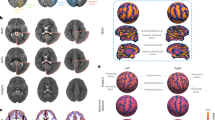
FBN-125 templates entail a set of multi-contrast template volumes of T1- and T2- weighted templates (Fig. 2A, B), corresponding DTI tensor templates of FA and MD average maps (Fig. 2C, D), and accompanying atlases with gross anatomical (Fig. 3A), symmetric (Fig. 3B) and asymmetric labels (Fig. 3C).
The FinnBrain Neonate FBN-125 templates for A T2-weighted, B T1-weighted, DTI-derived (C) fractional anisotropy, and D mean diffusivity. The grey colour scales depict intensity for (A, B) and DTI tensor scalar values for C (unitless) and D (mm2/s). Each axial slice has been tagged with a z coordinate of the adult MNI template space (in MRIcroGL software).
The anatomical labels for FBN-125 templates include A gross anatomical labels of grey and white matter as well as CSF, B asymmetric labels in FreeSurfer lookup table (LUT) compliant form, and C symmetric labels. Note the clear anatomical definition of the subcortical nuclei, especially the thalamus. Colour scales depict anatomical label numbers. Each axial slice has been tagged with a z coordinate of the adult MNI template space (in MRIcroGL software).
Consistency of manual labels used to create the atlases
The agreement of the manual segmentations for the 21 subtemplates was good for all structures: GCI ranged between 0.71 and 0.86 (Table 3). GCI scores of 0.7–1.0 are regarded as excellent28,29.
Novel means to transform neonatal functional MRI results to adult MNI standard space
We estimated standard transforms from the FBN-125 template to the adult MNI template space. We then transformed statistical maps obtained in our prior study reporting brain activations to social touch in neonates38 to adult MNI space. The registrations were accurate (Fig. 4) and worked equally well for unthresholded T maps (Supplementary Fig. 1).
Main effect of brushing vs. rest conditions in neonates A in the UNC neonate atlas template space as in Mariani Wigley et al., 2023, and B in the adult MNI space (mni_icbm152_t1_tal_nlin_sym_09a) after transforms to FBN-125 neonate template space and using the standard transforms from FBN-125 neonate to adult MNI space. The color bars visualise T values from thresholded cluster p < 0.005, FDR corrected for multiple comparisons (N = 18); see Mariani Wigley et al., 2023 for more information. Adult MNI space z coordinates appear on top of each axial slice. Note the different choice of coordinates in UNC/MNI template spaces to visualise the same regions of interest from the contrast in both images.
Surface-based Approach
We applied a surface-based approach, a FreeSurfer-based processing utilising the recon-all-clinical, recon-all and ciftify pipelines to our averaged T2-weighted image (Fig. 5). The standard surface transforms are potentially useful in surface-based applications. The outputs can be used in additional analyses by projection of statistical result maps, regions of interest, or any other pattern available in the standard HCP surface space to the FBN-125 surface space or aligning individual surface files to the FBN-125 surface space and from there to the HCP standard surface space. It is worth noting that the volumetric segmentations have variable agreement with JFL labels (Supplementary Table 1).
A White and pial surfaces marked with blue and red edge color, respectively, presented on the T2-weighted average image in multiple slices covering axial, coronal, and sagittal views. B 3D representation of white and pial surfaces of both hemispheres (left hemisphere on the left and right hemisphere on the right). The gyri are marked in green, and the sulci are marked in red. C The Desikan-Killiany parcellation represented on the synthetic T1-weighted image.
Discussion
We created a novel set of neonatal templates with a spatial resolution of 0.5 × 0.5 × 0.5, as do the Developing Human Connectome Project and atlases. The main novel aspects of our template, and possible advantages to some studies, are the age (2–7 weeks) of the infants in our sample, anatomically precise manual segmentation of subcortical structures (especially the thalamus is not subdivided into myelinated and unmyelinated parts), and the standard mapping to the MNI-152 template space that enables standard spatial transformations between neonatal and adult MNI space. This is potentially an important step in standardising the use of template spaces, which, according to a recent review is much needed6, and also enabling comparisons between neonates and adults. Second, a related contribution is that we created multimodal templates for structural and diffusion MRI, which are rare in the field (Table 1). We make the FBN-125 neonate templates, atlases, and adult MNI coregistration files publicly available for the scientific community (see Data availability). We provide volumetric templates that enable volumetric spatial normalisation and transform to adult MNI-152 space. This approach is straightforward and standardly used in fMRI studies. In some cases, surface-based methods are known to be more precise41,42,43. However, we would like to stress that the volumetric templates are recommended for volumetric analyses and processing as they are based on manual segmentation and joint label fusion (JLF) segmentation. The standard surface transforms are based on FreeSurfer processing and are potentially useful in surface-based applications, but the volumetric segmentations have variable agreement with JLF labels.
