Abstract
Alternatively activated M2-like macrophages have a profound impact on asthma pathogenesis. Basic Helix-Loop-Helix Family Member E40 (BHLHE40), a dimeric transcriptional factor, plays a key regulation in macrophage functions. Here we show that ovalbumin (OVA)-challenged mice exhibited greater expression of BHLHE40 in lung tissues. Bhlhe40 knockdown reduced the pulmonary lesions and allergy-induced inflammation in asthmatic mice. Moreover, an inhibitory effect of Bhlhe40 knockdown on alternative activation was observed in vivo and in vitro. We show a downstream target Neurturin (Nrtn) of Bhlhe40. Dual luciferase assay and ChIP-qPCR assay indicated that BHLHE40 bound to Nrtn promoter and reduced its transcriptional activity. Simultaneous knockdown of Bhlhe40 and Nrtn recovered the alternative activation of macrophages and rescued the OVA-elicited asthma phenotype. NRTN downregulation offset the alleviative effects of Bhlhe40 knockdown on asthma. This study demonstrates that BHLHE40 promotes allergic asthma, and contributes to the alternative activation of macrophages in asthma by inhibiting Nrtn transcription.
Similar content being viewed by others
Introduction
Bronchial asthma is a chronic airway inflammation disease characterized by bronchial hyperreactivity and airway inflammation and participated by various inflammatory cells and cytokines1. Macrophages are the most abundant immune cells in normal lungs and macrophage polarization plays an important role in asthma2. Alveolar macrophages are in direct contact with the environment as they are located on the epithelial surface of the lung3. Under stimulation with different microenvironments, macrophages can be polarized into classically activated M1-like or alternatively activated M2-like macrophages4. M1-like macrophages, induced by IFN-γ and lipopolysaccharide (LPS), are responsible for inflammation and protection against invading pathogens5. M2-like macrophages are stimulated by interleukin (IL)-4 and IL-13, and it contributes to anti-inflammatory responses and tissue remodeling6. Asthma can be categorized as non-allergic or allergic, with the latter being more common. Allergic asthma is characterized with T helper (Th2) immune response and airway hyperresponsiveness (AHR), and airway remodeling7. Th2 cytokines can not only promote IgE production, but also activate M2-like macrophages. Robbe et al.8 showed that M2-like macrophages are the major effector macrophages in allergic asthma whereas M1-like macrophages predominate in non-allergic asthma. The alternatively activated macrophages are associated with the severity of asthma, and it promotes the development of an allergic inflammatory response9. In turn, M2-like macrophages can enhance the Th2 immune response to aggravate allergic asthma10. The pathogenesis of allergic asthma is complex, and it is important to explore the underlying mechanism of allergic asthma.
Basic helix-loop-helix proteins are dimeric transcriptional factors, which form a large superfamily and play important roles in asthma development, such as hypoxia-inducible factor, aryl hydrocarbon receptor, and transcription factor EB11,12,13,14. BHLHE40 is reported to act as a key regulator in inflammatory and autoimmune diseases15,16. In acute lung injury, Bhlhe40 is required for LPS-induced injury, and Bhlhe40 can inhibit GSDMD-mediated pyroptosis17. Rauschmeier et al.18 showed that BHLHE40 transcription factor regulates alveolar macrophage self-renewal. In autoimmune neuroinflammation, Bhlhe40 is reported to be required for the induction of experimental autoimmune encephalomyelitis19. Besides, BHLHE40 expression was upregulated in the bone marrow derived macrophages (BMDMs) that were treated with IL-420. However, its roles in asthma have not been reported yet.
Neurturin (NRTN) is widely recognized as an essential neurotrophic factor in neuronal development21. Recent studies have suggested that NRTN involves in regulating the physiological process of asthma. A clinical examination of asthmatic patients showed that sputum NRTN level is associated with Type 2 airway inflammation in patients with asthma22. Nrtn−/− mice showed higher levels of neutrophils, cytokine-induced neutrophil chemoattractant, and a stronger tendency to activate the Th2 response23. Michel et al.24 reported that knockout of Nrtn enhanced the pronounced worsening of OVA-induced airway inflammation, Th2 response, and airway hyperresponsiveness. Besides, Nrtn inhibited the inflammatory response of lung macrophages caused by viral infection25.
In the present study, high-throughput mRNA sequencing was conducted to evaluate the transcriptome of mouse model of asthma. The key gene Bhlhe40 was upregulated in OVA-challenged mice. It was speculated that Bhlhe40 might regulate the development of allergic asthma. We investigated the roles of BHLHE40 in the development of asthma, explored its effects on activation of macrophages in vivo and in vitro. Additionally, BHLHE40 usually acts as a transcriptional suppressor to modulate gene expression. We explored the regulation between BHLHE40 and NRTN, and whether NRTN mediates the roles of BHLHE40 in allergic asthma.
Results
BHLHE40 is upregulated in asthma induced by OVA
We constructed a mouse model of asthma by OVA challenge as shown in Fig. S1A, and explored the mRNA profile of lung tissue with asthma. Histological results confirmed that mouse model of asthma was successfully established (Fig. S1B). Compared to the mice in the sham group, OVA-challenged mice showed thickened airway walls, extensive inflammatory cell infiltration, goblet cell metaplasia, and deformed alveolar structure. From principal component analysis (PCA), there was a clear separation between asthmatic mice and sham mice (PCA1 = 52.14%, PCA2 = 10.85%) (Fig. S1C). The differentially expressed genes (DEGs) were identified by a threshold of |Log2FC| >1 and adj p value <0.05 (Supplementary Data 1) and displayed it in a volcano map (Fig. S1D). In addition, GSEA was performed to identify pathways enriched among differentially expressed genes. The results revealed that these differentially expressed genes participate in many important signaling pathways (Supplementary Data 2). The GO pathways “leukocyte chemotaxis”, “macrophage migration”, “mast cell activation”, and “inflammatory response” were upregulated in asthmatic mice. Subsequently, we downloaded the GSE49705 dataset from the GEO database for further analysis. GSE49705 is a transcriptome analysis of Prdm11 knockout mice after OVA challenge. We compared the mRNA profiles in lung samples between wild-type and wild-type mice with OVA challenge and obtained the DEGs between healthy and asthmatic mice in GSE49705 dataset (Supplementary Data 3). Next, the 24 shared genes that upregulated in asthmatic mice was showed in a Venn diagram (Fig. S1E). To the best of our knowledge, among all the shared genes, 16 have been reported in asthma and 8 are novel (Bhlhe40, AA467197, Gpr171, H2-DMb1, Aqp9, Cyp4f18, Rpl3l, and Arhgdib) (Fig. S1F). Through further literature review and analysis, we narrowed down the candidates to Bhlhe40, Aqp9, and Arhgdib as the most suitable for investigation. Among these, Bhlhe40 stood out with the highest log2FC value (Supplementary Data 1), demonstrating significant upregulation in asthma (Fig. S1G). Besides, Bhlhe40 is reported to involved in inflammation regulation. Therefore, the significantly upregulated Bhlhe40 gene was identified as the target gene.
