Abstract
Understanding the robustness of ecological networks against sustained species losses is paramount to devising effective biodiversity conservation strategies. To explore the impacts of species losses on network robustness (the capacity of food webs to withstand primary extinctions), we used a trophic metaweb of 7808 vertebrates, invertebrates and plants and 281,023 interactions across Switzerland. We inferred twelve regional multi-habitat food webs and simulated non-random species extinction scenarios on these webs, focusing on broad habitat types and regional species abundances. Here, we show that targeted removal of species associated with specific habitat types, particularly wetlands, resulted in greater network fragmentation and accelerated network collapse compared to random species removals. Networks were more vulnerable to the initial loss of common rather than rare species. These findings underscore the critical need for integrated conservation strategies maintaining a diverse mosaic of habitats in a landscape to mitigate the cascading effects of species loss.

Similar content being viewed by others
Introduction
Species extinctions can trigger bottom-up extinction cascades driven by the loss of resources for consumers higher up in the food web, both in terrestrial and freshwater systems1,2,3,4. Loss of plant diversity in multi-trophic food webs, for example, can lead to strong direct negative effects on herbivores, and mediate indirect effects on taxa in even higher trophic levels through bottom-up trophic cascades5. Cascading extinctions lead to the loss of species (nodes) as well as the interactions (links) they share within the food web, which portrays the flow of energy and material responsible for maintaining myriad ecosystem functions6 within and between habitats in a landscape7.
Ongoing habitat loss and degradation, land-use intensification and climate change drive species extinctions8. The extent of these extinctions is at least partly dependent on community structure and network robustness of food webs2,8. Over the past few decades, land-use changes have resulted in the degradation and even loss of freshwater9,10,11, terrestrial, and transitional (e.g., wetland)12 habitats and species dependent on them13,14. Species can link these different habitat types through their dispersal and movement as well as through resource use across multiple habitat types or of different habitat types by juvenile and adult stages15,16,17. Thus, the loss of one type of habitat can lead to the loss of species within as well as between different habitat types connected by the flow of species and energy18. As global change drivers and ecosystem functions operate at local and regional scales19, these extinction cascades can ripple across the mosaic of habitat types in a landscape20 and potentially impact the food webs of entire regions21,22.
The robustness of a food web can be defined as its capacity to withstand primary species extinctions without undergoing significant changes in its structure and function as a result of secondary species losses23,24,25. By modelling the food web as a network, its robustness can be inferred using topological metrics as proxies for structural and functional stability. Certain topological characteristics of food webs are associated with higher robustness. While small-scale food webs can remain robust while being highly connected, as food webs grow larger and more complex, robustness requires low connectance (proportion of realised interactions compared to all possible interactions26) to avoid food webs collapsing under their own complexity26. Topological metrics are useful to describe and compare static food web properties related to the network’s ability to withstand perturbations, but they do not provide a comprehensive understanding of network robustness. A more extensive approach to measuring robustness to secondary extinctions is to conduct a perturbation analysis, where species loss is simulated, and changes in network structure are measured along the extinction sequence.
The inevitable effect of species loss through primary and secondary extinctions in a food web is network fragmentation, when one fully connected network breaks apart into fully isolated sub-networks, known as weakly connected components (henceforth WCCs). While breaking into loosely connected sub-networks can make the food web more modular, ensuring compartmentalisation of perturbations27, network fragmentation may restrict energy flow between species and limit species interactions to the small subsets of the former network28,29,30. As many ecosystem functions are outputs of these energy fluxes, increased network fragmentation can cause a reduction in the efficiency of ecosystem functions. The extent of these consequences depends on the number and size of the WCCs, providing a different aspect of food web robustness. Though the number of such components alone have not been shown to be related to food web stability30, measuring the change in size of the largest remaining component (henceforth: robustness coefficient) has been demonstrated to be a solid measure to evaluate robustness to sustained extinctions across many types of real-world networks, including empirical food webs31. Classically, studying the robustness of food webs to species extinctions has focused on the extent of secondary extinctions23. Yet the addition of a network fragmentation framework based on WCC-based metrics can be instrumental in studying the robustness of food webs experiencing continued species losses31.