Potential for better comparability for neonatal MRI studies
When reporting findings from a neuroimaging study, it is important to specify which template and coordinate space was used for spatial normalisation so that data collected using different methods can be compared across studies44. For adults, the MNI-152 template is the most frequently used standardised template space for spatial normalisation45,46,47. However, infant neuroimaging research predominantly processes infant data in a single subject space due to a lack of a standardised template6. Usage of off-the-shelf infant templates followed by study-specific templates was most common in studies using fMRI6. Specifically, in term-born populations, 81 studies used off-the-shelf atlases, 29 studies used a study-specific common space, and 16 studies used a single subject space, indicating strong preferences for off-the-shelf atlases. The most commonly used template/atlas across modalities were the UNC-infant atlases (24%) and the JHU-neonate atlases (13%)6.
In the case of task-based fMRI studies, reporting the coordinates of neural activity related to a specific task is useful for comparability and replication across studies. However, only some task fMRI studies with infant populations report the corresponding MNI coordinates of the activated brain areas48,49,50. For instance, while some studies reported the region of interest (ROI) coordinates from an off-the-shelf infant template51,52, some reported the coordinates of peak activity in Talairach space53. Further, in task fMRI studies comparing infant and adult samples, MNI space coordinates are reported for the adult participants, while the infant coordinates are reported in correspondence to an off-the-shelf infant atlas36. Some studies have reported the locations so that coordinates from the adult sample are given in Talairach space and coordinates in millimetre points from the anterior commissure point for the infant sample34. Many studies have opted to report the results in the predetermined ROIs33,35,54,55,56,57,58,59, but it is clear that, at the moment, the benefits of standard space and coordinates cannot be fully established in neonatal and infant MRI studies.
The FBN-125 templates could provide a standard waypoint template for all existing and future studies to enable investigators to compare locations of their activations through adult MNI coordinates (https://neurosynth.org/), report standard MNI coordinates that enable meta-analyses (https://www.brainmap.org/ale/), and store unthresholded T maps for later use (https://neurovault.org/). On a related note, recent advances in available longitudinal atlases spanning ages from gestation to the neonatal period60 (Serag et al., 60) as well as from birth to age 2 years61 can be integrated with our templates through serial registration across the longitudinal template series to the neonatal template, registration to FBN-125, and standard transform to the MNI space.
The typical features of the neonatal MRI, limitations and future directions
The neonate brain is roughly one third of the adult brain, which makes the proportional resolution worse by a factor of\(\quad \root 3 \of {1}/3\) - e.g. 1 mm3 resolution in the neonate brain is equivalent to 1.5 mm3 in an adult image. Overall, this makes the partial volume issues more pronounced. Second, the infant brain morphology usually has a lot more variance than in older ages1, e.g. the bones of the neonatal skull are not fused, and there may be marked left-right asymmetries, flattening in either anterior-posterior or superior-inferior direction, or even bulging of the brain out of the superior foramen–all this reflecting perfectly normal anatomy. Minor birth-related haemorrhages and incidental findings are also important to consider11, although they can often be dealt with via corrections in brain masks and selected exclusions of study participants. We performed the manual segmentations on the warped copies of the average template, which made the manual segmentation easier due to the relatively 'sharp' tissue borders. The initial averaging in template creation enabled both an increase in signal-to-noise ratio and up-sampling of the resolution from the initial scan resolution (here from 1.0 \(\times\) 1.0 \(\times\) 1.0 to 0.5 \(\times\) 0.5 \(\times\) 0.5). Even small structures, such as the claustrum, are visible in the templates, and the images could be used to manually label additional, smaller structures in the future.