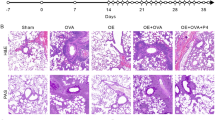
To further verify the results of high-throughput sequencing, we repeated the mouse model of asthma again. HE staining and PAS staining of lung tissues suggested that OVA administration induced airway inflammation, goblet cell metaplasia, and airway remodeling (Fig. 1A–C). Next, the expression of three asthma-associated genes, Ppard, Hpgd, and Npr3, was tested to verify the accuracy of the sequencing results. PPARD involves the pathogenesis of severe asthma26. Hpgd, and Npr3 are the hit on differentially expressed gene analysis. RT-qPCR analysis suggested that Ppard was upregulated in asthmatic mice, and the expression of Hpgd and Npr3 was downregulated (Fig. S1H), which was consistent with the high-throughput sequencing results. Besides, compared with sham mice, the mRNA level of Bhlhe40 was higher in asthmatic mice (Fig. 1D). Western blot and IHC staining for BHLHE40 indicated a high protein level of BHLHE40 in lung tissues of asthmatic mice (Fig. 1E, F). By fluorescent microscope, increased colocalization of BHLHE40 with F4/80 (macrophage marker) was found in the lung tissues of asthmatic mice (Fig. 1G), suggesting that BHLHE40 was highly expressed in alveolar macrophages in asthmatic mice.
A Schematic graph of animal sensitization and challenge. B Histological analysis of lung tissues stained with HE staining or PAS staining. C Histology score of HE staining and PAS staining. Statistical significance was determined with Mann-Whitney test. D mRNA expression of Bhlhe40 was assayed by RT-qPCR. Statistical analysis was performed by two-tailed Student’s t-test. E Western blot analysis of BHLBE40 in lung tissue. Statistical analysis was performed by two-tailed Student’s t-test. F Immunohistochemistry staining was performed to detected the level of BHLHE40 in lung tissue. Statistical analysis was performed by two-tailed Student’s t-test. G Double immunofluorescence staining for BHLHE40 (red) and F4/80 (green) in lung tissues. The number of double positive (F4/80 + BHLHE40 + ) cells was quantified. Statistical analysis was performed by two-tailed Student’s t-test. Error bars represent standard deviation. N = 8 biological replicates.
Bhlhe40 knockdown suppresses the development of asthma in mice
Furthermore, to interfere Bhlhe40 in vivo, mice in asthma group received lentiviral vector LV-shBhlhe40 or LV-shNC (Fig. 2A). As expected, Bhlhe40 was clearly reduced at both the mRNA and protein levels (Fig. 2B, C). From histological analysis, more inflammatory cell infiltration around airway, evident narrowing of the airways, goblet cell hyperplasia, and mucus production was induced by OVA. After Bhlhe40 knockdown, we found inhibition of inflammatory cell infiltration, along with decreased goblet cell hyperplasia and mucus production in the lung tissues (Fig. 2D, E). Besides, the protein concentration in BALFs was increased after OVA challenge, while knockdown of Bhlhe40 inhibited it (Fig. 2F). We also counted the number of inflammatory cells in BALF by Wright-Giemsa staining (Fig. 3A). OVA challenge increased the number of total inflammatory cells in BALF, but this increasing was offset after Bhlhe40 silencing (Fig. 3B). Specifically, accumulation of eosinophil, neutrophil, and macrophages was observed in mice with OVA inhalation (Fig. 3C–E). There were less inflammatory cells in mice with BHLHE40 knockdown. OVA induced aberrant T helper 2 (Th2) inflammatory immune response in lung, which was evident by increased level of cytokines, IL-4, IL-13, IL-5, and IL-10 in BALF and massive IgE production in serum (Fig. 3F, G). However, Bhlhe40 knockdown inhibited the production of these cytokines and IgE. Allergic asthma is characterized by inflammatory infiltration, airway hyperresponsiveness, and Th2 cell-mediated immune response. All results of this part showed that Bhlhe40 knockdown suppressed the typical symptoms of asthma, indicating that Bhlhe40 involves in the development of asthma.
A Mice in asthma group received lentiviral delivery of LV-shBhlhe40 or LV-shNC. B RT-qPCR tested the mRNA expression of Bhlhe40 in lung tissues. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. C Representative western blot and quantitative analysis of BHLHE40 expression in lung tissues. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. D Representative HE images in lung tissue sections and quantitative analysis of histologic scoring. Statistical analysis was performed by Kruskal-Wallis test. E Representative PAS images in lung tissue sections and quantitative analysis of histologic scoring. Statistical analysis was performed by Kruskal-Wallis test. F Total protein concentrations in BALF in OVA-induced asthma. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation. N = 8 biological replicates.
A Representative pictures from Giemsa staining for BALF cell. B The total number of cells in BALF. C The number of eosinophils in BALF. D The number of neutrophils in BALF. E The number of macrophages in BALF. F ELISA assay detected the level of IL-4, IL-5, IL-13, and IL-10 in BALF. G ELISA assay for IgE in serum. N = 8 biological replicates. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation.
Bhlhe40 knockdown inhibits the polarization of M2-like macrophage markers in asthmatic mice
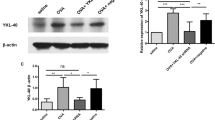
Macrophage polarization is heavily associated with asthma pathogenesis. M2 macrophage polarization is essential in allergic asthma. In the following study, we explore whether and how Bhlhe40 affected alternatively activated macrophages in asthma. YM1, FIZZ1, CD206 and ARG1 are the canonical markers of alternative activation. OVA challenge increased the concentration of YM1 in BALF and its mRNA expression in lung tissue (Fig. 4A, B). Fizz1 expression shared the similar trends (Fig. 4C). Moreover, RT-qPCR showed an upregulation of Cd206 and Arg1(Fig. 4D, E). Western blot analysis supported it (Fig. 4F–H). Importantly, the expression of all these alternative activation markers was decreased after Bhlhe40 knockdown, which indicates that Bhlhe40 contributes to the alternative activation of macrophages. Macrophages in the alveoli are divided into tissue-resident alveolar macrophages (TR-AMs) and monocyte-derived alveolar macrophages (Mo-AMs). Under certain pathological conditions, such as allergic asthma, blood monocytes migrate to the lungs and differentiate to Mo-AM, and replace TR-AMs27,28,29. In allergic asthma, exposure to allergens results in rapid monocyte recruitment and an increase in Mo-AMs, which promotes an acute inflammatory response. By contrast, TR-AMs play a critical role in the maintenance of lung tissue homeostasis30. In this work, the effects of Bhlhe40 on Mo-AMs were explored. Mo-AMs were isolated from BALF by flow cytometry analysis (Fig. S2A). We also found few Mo-AMs in sham mice, while the cell proportion of Mo-AMs was higher in OVA-challenged mice compared with the sham mice (Fig. S2B). Bhlhe40 downregulation decreased the recruitment of mononuclear macrophages. Besides, tracheal delivery of LV-shBhlhe40 decreased the expression of BHLHE40 in Mo-AMs (Fig. 4I). The status of Mo-AMs was explored by testing the expression of alternative activation markers. Bhlhe40 knockdown reduced the protein level of CD206 and Arg1 in Mo-AMs (Fig. 4I–L), suggesting that Bhlhe40 is conducive to the alternative activation of Mo-AMs.