The extinction probability of a species in response to habitat loss can be driven by factors such as geographic range size and abundance32,33. Rare species are more vulnerable to disturbances due to their small population sizes and/or restricted geographic ranges and/or occurrence in restricted habitats34,35. Simultaneously, rare species have also been shown to possess unique functional traits and contribute to ecosystem functioning3,35,36,37,38. Nevertheless, the abundance of common species has been shown to play a more central role than species richness in the maintenance of ecosystem functions39, through more persistent contributions compared to rarer species that are less connected within food webs40. Thus, the loss of rare species is expected to be less detrimental than the loss of common species. Simulations of species extinctions have demonstrated that food webs can be robust against random but fragile against non-random species extinction sequences, such as removing generalist29 or highly connected species23,41. Yet, such species are often highly abundant42, and while they can still experience a reduction in their abundance43, are less likely to become extinct under current anthropogenic stressors than rarer species. Thus, there is a need to understand how food webs respond to ecologically meaningful extinction scenarios, especially at the larger spatial scales at which global changes operate.
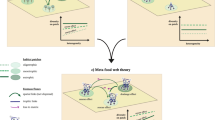
Simulating potential extinction scenarios at the regional scale requires comprehensive and spatially explicit knowledge of species occurrences and their interactions, as well as habitat associations. The use of a metaweb, representing all known potential interactions within a defined area44, provides a promising approach to streamlining existing trophic information. By assuming that the ability of a consumer species to feed on potential resources is fixed and combining this with information on local community composition, sub-networks of smaller scales can be inferred from the metaweb45 (Fig. 1a). A metaweb contains many potential interactions, which may not be realised in nature44, but local co-occurrence trims the potential of all ever-known interactions to those potentially occurring in a subset of the full extent of the metaweb. The spatial scale of the sub-networks may range from sites covering a few metres to kilometres46, to entire biogeographic regions47—albeit with increasing spatial scale, there is also an increase in the uncertainty of potentially “realised” interactions. Nonetheless, this approach provides the possibility to utilise existing data to create potential subsets of the metaweb which are standardised and comparable across space.
a The metaweb approach. All potential documented interactions between species in a region (here, Switzerland) were archived in a knowledge-based metaweb. By combining this with empirical knowledge of site-level co-occurrences (here, biogeographic regions and elevational groupings), site-level food webs can be inferred. b Building a literature-based metaweb. Trophic interaction data were collected from existing primary and secondary sources, expert opinion and online voluntary science repositories. For generalist taxa, trophic interactions that were recorded at taxonomic low resolution (genus or family) were inferred to the species level. Interactions were trimmed to remove unlikely interactions in which the two species did not share environmental associations. Both the empirical and inferred interactions were combined to form a knowledge-based metaweb. The animal and plant icons were sourced from Flaticon.com.
Here, we used a comprehensive metaweb compiled for the entirety of Switzerland to assess the impact of realistic extinction scenarios on regional food web robustness. By incorporating a wide range of animal and plant species alongside their habitat-type associations and national-scale occurrences as a proxy for abundance48, this represents a significant advancement in the study of regional food webs. By integrating this with historical data on species distributions in biogeographic regions and different elevational groupings, we can construct detailed potential regional sub-networks that can be used to simulate ecological scenarios of species loss. This approach allows us to explore how species abundances and their dependencies to multiple habitats may influence the robustness of regional food webs through their biotic dependencies, particularly in the face of realistic and sustained species extinction sequences. It additionally allows us to find potential large-scale spatial patterns in their responses. Specifically, we asked: (1) how the connectance of regional food webs vary between biogeographic regions and by elevation, (2) how regional food webs respond to a targeted removal of species associated with specific habitats, and (3) how regional food webs respond to a targeted removal of species by their relative regional abundance.
Here, we demonstrate that in comparison to random scenarios, regional multi-habitat food webs are more vulnerable to scenarios in which species associated with certain habitat types (especially wetland-associated species) experienced higher probabilities of extinction. These findings highlight how species loss in one habitat can have cascading effects across entire regions due to trophic connections linking multiple habitats. Furthermore, the removal of common species had a more severe negative impact on food web robustness than the removal of rarer species, indicating that common species contribute more strongly to maintaining regional robustness. As global change continues to drive non-random species losses across spatial and temporal scales, conservation efforts must account for the complex responses arising from habitat associations and species abundances.