It is imperative to note that the infant brain tissue contrast changes in at least three different phases1: '(1) the infantile phase (≤3 months), in which the grey matter shows a relatively higher signal intensity than the white matter in T1-weighted images, and the tissue contrast in T2-weighted images is better than in T1-weighted images; (2) the isointense phase (5–9 months), in which the signal intensity of the white matter is increasing during the development due to the myelination and maturation process; in this phase, grey matter and white matter have the lowest signal differentiation in both T1-weighted and T2-weighted images; (3) the early adult-like phase (≥12 months), where the grey matter intensity is much lower than the white matter intensity in T1-weighted images, largely similar to the tissue contrast pattern in adult T1-weighted images.' Our atlas has been built from a Finnish (Scandinavian Caucasian) term-born population scanned at the gestation-corrected age of 1–5 weeks (age from birth 2–7 weeks) and is best suited for analyses on neonatal/early infancy data. Unfortunately, we have only a small number of participants with a follow-up scan after their neonatal scan, and we are thus not able to contribute to longitudinal atlas development across infancy. Consequently, our templates may not fit the needs of studies carried out in preterm populations that have also used a mixed set of templates54,55,58. The joint efforts of large-scale projects such as the Developing Human Connectome Project, Baby Human Connectome Project, and Healthy Brain Child Development will provide high-quality data and related software to support 4D atlas development from infancy to early childhood and beyond60,61. Future work from our group and others could focus on creating surfaces on both the cortical and subcortical structures and the use of the fMRI to parcellate the surfaces; for instance, the methods of the study where Lewis et al. developed and validated a surface-based approach to create a functional parcellation of both cortical and subcortical structures in adults62 could be replicated with neonate data, and the creation of standard transforms between neonatal and adult surface spaces. We propose that future studies that introduce new templates and atlases would include standard transforms to the adult MNI space, as was done in the current study.
Conclusions
Neonatal brain segmentation remains a key challenge for developmental neuroscience. Advances in the field may rely on producing better templates, atlases, and segmentation tools. We contribute to this endeavour here by creating and sharing our FBN-125 neonatal templates, atlases, and standard registrations between neonatal and adult standard spaces. The created labels are amenable to coregistration to diffusion or functional scans, e.g. for tractography and seed-based connectivity analyses. Finally, other groups can contribute to the manual labelling of additional structures, hopefully in time producing increasingly detailed atlas labelling.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
We make the FBN-125 templates and atlases publicly available for the scientific community, and also provide the standard coregistration files between the FBN-125 and adult MNI spaces. NITRC: http://www.nitrc.org/projects/fbn125/
References
Li, G. et al. Computational neuroanatomy of baby brains: a review. Neuroimage 185, 906–925 (2019).
Makropoulos, A., Counsell, S. J. & Rueckert, D. A review on automatic fetal and neonatal brain MRI segmentation. Neuroimage 170, 231–248 (2018).
Devi, C. N., Chandrasekharan, A., Sundararaman, V. K. & Alex, Z. C. Neonatal brain MRI segmentation: a review. Comput. Biol. Med. 64, 163–178 (2015).
Oishi, K., Chang, L. & Huang, H. Baby brain atlases. Neuroimage 185, 865–880 (2019).
Zöllei, L., Iglesias, J. E., Ou, Y., Grant, P. E. & Fischl, B. Infant FreeSurfer: an automated segmentation and surface extraction pipeline for T1-weighted neuroimaging data of infants 0–2 years. Neuroimage 218, (2020).
Dufford, A. J. et al. (Un)common space in infant neuroimaging studies: a systematic review of infant templates. Hum. Brain Mapp. 43, 3007–3016 (2022).
Acosta, H. et al. Partial support for an interaction between a polygenic risk score for major depressive disorder and prenatal maternal depressive symptoms on infant right amygdalar volumes. Cereb. Cortex 30, 6121–6134 (2020).
Lehtola, S. J. et al. Associations of age and sex with brain volumes and asymmetry in 2–5-week-old infants. Brain Struct. Funct. 224, 501–513 (2019).
Merisaari, H. et al. Effect of number of diffusion encoding directions in neonatal diffusion tensor imaging using tract-based spatial statistical analysis. Eur. J. Neurosci. 58, 3827–3837 (2023).
Merisaari, H. et al. Test-retest reliability of diffusion tensor imaging metrics in neonates. Neuroimage 197, 598–607 (2019).
Kumpulainen, V. et al. Prevalence and risk factors of incidental findings in brain MRIs of healthy neonates—the FinnBrain birth cohort study. Front. Neurol. 10, 1347 (2020).
Copeland, A. et al. Infant and child MRI: a review of scanning procedures. Front. Neurosci. 15, 1–16 (2021).
Fonov, V. et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54, 313–327 (2011).
Fonov, V. S., Evans, A. C., McKinstry, R. C., Almli, C. R. & Collins, D. L. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 47, S102 (2009).