A ELISA forYM1 in BALF. B–E RT-qPCR assay showed the mRNA expression of Ym1, Fizz1, Cd206, Arg1 in lung tissues. F The protein level of CD206 and ARG1 in lung tissues. G Quantitative analysis of CD206 protein level.H Quantitative analysis of ARG1 protein level. I Western blot assessed the level of BHLHE40, CD206, and ARG1 in monocyte-derived alveolar macrophages (Mo-AMs). J Quantitative analysis of BHLHE40 protein level in Mo-AMs. K Quantitative analysis of CD206 protein level in Mo-AMs. L Quantitative analysis of ARG1 protein level in Mo-AMs. N = 8 biological replicates. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation.
Bhlhe40 knockdown impaired IL-4–induced alternative activation of BMDMs
For further confirming the function of Bhlhe40 in M2-like macrophage, we isolated BMDMs from mice, and it was subjected to IL-4 treatment for alternative activation of macrophages in vitro (Fig. 5A). Bhlhe40 was silenced in the cells by lentiviral infection. RT-qPCR showed that IL-4 treatment increased Bhlhe40 expression, and LV-shBhlhe40 decreased the expression of Bhlhe40 in BMDMs (Fig. 5B). Also, the protein level of BHLHE40 shared the same trends (Fig. 5C). The mRNA expression of Ym1, Fizz1, Cd206, and Arg1 was raised in BMDMs by IL-4 administration, indicating that IL-4 induced BMDMs towards M2-like macrophages (Fig. 5D–G). Compared with untreated cells, IL-4-stimulated BMDMs showed increased protein level of CD206 and ARG1 (Fig. 5H). However, Bhlhe40 downregulation counteracts this induction. From IF staining for Arg1, we also found that IL-4 increased Arg1 protein level, while it was diminished by Bhlhe40 knockdown (Fig. 5H). Consistent with the results in vivo, Bhlhe40 knockdown exerted an inhibitory effect on alternative activation in vitro. Besides, the role of Bhlhe40 in M1 response was evaluated by IF staining of INOS. It was found that Bhlhe40 did not affect iNOS expression in IL-4-stimulated BMDMs (Fig. S4A).
A Bone marrow derived macrophages (BMDMs) were isolated from BALB/c mice, and then stimulated with IL-4 to induce M1 polarization. Lentivirus infection was conducted before IL-4 treatment. B The expression level of Bhlhe40 was determined by RT-qPCR. C Western blot analysis of BHLBE40 in BMDMs. D–G The mRNA expression level of Ym1, Fizz1, Cd206, and Arg1 in BMDMs. H The protein level of CD206 and ARG1 in BMDMs. I Immunofluorescence analysis of ARG1 expression in BMDMs, and the quantification of ARG1 positive cell count. N = 4 biological replicates. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation.
NRTN mediates the BHLHE40 regulation for IL-4–induced alternative activation
NRTN is a neurotrophin, which is involved in the pathogenesis of asthma23. In OVA-challenged model, Nrtn knockout mice showed aggravated inflammation and airway hyperresponsiveness24. According to JASPAR prediction, there are several transcription factors binding sites of BHLHE40 on the Nrtn promoter. Therefore, we speculated that BHLHE40 might regulate the transcription of Nrtn. In BMDMs, IL-4 administration decreased the expression level of Nrtn, while Bhlhe40 knockdown decreased it (Fig. 6A). Similar results were also found at the translation level (Fig. 6B). Furthermore, luciferase assay and ChIP-qPCR analysis were performed to validate that potential regulation. As shown in Fig. 6C, we used three reporter constructs containing fragments of the Nrtn promoter that drive luciferase expression when transcriptionally active. The luciferase activities were lower in Bhlhe40-transfected cells than in vector-transfected cells, indicating that BHLHE40 inhibits the transcriptional activity of Nrtn. ChiP-qPCR results showed that BHLHE40 indeed binds to Nrtn, as evident by increased enrichment of the promoter of Nrtn in BHLHE40-ChIP samples (Fig. 6D). Subsequently, we explore whether Nrtn mediates the roles of BHLHE40 in M2-like macrophages. The above results suggested that BHLHE40 negatively regulated the transcription of Nrtn. Lentivirus for Nrtn knockdown efficiency was examined by western blot (Fig. S3A). We silenced BHLHE40 in BDMDs and a rescue experiment for Nrtn was performed. RT-qPCR analysis presented that Nrtn knockdown reversed the Bhlhe40 silencing-induced downregulation of Fizz1, Ym1, Arg1, and Cd206 in BDMDs (Fig. 6E). Similar changes of CD206 and ARG1 was observed at protein level (Fig. 6F). Besides, IF staining showed high expression of ARG1 in BMDMs transfected with shBhlhe40 and shNrtn (Fig. 6G). Bhlhe40 downregulation increased the expression of Nrtn, resulting in an inhibition of alternative activation, while it was activated after co-silencing Bhlhe40 and Nrtn.
A, BNrtn expression level was determined by RT-qPCR and western blot. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. C Dual-luciferase reporter assay. N = 3 biological replicates. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. D Validation by ChIP-qPCR analysis of BHLHE40 binding to Nrtn in BMDMs. Statistical analysis was performed by two-tailed Student’s t-test. E RT-qPCR showed the mRNA expression of Fizz1, Ym1, Arg1, and Cd206. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. F The protein level of CD206 and ARG1 was assayed by western blot. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. G Immunofluorescence analysis of ARG1 expression in BMDMs, and the quantification of ARG1 positive cell count. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation. N = 3-4 biological replicates.
Moreover, the potential mechanism of Bhlhe40-Nrtn axis was validated in physiological peritoneal macrophages. As shown in Fig. 7A, physiological peritoneal macrophages were obtained from mice stimulated with thioglycolate and IL-4. BHLHE40 binding to Nrtn was confirmed in physiological macrophages via ChIP-qPCR. As BHLHE40 is reported to bind the IL-10 promotor, IL-10 was selected to serve the positive control. It revealed similar enrichment profiles in Nrtn and IL-10 (Fig. 7B). Infection of LV-shBhlhe40 decreased the Bhlhe40 expression both at mRNA and protein level (Fig. 7C, D). Besides, IF staining found that Bhlhe40 decreased the expression of ARG1 in peritoneal macrophages (Fig. 7E). Knockdown both Bhlhe40 and Nrtn recovered the expression of ARG1 compared to knockdown Bhlhe40 alone (Fig. 7F), indicating that Bhlhe40 regulates the macrophage polarization possibly by downregulating Nrtn. IF staining for INOS was conducted. We showed similar results in peritoneal macrophages as in BMDMs (Fig. S4B, C). Bhlhe40 did not affect iNOS expression in physiological peritoneal macrophages.