Methods
Study area and network data
We conducted our study in Switzerland (ca. 41,000 km2) in central Europe, a region characterised by high variability in environmental conditions, with elevation ranges from 196 to 4634 m a.s.l49., in landscapes ranging from densely populated lowlands to remote alpine peaks. Switzerland can be classified into six distinct biogeographic regions (henceforth BGRs): Jura Mountains, Central Plateau, Northern Alps, Eastern Central Alps, Western Central Alps, and Southern Alps50 (Fig. 2a). Around 86,000 multicellular species are estimated to occur within these regions, of which around 56,000 have been identified51.
a a map of the six biogeographic regions of Switzerland. A mean digital elevation raster map (25-m resolution) is overlaid, in which brown values represent lower elevations, whiter values represent intermediate elevations, and grey values represent high elevations. The ranges represent the ranges of the raster map. b a network diagram visualising interactions between major groups in the trophiCH metaweb. The nodes represent the major groups, while the arrows represent their interactions. The width of the arrows represents the number of interactions shared between those groups. c lollipop plots of species richness (left) and interaction richness (left) of the 12 regional food webs in this present study. Grey lollipops represent the montane regions and brown lollipops represent the lowland regions. d a chord diagram illustrating the shared habitat associations of the species in the metaweb. The shared bands represent the number of species, increasing with bandwidth. The animal and plant icons were sourced from Flaticon.com.
Due to a long natural history tradition, the spatial distribution of around 10,000 of these species is well-documented52. Moreover, the recently compiled trophiCH metaweb contains information on 1,112,877 potential trophic interactions (i.e., feeding interactions) between 23,022 plant and animal species, including 126 feeding guilds for Switzerland48. The feeding guilds correspond to non-species level groups treated as single nodes in the network. These nodes consist of large resource pools such as detritus and algae as well as groups that could not be resolved at a higher taxonomic resolution and thus grouped into single nodes, e.g., fungi, isopods, some dipteran and beetle families53. The potential interactions presented in this metaweb are primarily based on a compilation of empirical data on trophic interactions, ranging from direct observations, existing data sets, primary and grey literature, to expert knowledge53 (Fig. 1b). For many species with generalised feeding behaviours, their diet information was often only present at the family or genus levels. As such, taxonomically low-resolution trophic data had been used to infer species-level interactions53,54. For example, if a spider species was known to feed on a family of beetles, the spider species was assumed to potentially feed on all beetle species within that family53 (Fig. 1b). These inferred interactions were further trimmed using a geographic distribution model55 based on matching the associations of potentially interacting species to habitat types and their vertical stratification within the habitat (Fig. 1b). Full details on the methods used to build and validate the trophiCH data set are available within the associated data paper53.
In this study, we used the complete trophiCH data set, trimmed to include only species for which spatial data was readily available in our region (Fig. 2a). The final metaweb included 281,023 interactions between 7692 species (3205 vascular plants, 425 vertebrates, 740 arachnids, 2942 insects, 237 molluscs, 143 other invertebrates) and 116 feeding guilds (Fig. 2b). Using this metaweb, we inferred twelve food webs (henceforth: regional multi-habitat food webs) based on the co-occurrence of taxa within a combination of the six BGRs and two elevation groupings: montane (above 1200 m a.s.l) and lowland (below 1200 m a. s.l.; Fig. 2a, c).
We assigned BGR associations as classified by Flora Indicativa for 2789 plant species56. For the remaining 416 plant and all 4487 animal species, we extracted BGR associations by intersecting a polygon of the BGR map57 with raw species occurrence data from InfoSpecies, the Swiss Information Centre for Species52. For the distribution of species between the two elevation groupings, we applied the existing classifications of the Flora of the European Alps, which classified plant species as montane if they were limited to above 1200 m a.s.l., as lowland if they are limited to 1200 m a.s.l. and below, and as both, if they occur in both elevational groupings58. This information was available for 3100 plant species. For the remaining 105 plant and all 4487 animal species, we intersected occurrence data points from the InfoSpecies data set with a digital elevation model raster at 25-m resolution59.
We classified species as montane, lowland, or both based on their elevational distributions using a percentile-based approach. For each species, we calculated the fraction (F) of occurrences above 1200 m a.s.l. and the absolute number (A) of montane occurrences. Species exceeding a chosen fraction threshold (e.g., 97.5th percentile) were initially classified as montane, while all others were assigned to the lowland category. Species classified as lowland were subsequently reassigned to both if their absolute number of montane occurrences exceeded a second, abundance-based threshold (e.g., 75th percentile). We systematically tested 81 × 81 combinations of F and A thresholds for the species where occurrence data and Flora Alpina classifications were available, ranging from the 60th to 100th percentiles in 0.5% steps, totalling 6561 combinations. For the species with available data, each combination’s resulting classification was evaluated against the expert-based classifications from Flora Alpina. The most accurate classification (denoted F75 × A97.5) achieved 86.15% agreement and was used for the main analyses. To assess the robustness of the classification, we performed a sensitivity analysis using seven additional threshold combinations: F70 × A95, F70 × A97.5, F75 × A95, F75 × A97.5, F80 × A95, F80 × A97.5, and a control classification based on strict presence: species were assigned as montane or lowland if they occurred exclusively in one elevation zone, or both if present in both. Results from the linear models and robustness simulations were consistent across all thresholds (Supplementary Figs. 1, 2). Thus, we report the results based on the F75 × A97.5 classification, which aligned best with Flora Alpina’s classification. Where Flora Alpina classifications were available, they were used directly, overriding our percentile-based assignments.