Guimond, A., Roche, A., Ayache, N. & Meunier, J. Three-dimensional multimodal brain warping using the demons algorithm and adaptive intensity corrections. IEEE Trans. Med. Imaging 20, 58–69 (2001).
Miller, M. et al. Statistical methods in computational anatomy. Stat. Methods Med. Res. 6, 267–299 (1997).
Ward Jr, J. H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58, 236–244 (1963).
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17, 143–155 (2002).
Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
Oguz, I. et al. DTIPrep: quality control of diffusion-weighted images. Front. Neuroinform. 8, 1–11 (2014).
Andersson, J. L. R. R. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016).
Lewis, J. D., Fonov, V. S., Collins, D. L., Evans, A. C. & Tohka, J. Cortical and subcortical T1 white/gray contrast, chronological age, and cognitive performance. Neuroimage 196, 276–288 (2019).
McCarthy, P. FSLeyes (1.2.0). Zenodo https://doi.org/10.5281/zenodo.5504114 (2021).
Hashempour, N. et al. A novel approach for manual segmentation of the amygdala and hippocampus in neonate MRI. Front. Neurosci. 13, 1–15 (2019).
Perlaki, G. et al. Comparison of accuracy between FSL’s FIRST and Freesurfer for caudate nucleus and putamen segmentation. Sci. Rep. 7, 2418 (2017).
Owens-Walton, C. et al. Increased functional connectivity of thalamic subdivisions in patients with Parkinson’s disease. PLoS ONE14, e0222002 (2019).
Douaud, G. et al. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain 130, 2375–2386 (2007).
Kouwenhoven, E., Giezen, M. & Struikmans, H. Measuring the similarity of target volume delineations independent of the number of observers. Phys. Med. Biol. 54, 2863–2873 (2009).
Visser, M. et al. Inter-rater agreement in glioma segmentations on longitudinal MRI. Neuroimage Clin. 22, 101727 (2019).
Wang, S. et al. Assessment of neonatal brain volume and growth at different postmenstrual ages by conventional MRI. Medicine 97, e11633 (2018).
Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011).
Tustison, N. J. & Avants, B. B. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 23, 39 (2013).
Wild, C. J. et al. Adult-like processing of naturalistic sounds in auditory cortex by 3- and 9-month old infants. Neuroimage 157, 623–634 (2017).
Biagi, L., Crespi, S. A., Tosetti, M. & Morrone, M. C. BOLD response selective to flow-motion in very young infants. PLoS Biol. 13, e1002260 (2015).
Dehaene-Lambertz, G., Dehaene, S. & Hertz-Pannier, L. Functional neuroimaging of speech perception in infants. Science 298, 2013–2015 (2002).
Goksan, S. et al. fMRI reveals neural activity overlap between adult and infant pain. Elife 2015, 1–13 (2015).
Shi, F. et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS ONE 6, e18746 (2011).
Mariani Wigley, I. L. C. et al. Infants´ sex affects neural responses to affective touch in early infancy. Dev. Psychobiol. 65, e22419 (2023).
Gopinath, K. et al. Recon-all-clinical: cortical surface reconstruction and analysis of heterogeneous clinical brain MRI. Preprint at arXiv https://doi.org/10.48550/arXiv.2409.03889 (2024).
Dickie, E. W. et al. Ciftify: a framework for surface-based analysis of legacy MR acquisitions. Neuroimage 197, 818–826 (2019).
Klein, A. et al. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage 51, 214–220 (2010).
Coalson, T. S., Van Essen, D. C. & Glasser, M. F. The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proc. Natl. Acad. Sci. USA 115, E6356–E6365 (2018).
Ghosh, S. S. et al. Evaluating the validity of volume-based and surface-based brain image registration for developmental cognitive neuroscience studies in children 4 to 11years of age. Neuroimage 53, 85–93 (2010).
Poldrack, R. A. et al. Guidelines for reporting an fMRI study. Neuroimage 40, 409–414 (2008).
Mazziotta, J. C., Toga, A. W., Evans, A., Fox, P. & Lancaster, J. A probabilistic atlas of the human brain: theory and rationale for its development. Neuroimage 2, 89–101 (1995).
Mazziotta, J. et al. A four-dimensional probabilistic atlas of the human brain. J. Am. Med. Inform. Assoc. 8, 401–430 (2001).
Mazziotta, J. et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. B Biol. Sci. 356, 1293–1322 (2001).
Ellis, C. T., Skalaban, L. J., Yates, T. S. & Turk-Browne, N. B. Attention recruits frontal cortex in human infants. Proc Natl Acad Sci USA 118, e2021474118 (2021).