A Mice were stimulated with thioglycolate and IL-4 by intraperitoneal injection, and then physiological peritoneal macrophages were obtained. B Validation by ChIP-qPCR analysis of BHLHE40 binding to Nrtn in peritoneal macrophages. Statistical analysis was performed by two-tailed Student’s t-test. C RT-qPCR examined the mRNA level of Bhlhe40 in peritoneal macrophages. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. D Representative immunoblot and quantitative analysis of BHLHE40 expression. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. E Immunofluorescence analysis of ARG1 expression in peritoneal macrophages, and the quantification of ARG1 positive cell count. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. F Peritoneal macrophages were infected with LV-shBhlhe40 and LV-shNrtn, and then were subjected to immunofluorescence staining for ARG1. The number of ARG1 positive cells was quantified. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation. N = 4 biological replicates.
Nrtn knockdown aggravates the development of asthma in mice
To further validate the potential mechanism in vivo, mice in asthma group received lentiviral delivery of LV-shBHLHE40 and LV-shNrtn (Fig. 8A). RT-qPCR results showed a downregulation of Nrtn at mRNA level in mice received LV-shNrtn (Fig. 8B). NRTN protein level shared the same trends (Fig. 8C). Histologic assessment found that Nrtn knockdown exacerbated lung pathology in asthmatic mice ((Fig. 8D, E). From the representative HE images of lung tissues, co-silencing of Bhlhe40 and Nrtn increased infiltrated inflammatory cells and enhanced airway wall thickening compared to Bhlhe40 silencing alone. Results of PAS staining displayed goblet cell proliferation and mucus over-secretion in mice with Bhlhe40 and Nrtn knockdown. Additionally, Nrtn downregulation recovered the Th2 inflammatory immune response in asthma mice, which was evident by increased levels of Th2 cytokines (Fig. 8F). Nrtn knockdown offset the inhibitory impact of Bhlhe40 downregulation on Mo-AM recruitment (Fig. S5). Lentiviral delivery of LV-shNrtn decreased NRTN protein level in Mo-AMs (Fig. 8G), while increased the expression of ARG1 marker (Fig. 8H). These findings suggest that NRTN is the key mediator for BHLHE40 function in asthma.
A Mice in asthma group received lentiviral delivery of LV-shBhlhe40 or LV-shNrtn. B The mRNA expression of Nrtn in lung tissue. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. C Representative immunoblot and quantitative analysis of NRTN expression in lung tissues. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. D Histological analysis of lung tissues stained with HE staining or PAS staining. E Histology score of HE staining and PAS staining. Statistical analysis was performed by Kruskal-Wallis test. F ELISA assay detected the level of IL-4, IL-5, IL-13 and IL-10 in BALF. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. G Western blot assessed the level of NRTN in monocyte-derived alveolar macrophages (Mo-AMs). Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. H Representative immunoblot and quantitative analysis of ARG1 expression in Mo-AMs. Statistical analysis was performed by One-way ANOVA with Tukey’s post-test. Error bars represent standard deviation. N = 8 biological replicates.
Discussion
In the present study, we analyzed the mRNA profile of lung tissue from OVA-induced allergic mice and identified Bhlhe40 as a key gene that is involved in the pathogenesis of the asthma. mRNA sequencing data showed the upregulation of Bhlhe40 in lung tissue of asthmatic mice. What’s more, Bhlhe40 knockdown remarkably reduced the pulmonary lesions and Th2 immune response, hampering asthma progression.
Allergic asthma is an IgE-mediated sensitization to inhaled allergens, which is characterized by Th2 cell response and inflammatory cell infiltration31. Th2 cells can generate a canonical set of cytokine IL-4, IL-5, IL-13 and IL-10, which mediates a pathologic role in asthma32. IL-4 and IL-10 induces B cell activation and leads to excessive IgE secretion33. IL-5 markedly regulates the proliferation, differentiation, and activation of eosinophils, while IL-13 is responsible for mucus production and airway hyperresponsiveness34,35. Previous studies have highlighted that BHLHE40 involves in the regulation of immune diseases. In experimental autoimmune encephalomyelitis (EAE), BHLHE40 is required for Th cells to mediate pathogenicity19,36. Jarjour et al.16 reported that BHLHE40 is indispensable in T cells for a protective Th2 cell response. Therefore, BHLHE40 may facilitate inflammatory response in asthma progression, and Th2 response is one of the important targets. However, Huynh et al.37 showed that absent of Bhlhe40 in mice results in higher susceptibility to Mycobacterium tuberculosis. BHLHE40 exerts different role in these two lung diseases. In tuberculosis, BHLHE40 inhibits IL-10 expression, thereby decreasing early susceptibility. In contrast, in asthma, we found that BHLHE40 knockdown was accompanied by a decrease in IL-10 secretion in BALF. The different pathogenic mechanisms in allergic asthma and tuberculosis may account for it. Mycobacterium tuberculosis infection activates innate and adaptive immunity in the body38. IL-10 limits the protective Th1 immune response to Mycobacterium tuberculosis, exacerbates infection39. Allergic asthma is a chronic disease characterized by Th2 immune response. Th2 cytokines stimulate B-cell activation, thus enhancing mucus hypersecretion and IgE production. Besides, loss of BHLHE40 in asthma mice showed decreased IL-10 secretion, which is contrary to previous reports. BHLHE40 is reported to inhibit the transcription of IL-10 in T cells19,40. Alveolar lavage fluid is a complex environment, and the secretion of IL-10 is related to a variety of cells. Qian et al.41 indicated that B-cell-derived IL-10 promotes allergic sensitization and asthma progression. Therefore, it can be speculated that Bhlhe40 knockdown may decrease the Th2 response, inhibit the B cell activation, thus reducing IL-10 secretion.
The alternative activated M2-like macrophages was increased in allergic asthma, while M1-like macrophages predominate in non-allergic inflammation model4,42. M2-like macrophages predominantly secrete high level of chemokines, which induces airway eosinophil infiltration and mucus hypersecretion43. Th2 cytokines, such as IL-4 and IL-13, are major inducers of M2 macrophage polarization, and the increase in alternative activated macrophage polarization in the lungs reflects allergic asthma severity44. In turn, M2-like macrophages can enhance the Th2 immune response, which forms positive feedback to aggravate allergic asthma10,45. We also found the alternative activation of macrophages in mice with OVA-induced asthma. In allergic asthma, allergen exposure induces a rapid recruitment of monocytes and increase in Mo-AMs that promote acute inflammatory responses28. BHLHE40 is required for macrophage alternative activation, especially Mo-AMs. Downregulation of BHLHE40 reduced the expression of markers of M2-like macrophages in vivo and in vitro. However, in myeloid cell–conditional Bhlhe40 knockout mice, it has been reported that Bhlhe40 has no effect on macrophage alternative activation46, which contradicts our findings. In the physiologic macrophages obtained from the abdominal cavity, we also found that Bhlhe40 regulates alternative activation. On the one hand, asthma model is quite different from the animal model that Jarjour et al.46 reported. The animal strains and genetic backgrounds used in the two models are also different. Besides, the pathological response of allergic asthma is complex, and the immune environment in the lung is also different from that in the abdominal cavity. These factors could therefore bias the results. On the other hand, in the allergic asthma model, there are positive feedback between Th2 immune response and M2-like macrophages, and Bhlhe40 may regulate alternative activation of macrophages by affecting Th2 cytokine secretion.