We filtered the original species list into twelve regional assemblages using the presence of the species in the BGRs and elevation groupings. Then, we created regional multi-habitat food webs for the twelve regions by filtering the metaweb by the twelve regional assemblages and retaining only the interactions between species co-occurring within the regions (Fig. 2c).
For the classification of species’ associations to habitat types, we used the habitat-association data set provided with the trophiCH metaweb48, which classifies the association of each species to one or more of the nine broad habitat types that correspond to the TypoCH habitat classification60 for Switzerland. This classification includes agricultural, aquatic, barren, grassland, forest, ruderal, shrubland, urban, and wetland habitat types. This information was based on the standardisation of habitat-type associations from existing literature and inferences based on a combination of occurrence data and the Habitat Map of Switzerland53,61. While 1173 species were habitat specialists in this broad classification, most species associations included multiple habitat types (median = 4, n = 7692, Fig. 2d). At the national scale, an aggregation at this coarse level of habitat type pools together species that may never overlap, such as species in lowland intensive low pH meadows and high alpine high pH meadows. However, we aimed to minimise this occurrence through the spatial segregation of the metaweb into both the BGRs and elevation groupings, such that highly unlikely co-occurrences are minimised.
To obtain species abundances, we used total observation counts for the species in Switzerland, provided by InfoSpecies52. As this data was unavailable for 45 (0.5%) species, the abundance-related simulations (see section: Simulations of species extinction scenarios) were performed using a second set of twelve regional food webs that excluded these species and their interactions, along with other species only associated with them.
Comparison of differences in network structure with changes in relative habitat cover
We calculated the proportional cover of each broad habitat type within the twelve regions (six BGRs and two elevation groupings). We intersected a rasterised version of the Habitat Map of Switzerland61 at a 1-m resolution with a 25-m digital elevation model59 and the BGR polygonal map57. We summed the pixel count of each habitat type per region and calculated the relative habitat cover from these pixel counts. We calculated the relative habitat cover for agricultural, aquatic, grassland, forest and wetland habitat types and the other less productive habitat types (shrubland, ruderal, barren and urban). Spatial analyses were conducted using the rasterio62 and geopandas63 libraries in Python. We conducted a principal component analysis (PCA)—using the prcomp function in R—to determine whether relative habitat compositional differences between the BGRs and elevations can be evidenced between the twelve regions. Then, we used a multiple linear regression with the elevational grouping, the first (PC1) and second (PC2) principal components as predictor variables and connectance as the fixed response variable. We used the lm function in R to fit models. We calculated connectance using the edge_density() from the igraph64 package.
Simulations of species extinction scenarios
For each of the regional multi-habitat food webs, we ran simulations of species extinction scenarios and monitored metrics related to robustness along the simulations. At each time step of the simulation, one species was removed (a primary extinction). Subsequently, any species for which the primary extinction led to a lack of other dietary sources were also removed (secondary extinctions; Fig. 3). Additionally, plant species that became fully isolated from the network (as no consumer remained) were removed, as they no longer played a functional role in maintaining the network’s structure. The sequence continued until only a few highly connected non-species-level nodes remained. These nodes, which form essential functional feeding guilds, were excluded from removal due to their low likelihood of complete extinction. Specifically, each of these guilds was treated as a single node, and not removed: Acari, algae, Annelida, bones, blood, carrion, collembola, Copepoda, detritus, fungi, faeces, honeydew, Isopoda, keratin, lichen, macrozoobenthos, microbes, mosses, Nematoda, plankton, particulate organic matter, Psocoptera, invertebrates, vertebrates, animals, and plants. The latter four nodes were connected to hypergeneralist species for which no species-level resources were available.