Ellis, C. T. et al. Evidence of hippocampal learning in human infants. Curr. Biol. 31, 3358–3364.e4 (2021).
Graham, A. M., Fisher, P. A. & Pfeifer, J. H. What sleeping babies hear: a functional MRI study of interparental conflict and infants’ emotion processing. Psychol. Sci. 24, 782–789 (2013).
Kuklisova-Murgasova, M. et al. A dynamic 4D probabilistic atlas of the developing brain. Neuroimage 54, 2750–2763 (2011).
Williams, G. et al. Functional magnetic resonance imaging can be used to explore tactile and nociceptive processing in the infant brain. Acta Paediatr. 104, 158–166 (2015).
Blasi, A. et al. Early specialization for voice and emotion processing in the infant brain. Curr. Biol. 21, 1220–1224 (2011).
Allievi, A. G. et al. Maturation of sensori-motor functional responses in the preterm brain. Cereb. Cortex 26, 402–413 (2016).
Anderson, A. W. et al. Neonatal auditory activation detected by functional magnetic resonance imaging. Magn. Reson. Imaging 19, 1–5 (2001).
Baldoli, C. et al. Maturation of preterm newborn brains: a fMRI–DTI study of auditory processing of linguistic stimuli and white matter development. Brain Struct. Funct. 220, 3733–3751 (2015).
Deen, B. et al. Organization of high-level visual cortex in human infants. Nat. Commun. 8, 13995 (2017).
Lee, W. et al. Visual functional magnetic resonance imaging of preterm infants. Dev. Med. Child Neurol. 54, 724–729 (2012).
Perani, D. et al. Functional specializations for music processing in the human newborn brain. Proc. Natl. Acad. Sci. USA 107, 4758–4763 (2010).
Serag, A. et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage 59, 2255–2265 (2012).
Ahmad, S. et al. Multifaceted atlases of the human brain in its infancy. Nat. Methods 20, 55–64 (2023).
Lewis, J. D., Bezgin, G., Fonov, V. S., Collins, D. L. & Evans, A. C. A sub+cortical fMRI-based surface parcellation. Hum. Brain Mapp. 43, 616–632 (2022).
Acknowledgements
We would like to warmly thank all FinnBrain families who participated in the study. We would also like to thank the research team that had a supportive role in the current study: Satu Lehtola for her help in data collection, Maria Lavonius for her help in recruiting the participants, Jani Saunavaara for implementing the MRI sequences, Riitta Parkkola for reviewing the MR images for incidental findings.
Author information
Authors and Affiliations
Contributions
J.J.T., Conceptualisation, creation of manual segmentation guidelines, supervising the manual segmentation work, creation of the DTI templates, creating the atlas labels, creating and validating the standard coregistrations, statistical analyses, drafting the initial version of the manuscript and leading the manuscript writing. A.R., Performing manual segmentation, statistical analyses and drafting the initial version of the manuscript. E.P.P., Drafting the initial version of the manuscript and providing feedback to early versions of the manuscript. N.H., Performing manual segmentation, creation of manual segmentation guidelines, co-supervising the manual segmentation work of the subcortical structures and drafting the initial version of the manuscript. E.U. and K.L., Performing manual segmentation and drafting the initial version of the manuscript. A.J., S.L., H.K.A., E.V., W.B. and I.S., Drafting the initial version of the manuscript. I.W., Validating the standard coregistrations. V.S.F. and D.L.C., Developed the tools used in the template creation and segmentation of the data. H.M., Creation of the DTI templates and drafting the initial version of the manuscript. L.K. and H.K., Planned and funded the MRI measurements, established the FinnBrain Birth Cohort and built the infrastructure for carrying out the study. J.D.L., Conceptualisation, creation of the MRI sequences, creation of manual segmentation guidelines, supervising the manual segmentation work, performing the segmentation of the ‘base template’ creating the atlas labels, validating the standard coregistrations, drafting the initial version of the manuscript and leading the manuscript writing. All authors participated in writing the manuscript and accepted the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Pew-Thian Yap, Dustin Scheinost and Nagehan Demirci for their contribution to the peer review of this work. Primary Handling Editor: Benjamin Bessieres.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tuulari, J.J., Rosberg, A., Pulli, E.P. et al. The FinnBrain multimodal neonatal template and atlas collection. Commun Biol 8, 600 (2025). https://doi.org/10.1038/s42003-025-07963-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07963-7