Nrtn is originally identified in neuronal cells and its role in inflammation has been reported47. Besides, Nrtn showed an inhibitory effect on the progression of asthma23,24. In the present work, we proved that BHLHE40 bound to the promotor region of Nrtn and repressed its transcription. Besides, simultaneous knockdown of Bhlhe40 and Nrtn induced the alternative activation of macrophages, suggesting that NRTN inhibited the alternative activation and BHLHE40 might target NRTN to regulate macrophage polarization. Nrtn knockdown offset the effects of Bhlhe40 knockdown on OVA-elicited asthma phenotype. BHLHE40 might promote asthma progression via targeting Nrtn transcription.
Nevertheless, this study has also some limitations which deserve discussion. In the current study, we focused on the impact of Bhlhe40 on Mo-AM, whereases TR-AM were not investigated at the same level of detail. Future studies should address this gap to provide a more comprehensive understanding of the underlying mechanisms. Besides, there may be a combination of two aspects of Bhlhe40 in asthma. We have explored the direct roles of Bhlhe40 in alternative activation of macrophages but lacking the data about the roles of Bhlhe40 in Th2 cells. In the future, we should use the cell co-culture model of Th2 cells and macrophages to explore whether loss of Bhlhe40 in Th2 cells will affect the alternative activation of macrophages.
Together, our findings illustrate the functions of BHLHE40 in allergic asthma. BHLHE40 is required for the progression of asthma. Knockdown of Bhlhe40 reduced Th2 immune response, alternative activation of macrophages, accumulation of eosinophils, and airway inflammation response, thus relieving the progression of allergic asthma. Besides, as a transcription factor, BHLHE40 negatively regulates the transcription of downstream target Nrtn and thereby enhancing alternative activation of macrophages. These findings suggested that BHLHE40 contributes to allergic asthma development, and the potential mechanism is involved to Nrtn. Thus, the present work offers an insight into pathogenesis of asthma, and targeting BHLHE40 may be a potential strategy for intervening allergic asthma.
Material and methods
Animal
Female BALB/c mice (6–8 weeks old) were used for asthma model, as females are more susceptible to the development of allergic airway inflammation than males. Mice were housed at 22 ± 2 °C and humidity 55 ± 5% under a 12/12 h dark/light cycle with free water and food. Total 163 animals were used and randomly assigned to each experimental group using a random sequence generator. Nine mice in OVA group died after surgery. The experimental unit is a single animal.
Animal model
As shown in Fig. 1A, the mice in asthma group were intraperitoneally injected with 20 μg OVA and 2 mg Al(OH)3 on days 0 and days 14. One week after the final injection, the mice received inhalation with 1% OVA for 30 min on day 21, day 22, day 23. The mice in sham group were injected an equal volume of saline, and then inhaled with PBS. For mRNA-seq, the mice in Sham and OVA challenge groups (6 animals per group) were sacrificed 24 h after the last inhalation, and the lung tissues were collected for analysis. Another Sham and OVA challenge groups (8 animals per group) were set to verify the sequencing results. Additionally, to explore the role of BHLHE40 in asthma in vivo, mice were randomly allocated to four group (16 animals per group) and subjected to the following treatments: 1) Sham, 2) OVA challenge, 3) OVA + LV-shNC, 4) OVA + LV-shBhlhe40. Bhlhe40 shRNA lentiviral vectors (50 μL, 1 × 108 TU/mL) were IT-delivered into the anesthetized animals 2 days before the first inhalation with OVA. A mock virus (negative control shRNA) was used as control. For rescue animal experiments, as depicted in Fig. 8A, asthma mice were injected with Bhlhe40 shRNA lentiviral vectors and Nrtn shRNA lentiviral vectors (8 animals per group). The construction of the lentivirus vectors was entrusted to General Biosystems (Anhui) Co., Ltd. Mice were sacrificed 24 h after the last inhalation, and peripheral blood, bronchoalveolar lavage fluid (BALF), and lung tissues were harvested for further studies. Animal sacrifice was performed by exsanguination under deep isoflurane anesthesia at the end of the experiments. When mice exhibit severe respiratory distress, lethargy, unresponsiveness, or an inability to eat and drink normally, it is considered a humane endpoint. We have complied with all relevant ethical regulations for animal use. An experimental protocol was prepared and approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University.
Cell line and cell culture
A total of 38 mice were used for primary cell isolation. Bone marrow–derived macrophages (BMDMs) were isolated from femurs and tibias of mice as described previously48. Basically, tibia and femur were collected, the epiphysis was cut, and the bone marrow cavity was flushed by phosphate buffer saline (PBS) to obtain the bone marrow. Next, bone marrow cells were cultured in DMEM containing 10% fetal bovine serum (FBS) supplemented with 10 ng/mL M-CSF for 7 days to differentiate into BMDMs. BMDMs were stimulated with IL-4 (20 ng/mL) for 24 h to induce M2-like macrophages. Lentivirus transfection was conducted before IL-4 treatment. BMDMs were infected with lentivirus at an MOI of 10. After 48 h, infected BMDMs were treated with IL-4. For the isolation of peritoneal macrophages, according to the previously reported49, mice receive a combination of thioglycolate and IL-4c treatment to induce peritoneal macrophage recruitment and differentiation. In detail, 5 μg of murine IL-4 was mixed with 25 μg of anti-IL-4 to prepare fresh IL-4c. IL-4c and thioglycolate was administered via intraperitoneal injection to each mouse at day 0 and day 2. Mice were euthanized on day 4, and 5 mL of PBS plus 5 mM Ethylene Diamine Tetraacetic Acid (EDTA) was injected intraperitoneally. After a short soft massage of the abdomen, the lavage fluid was collected for centrifugation. Next, the cell pellet was resuspended and seed in RPMI medium (supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 μM non-essential amino acids, 100 U/mL penicillin, 100 μg/mL streptomycin, and 50 μM β-mercaptoethanol). The non-adherent cells were then removed and the adherent cells were considered as peritoneal macrophages. Macrophages were infected with lentiviral vector, and simultaneous administration of IL-4 exogenous stimulation maintains the cellular alternative activation state.