Circles represent nodes (species), and the black lines between them represent links (interactions). The initial network on the far left undergoes three types of node removal sequences with up to five primary node removal steps. a the ideal scenario in which primary losses (green nodes) result in no secondary losses or the loss of secondary interactions (dark grey dashed lines), while the other two (b, c) scenarios represent less ideal sequences in which primary losses (dark and light pink, respectively result in secondary losses (light grey nodes). The nodes with a black outline represent the nodes that form the largest WCC. d the function for the robustness coefficient is present, and on the bottom right, the graph shows the evolution of size of the largest WCC (y-axis) as nodes are removed (x-axis).
We simulated five scenarios in which we increased the initial removal probability of species associated with one of five habitat types (agroecosystems, aquatic ecosystems, forests, grasslands and wetlands) such that species associated with the chosen habitat type to any degree were more likely to be first removed. For each species, we computed the proportion of the habitat type to the total number of associated habitats in which the species may occur to determine the degree to which the species was a habitat specialist. Then, we built probability subgroups based on all the unique combinations of habitats that all species could be associated with, as follows. For example, in a simulation targeting grassland-associated species, the subgroup of species associated with grasslands alone would have the highest extinction probability. In contrast, the subgroup of species associated with grasslands and one other habitat type would have half the likelihood of the first. Species associated with grasslands and two other habitat types would have a third of the likelihood of the first, and so on. Additionally, we set the probability of the subgroup of species not associated with the targeted habitat type to an estimated background extinction rate of 0.1 extinctions per million species per year65, multiplied by the number of species per region, to determine a background minimum probability of extinctions per year. Then, the other probabilities for each subgroup were back-calculated from this value, such that they formed a valid probability distribution. At each time step of the simulation, a subgroup was first chosen according to these probabilities, and a species chosen randomly from within the subgroup was removed (Further details: Supplementary Methods Note 1). We calculated all probabilities using the NumPy package66 in Python using rng.choice.
In parallel, we simulated two scenarios in which we increased the initial removal probability of rare species or, alternatively, common species. The removal probability for the rare species extinction scenario was set as proportional to the inverse of its abundance in Switzerland. Conversely, in the scenario where common species were more likely to be removed first, we set the species’ removal probability as proportional to the species’ normalised abundance count. We also simulated scenarios in which species were removed randomly (i.e., all species are assigned the same removal probability) as a control. We ran 1000 simulations per region and scenario for a total of 108,000 simulations.
To monitor network robustness over the removal time step, we measured metrics related to total network fragmentation, specifically the size of the largest WCC (Weakly Connected Component: the most species-rich sub-network of the ensemble of isolated connected sub-networks constituting the food web), and the cumulative number of secondary extinctions. Due to computer calculation limitations, we measured network fragmentation metrics at every tenth primary extinction step, corresponding to the removal of 10 species at a time. This equates to a step size of approximately 0.20% species loss in the largest food web (4978 species, the lowland central plateau food web, Fig. 2c) and up to 0.99% in the smallest (1012 species, the montane central plateau food web, Fig. 2c). For every primary extinction step, we calculated the size of the largest WCC, and the species richness. We used the weakly_connected_components and number_of_nodes functions from the NetworkX package67 in Python to extract the size of the largest WCC and the total species richness. To calculate the cumulative number of secondary extinctions at each primary extinction step, we subtracted the observed species richness at each step from the expected species richness (i.e. assuming no secondary extinctions).
For each region and scenario, we calculated the median, interquartile ranges and the 95th percentile confidence intervals of the metrics based on the 1000 simulations. For each curve, we additionally calculated the robustness coefficient31. In an ideally robust network, the size of the largest WCC should decrease linearly under sustained primary extinctions25 (Fig. 3). However, the more vulnerable a network, the faster the size will decrease31. The robustness coefficient is defined as the ratio of the area under the curve of the size of the largest WCC between observed and ideal extinction sequences31. It has been proposed that the robustness coefficient could define the topological robustness of any type of network under sustained attacks31. We subsequently calculated robustness coefficients for all simulations and compared them to the ideal values. We performed non-parametric Kruskal-Wallis and post-hoc Dunn tests to determine significant differences in robustness coefficients between the targeted scenarios and the null model and calculated pairwise Cliff’s delta effect sizes to measure the magnitude of the differences.