Histological evaluation
Histological changes were evaluated by hematoxylin and eosin (HE) staining and Periodic Acid-Schiff (PAS) staining. The lung tissues were embedded in paraffin and cut into 5 μm sections. HE staining was carried out according to standard protocol. For PAS staining, the hydrated sections were immersed with Periodic acid solution for 10 min and then incubated with Schiff’s Solution for 15 min. Hematoxylin was used to counterstain. The images were captured with BX43 microscope (Olympus, Japan). Histology score was quantified according to the previous reports. Five randomly selected areas on each section were blindly evaluated by two experienced observers. Airway inflammation was scored on HE sections based on the following inflammatory conditions: 0, no inflammatory infiltrate around all airways and vessels. 1, some infiltrates are detectable around airways and vessels. 2, most of the area around the airway is surrounded by 1 layer of inflammatory cells. 3, the majority of airways and vessels show inflammatory infiltrates. Many layers are thicker than two layers, ranging from 2 to 4 layers. 4, most of the area around the airway is surrounded by 4 or more layers of inflammatory cells. Mucus production was quantified on PAS slides. The score is based on the following conditions: 0, no goblet cell. 1, 5% PAS circumferential goblet cell staining. 2, 30% PAS goblet cells. 3, 30–60% PAS goblet cells. 4, 60% PAS goblet cells.
Flow cytometry analysis
Mo-AMs were isolated from BALF according to the previous method50. Briefly, collected BALF was filtered through 70 μm cell filter, and, and centrifuged with 300 g, 5 min at 4 °C. The cell pellet was resuspended in buffer and incubated with anti-CD16/32 (E-AB-F0997A, Elabscience, China) to block non-specific binding. Next, cells were washed and incubated with anti-CD64 (E-AB-F1186E, Elabscience, China), anti-Siglec-F (53-1702-82, eBioscience, USA), and anti-CD11b (E-AB-F1081D, Elabscience, China) at 4 °C for 30 min. After that, 5 μL of 7-AAD (E-CK-A162, Elabscience, China) was added to further incubate the cells in dark at 4 °C for 20 min. Then the cells were detected by NovoCyte flow cytometer (Agilent, USA). Gating strategy was done as follows: SSC/FSC>live/dead>CD64 + >Siglec-F + >CD11b+ (Fig. S2A). The Mo-AMs (Siglec-F + / CD11b + ) were collected for further analysis.
RT-qPCR
Total RNA was isolated from tissues or cells by TRIpure method. Nucleic acid concentration was determined by Nanodrop. One microgram of RNA was reverse transcribed to cDNA and then subjected to qPCR using specific primers (Table 1). qPCR was performed using SYBR Green Master Mix and Exicycler 96 real-time PCR system (Bioneer, Korea). The gene expression was normalized by using the 2−ΔΔ CT method. Gapdh was used as an internal reference gene. All the primers were listed in Table 1.
Western-blot
Tissues or cells were lysed in ice-cold modified RIPA buffer. After centrifugation, the supernatants were collected and BCA method was used to assess the protein concentration. Next, proteins were separated on SDS-PAGE and transferred to polyvinylidene fluoride membrane. The membrane was blocked in solution and incubated at 4 °C overnight with the following primary antibodies: anti- BHLHE40 (1:1000, A6534, ABclonal), anti-ARG1 (1:1000, A1847, ABclonal), anti-CD206 (1:1000, DF4149, Affinity), anti-NRTN (1:1000, DF13481, Affinity). GAPDH served as a loading control. The bound primary antibody was detected by peroxidase-conjugated goat anti-mice IgG (1:3000, SE131, Solarbio) or peroxidase-conjugated goat anti-rabbit IgG (1:3000, SE134, Solarbio). Protein blot was visualized with the ECL analysis system. Molecular weight was indicated by using a pre-stained protein ladder (KF8006, Affinity).
Immunohistochemistry (IHC)
Antigen retrieval was conducted by boiling the sections in citrate-buffered antigen retrieval solution. After, lung sections were incubated with 3% H2O2 and then blocked with 1% BSA for 15 min. Next, sections were stained with primary antibodies against BHLHE40 (1:100, 17895-1-AP, Proteintech, China) and secondary antibodies HRP-labeled goat anti-rabbit IgG (1:500, #31460, thermoFisher, USA). After developing with diaminobenzidine solution, the sections were counterstained with hematoxylin. Stained sections were finally analyzed using BX43 microscope (Olympus, Japan).
Immunofluorescence
For single-staining immunofluorescence, cell slides were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.5% Triton X-100 for 30 min. Subsequently, slides were blocked with 1% BSA and incubated with primary antibody against ARG1 (1:100, DF6657, Affinity) or INOS (1:100, sc-7271, Santa cruz). The slides were then incubated with Cy3-labeled goat anti-rabbit IgG (1:200, A27039, invitrogen). For double immunofluorescence staining, slides were incubated with primary antibody against BHLHE40 (1:500, NB100-1800SS, Novus), and F4/80 (1:50, Sc-377009, Santa cruz) at 4 °C overnight. Next, corresponding fluorescent secondary antibodies (Cy3-labeled goat anti-rabbit IgG, FITC-labeled goat anti-mouse IgG (1:200, ab6785, Abcam)) were used for incubation. DAPI was used for nuclear staining. Images were obtained on a fluorescence microscope (Olympus, Japan). Image analysis was conducted through Image-Pro Plus (Media Cybernetics). Five visual fields were randomly selected from each section or slice to count the number of positive cells.
Wright-Giemsa staining
After centrifugation, the pelleted cells in BALFs were resuspended in PBS, and the total cell numbers were counted. Ten μL cell suspension was made into the cell smear. After Wright-Giemsa staining, eosinophils, neutrophils, and macrophages were differentially counted under microscope (Olympus, Japan).
ELISA
The concentrations of IL-4, IL-5, IL-13 and IL-10 in BALFs were measured by ELISA, according to the manufacturer’s protocols (Lianke, China). Total IgE in serum samples was measured using a mouse ELISA Kit (Lianke, China). The level of YM1 in BALFs were determined using a Mouse Chi3l3 ELISA Kit (Fine Biotech, China) in accordance with the manufacturer’s instructions. The ELX-800 plate reader (BioTek, USA) were used for ELISA.
Dual luciferase assay
According to JASPAR prediction, we found two BHLHE40 binding sites in Nrtn promoter. To measure promoter activity, reporter plasmids containing different lengths of the Nrtn promoter sequence and Bhlhe40 overexpression plasmids were constructed. 293 T cells were co-transfected with reporter plasmids and Bhlhe40 overexpression plasmids for 48 h. Next, cells were harvested and luciferase activity was measured using the dual luciferase assay kit according to the manufacturer’s instructions (Keygen, China). The luciferase signals were detected by a Synergy H1 multifunctional microplate reader (BioTek, USA).
ChIP-qPCR
The ChIP assay was conducted using chromatin immunoprecipitation (ChIP) Kit (Beyotime, China).
In brief, after IL-4 induction, BMDMs were incubated with 1% formaldehyde for 10 min to crosslink the target protein and the corresponding DNA. Next, it was mixed with 1.1 mL Glycine solution for 5 min. After washing with cold phosphate buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), the samples were resuspended in SDS lysis buffer containing 1 mM PMSF and incubated on an ice bath for 10 min to fully lyse the cells. Ultrasonic cell disruptor (DHS-1000D, Dehongsehng Biotech, China) was applied to shear the chromatin. The condition of ultrasonic treatment was set to 10 seconds each time for a total of 4 times, and the power was set to 30% of the maximum power 50 W, and a 2 mm ultrasonic head was used. Chromatin in supernatants was collected by centrifugation and then incubated with anti-BHLHE40 at 4 °C overnight. As a negative control, chromatin was incubated with mouse IgG. The immune complex was precipitated with protein G-Agarose beads. Precipitated complex was eluted from the beads. Crosslinks between proteins and DNA were reversed with 5 M NaCl at 65 °C for 4 h. Subsequently, DNA was purified, and quantification of the DNA target regions was assessed by qPCR. Data were calculated as enrichment of the ChIP samples relative to the input. Primers used for ChIP-qPCR were listed in Table 1.
High-throughput sequencing
mRNA sequencing work was commissioned by Lianchuan Biotechnology Co., Ltd. (Hangzhou, China). Total RNA was isolated from lung tissues and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The RNA amount and purity of each sample was quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). The RNA integrity was assessed by Agilent 2100 with RIN number >7.0. Next, the mRNA was fragmented into small pieces and the cleaved RNA fragments were then reverse-transcribed to create the final cDNA library in accordance with the protocol for the mRNA-Seq sample preparation kit (Illumina, San Diego, CA, USA). At last, the 150 bp paired-end sequencing was performed on an Illumina Novaseq 6000 (LC Bio Technology CO.,Ltd. Hangzhou, China) following the vendor’s recommended protocol. Evaluation of raw sequencing data was done with Fast-QC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Alignment of sequencing data was performed using Hisat2 (https://daehwankimlab.github.io/hisat2/).
Bioinformation analysis
Addition to local mRNA sequencing, Datasets GSE49705 were obtained from GEO database. The mRNA profiles between asthmatic mice and healthy mice were investigated. GSE49705 dataset is a transcriptome analysis of Prdm11 knockout mice after OVA challenge, and includes four groups (wild-type, wild-type challenged, mutant, and mutant challenged group). We compared the mRNA profiles in lung samples between wild-type and wild-type mice with OVA challenge. Differentially expressed gene (DEG) analysis was performed by R package DESeq2 with threshold of |Log2FC| >1 and adj p-value <0.05. Principal component analysis (PCA) plot, Volcano diagram, and Heatmap were generated by using R package (ggplot2 v3.5.0, and pheatmap v1.0.12). Gene Set Enrichment Analysis (GSEA) was performed by using R package (clusterProfiler v4.10.1).
Statistical and reproducibility
All data are presented as mean ± SD. Animal experiments were performed with N = 5-8 independent biological replicates per group. Cell experiments have three or four cell culture batches per group (N = 3-4). Sample size calculation was performed using an online calculation tool (http://powerandsamplesize.com/Calculators/). One-way ANOVA with Tukey’s post-test was conducted for multiple comparisons (except for histology score). Two-tailed Student’s t-test was used to compare two independent groups. All data were checked for normal distribution and homogeneity of variance before analysis. Confounders were not controlled. No exclusion criteria were set. For mRNA-seq, the sample OVA-5 was excluded as it was a clear outlier by PCA. The criterion of statistical significance was P <0.05. Statistical analyses of the data were conducted with GraphPad Prism software (8.0 version).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Numerical source data are included in the main text (Supplementary data file). mRNA sequencing files have been deposited into the GEO public database under accession number GSE295506. The dataset GSE49705 can be obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49705).
References
Chen, P. C. et al. Early-life EV-A71 infection augments allergen-induced airway inflammation in asthma through trained macrophage immunity. Cell Mol. Immunol. 18, 472–483 (2021).
Sun, X. et al. Anti-inflammatory mechanisms of the novel cytokine interleukin-38 in allergic asthma. Cell Mol. Immunol. 17, 631–646 (2020).
Ren, P. et al. Impairment of alveolar macrophage transcription in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med 175, 1151–1157 (2007).
Saradna, A., Do, D. C., Kumar, S., Fu, Q. L. & Gao, P. Macrophage polarization and allergic asthma. Transl. Res 191, 1–14 (2018).
Jiang, X., Du, M. R., Li, M. & Wang, H. Three macrophage subsets are identified in the uterus during early human pregnancy. Cell Mol. Immunol. 15, 1027–1037 (2018).
Huang, R., Wang, X., Zhou, Y. & Xiao, Y. RANKL-induced M1 macrophages are involved in bone formation. Bone Res 5, 17019 (2017).
Bosnjak, B., Stelzmueller, B., Erb, K. J. & Epstein, M. M. Treatment of allergic asthma: modulation of Th2 cells and their responses. Respir. Res 12, 114 (2011).
Robbe, P. et al. Distinct macrophage phenotypes in allergic and nonallergic lung inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L358–L367 (2015).
Lee, J. W. et al. The Role of Macrophages in the Development of Acute and Chronic Inflammatory Lung Diseases. Cells 10, https://doi.org/10.3390/cells10040897 (2021).
Han, X. et al. RNA m(6)A methylation modulates airway inflammation in allergic asthma via PTX3-dependent macrophage homeostasis. Nat. Commun. 14, 7328 (2023).
Jones, S. An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004).
Huerta-Yepez, S. et al. Hypoxia inducible factor promotes murine allergic airway inflammation and is increased in asthma and rhinitis. Allergy 66, 909–918 (2011).
Poulain-Godefroy, O. et al. The Aryl Hydrocarbon Receptor in Asthma: Friend or Foe? Int. J. Mol. Sci. 21, https://doi.org/10.3390/ijms21228797 (2020).
Theofani, E. et al. TFEB signaling attenuates NLRP3-driven inflammatory responses in severe asthma. Allergy 77, 2131–2146 (2022).
Cook, M. E., Jarjour, N. N., Lin, C. C. & Edelson, B. T. Transcription Factor Bhlhe40 in Immunity and Autoimmunity. Trends Immunol. 41, 1023–1036 (2020).
Jarjour, N. N. et al. BHLHE40 Promotes T(H)2 Cell-Mediated Antihelminth Immunity and Reveals Cooperative CSF2RB Family Cytokines. J. Immunol. 204, 923–932 (2020).
Hu, X. et al. Bhlhe40 deficiency attenuates LPS-induced acute lung injury through preventing macrophage pyroptosis. Respir. Res 25, 100 (2024).
Rauschmeier, R. et al. Bhlhe40 and Bhlhe41 transcription factors regulate alveolar macrophage self-renewal and identity. EMBO J. 38, e101233 (2019).
Lin, C. C. et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun. 5, 3551 (2014).
Chu, Y. B. et al. Irf1- and Egr1-activated transcription plays a key role in macrophage polarization: A multiomics sequencing study with partial validation. Int Immunopharmacol. 99, 108072 (2021).
Bigalke, J. M. et al. Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain. Sci. Adv. 5, eaau4202 (2019).
Sato, S. et al. Sputum Neurturin Levels in Adult Asthmatic Subjects. J. Asthma Allergy 16, 889–901 (2023).
Mauffray, M. et al. Neurturin influences inflammatory responses and airway remodeling in different mouse asthma models. J. Immunol. 194, 1423–1433 (2015).
Michel, T. et al. Increased Th2 cytokine secretion, eosinophilic airway inflammation, and airway hyperresponsiveness in neurturin-deficient mice. J. Immunol. 186, 6497–6504 (2011).
Connolly, E. et al. Neurturin regulates the lung-resident macrophage inflammatory response to viral infection. Life Sci Alliance 3, https://doi.org/10.26508/lsa.202000780 (2020).
Salameh, L. et al. In Silico Bioinformatics Followed by Molecular Validation Using Archival FFPE Tissue Biopsies Identifies a Panel of Transcripts Associated with Severe Asthma and Lung Cancer. Cancers (Basel) 14, https://doi.org/10.3390/cancers14071663 (2022).
Machiels, B. et al. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat. Immunol. 18, 1310–1320 (2017).
Zaslona, Z. et al. Resident alveolar macrophages suppress, whereas recruited monocytes promote, allergic lung inflammation in murine models of asthma. J. Immunol. 193, 4245–4253 (2014).
Misharin, A. V. et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J. Exp. Med 214, 2387–2404 (2017).
Draijer, C. & Peters-Golden, M. Alveolar Macrophages in Allergic Asthma: the Forgotten Cell Awakes. Curr. Allergy Asthma Rep. 17, 12 (2017).
Kim, Y. M., Kim, Y. S., Jeon, S. G. & Kim, Y. K. Immunopathogenesis of allergic asthma: more than the th2 hypothesis. Allergy Asthma Immunol. Res 5, 189–196 (2013).
Lee, C. C., Huang, H. Y. & Chiang, B. L. Lentiviral-mediated GATA-3 RNAi decreases allergic airway inflammation and hyperresponsiveness. Mol. Ther. 16, 60–65 (2008).
Wang, M. C. et al. Sophoraflavanone G from Sophora flavescens Ameliorates Allergic Airway Inflammation by Suppressing Th2 Response and Oxidative Stress in a Murine Asthma Model. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms23116104 (2022).
Liou, C. J. et al. Sesamol Alleviates Airway Hyperresponsiveness and Oxidative Stress in Asthmatic Mice. Antioxidants (Basel) 9, https://doi.org/10.3390/antiox9040295 (2020).
Lu, C. et al. Piperlongumine reduces ovalbumin‑induced asthma and airway inflammation by regulating nuclear factor‑kappaB activation. Int J. Mol. Med 44, 1855–1865 (2019).
Lin, C. C. et al. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J. Exp. Med 213, 251–271 (2016).
Huynh, J. P. et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med 215, 1823–1838 (2018).
Divangahi, M., Desjardins, D., Nunes-Alves, C., Remold, H. G. & Behar, S. M. Eicosanoid pathways regulate adaptive immunity to Mycobacterium tuberculosis. Nat. Immunol. 11, 751–758 (2010).
Redford, P. S. et al. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 40, 2200–2210 (2010).
Uyeda, M. J. et al. BHLHE40 Regulates IL-10 and IFN-gamma Production in T Cells but Does Not Interfere With Human Type 1 Regulatory T Cell Differentiation. Front Immunol. 12, 683680 (2021).
Qian, G. et al. B-cell-derived IL-10 promotes allergic sensitization in asthma regulated by Bcl-3. Cell Mol. Immunol. 20, 1313–1327 (2023).
Girodet, P. O. et al. Alternative Macrophage Activation Is Increased in Asthma. Am. J. Respir. Cell Mol. Biol. 55, 467–475 (2016).
Hammad, H. & Lambrecht, B. N. The basic immunology of asthma. Cell 184, 2521–2522 (2021).
Xue, J. et al. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun. 6, 7158 (2015).
Liu, Y., Wu, M., Zhong, C., Xu, B. & Kang, L. M2-like macrophages transplantation protects against the doxorubicin-induced heart failure via mitochondrial transfer. Biomater. Res 26, 14 (2022).
Jarjour, N. N. et al. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat. Immunol. 20, 687–700 (2019).
Kotzbauer, P. T. et al. Neurturin, a relative of glial-cell-line-derived neurotrophic factor. Nature 384, 467–470 (1996).
Zhang, H. et al. AMFR drives allergic asthma development by promoting alveolar macrophage-derived GM-CSF production. J. Exp. Med. 219, https://doi.org/10.1084/jem.20211828 (2022).
Gundra, U. M. et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123, e110–e122 (2014).
Singh, A. et al. Nanoparticle targeting of de novo profibrotic macrophages mitigates lung fibrosis. Proc. Natl Acad. Sci. USA 119, e2121098119 (2022).
Acknowledgements
This work was supported by the Basic Scientific Research Project of Colleges and Universities of Liaoning Province (Key Program, No. LJKZ0746) and the Basic Scientific Research Project of Colleges and Universities of Liaoning Province (General Program, No. LJKZ0746) and the Natural Science Foundation of Liaoning Province of China (No. 2020-MS-184) and the Science and Technology Plan of Liaoning Province (No. 2023JH2/101700187).
Author information
Authors and Affiliations
Contributions
Conceptualization: Bing Dai, Li Chen, Si Liu, and Lishen Shan. Methodology: Xiaowen Li, Qianlan Zhou, and Qinzhen Zhang. Investigation: Bing Dai, Li Chen, and Xiaowen Li. Validation: Lina Han, Wenxin Shen, and Qing Chang. Data curation: Li Chen, Lina Han, and Wenxin Shen. Writing - Original Draft: Bing Dai, Li Chen, and Xiaowen Li; Writing - Review & Editing: Bing Dai, Li Chen, Si Liu, and Yuhong Zhao. Supervision: Si Liu and Yuhong Zhao; Funding acquisition: Lishen Shan, and Qing Chang. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
No human participants involved in this article. All experimental procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University.
Peer review
Peer review information
Communications Biology thanks Chih-Chung Lin, Anna Podlesny-Drabiniok and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Si Ming Man and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dai, B., Chen, L., Li, X. et al. BHLHE40 contributes to allergic asthma progression in mice through NRTN downregulation in macrophages. Commun Biol 8, 863 (2025). https://doi.org/10.1038/s42003-025-08288-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08288-1