Software
All simulations were conducted in Python 3.11.468; food webs were manipulated using the NetworkX package67. All statistical analyses were conducted in R 4.3.269 and RStudio 2023.12.1 + 40269 using the igraph64, dunn.test70 and effsize71 packages.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Comparison of differences in network structure with changes in relative habitat cover
We performed a PCA to explore the relationships among the different proportions of habitat types within the twelve regions. The first two principal components (PCs) explained 68.55% of the total variance (Fig. 4a). The first PC (PC1: 50.79% explained variance) primarily differentiated larger proportions of anthropogenic or disturbed (urban, ruderal, agroecosystem) or aquatic habitat, which showed strong positive loadings, from larger proportions of grasslands and shrublands, which had stronger negative loadings. The second PC (PC2: 17.76% explained variance) differentiated a strong positive loading for barren cover against strong negative loadings for wetlands and forests. The montane regions showed a trend towards more negative values for PC1, while the lowland regions showed a trend towards more positive values for PC1. The more negative loadings for PC2 were driven by the “montane” regions in biogeographic regions with relatively low elevation (central plateau, jura mountains). In general, all regions contained larger proportions of forest and grassland habitats in comparison to other habitats (Fig. 4b).
a principal component analysis showing the changes in relative fractions of habitat types (arrows) within the 12 regions. Brown points represent lowland regions, while grey points represent montane regions. The size of the point represents connectance. b bar plots representing the relative fractions of habitat types per biogeographic region for the montane (left) and the lowland (right) regions. The biogeographic regions are Eastern Central Alps (EA), Jura Mountains (JU), Central Plateau (CP), Northern Alps (NA), Southern Alps (SA) and Western Central Alps (WA).
We fitted linear models to examine the effect of differences in habitat proportions on connectance in the regional multi-habitat food webs (Supplementary Table 1). The fixed effects included the categorical variable concerning the elevation grouping (montane vs lowland), PC1, and PC2. Connectance showed a significant negative relationship with PC2 (β = −0.001, p = 0.002, n = 12), indicating that connectance tends to decrease along the PC2 gradient, towards regions with higher cover of barren regions (Fig. 4a). Connectance showed a slight trend towards increasing along the PC1 gradient (β = 0.001, p = 0.077, n = 12). The elevational grouping also had a significant negative effect (β = −0.012, p < 0.001, n = 12); lowland food webs demonstrated lower connectance than montane food webs. The model explained 93.8% of the variance in connectance (Adjusted R² = 0.815, F (3,8) = 56.42, p < 0.001, n = 12).
Simulations of species extinction scenarios by habitat type
As the BGRs showed similar overall results for all simulations, we only present the results of the region with the most even distribution of montane and lowland regions, the Northern Alps (Fig. 5; for the other regions, see Supplementary Figs. 3–12). Moreover, we focus on the early part of the simulation, before all species in the relevant habitat have been removed. After this, the species were removed fully randomly. All scenarios targeting species in specific habitat types first experienced steeper increases in cumulative secondary extinctions in comparison to the random model (Fig. 5a–e). In the montane regions, all non-random simulations experienced an earlier network collapse than the random scenario (after 1627 ± 15 primary extinctions, n = 1000), but the scenarios targeting wetlands experienced the fastest network collapse after 1531 ± 5 primary extinctions (n = 1000, Fig. 5a). Concurrently, the scenarios targeting montane wetlands experienced the steepest increases in cumulative secondary extinctions (Fig. 5a). In the lowlands, the fastest increase in cumulative secondary extinctions were observed in the scenarios in which lowland agroecosystem species were first removed, and earliest network collapse (3791 ± 35, n = 1000) in comparison to the random scenarios (3862 ± 38, n = 1000, Fig. 5e).
a–e represent simulations for singular habitat type extinction scenarios (agroecosystem: yellow, grassland: bright green, aquatic: blue, forest: dark green, wetland: purple) and the random model (black). In each subfigure, the left panels represent the number of cumulative secondary extinctions as a function of the number of primary extinctions. On these panels, curves represent the median of 1000 simulations, the darker ribbons represent the interquartile range, and the lighter ribbons the 95th percentile confidence interval for montanes (above) and lowlands (below) in the Northern Alps biogeographic region. The grey boxes represent the part of the simulation in which species within the targeted habitat type have all been removed. From this point onwards, the simulation is fully random until all species have been removed from the food web. The right panels of the subfigures represent violin boxplots of the robustness coefficient for each extinction scenario and the adjusted p-values for the pairwise Dunn tests and Cliff’s delta effect sizes. The violin plots visualise the distributions of the data points, each representing the outcome of one simulation.
In both montane and lowland multi-habitat food webs, all five scenarios targeting the different habitat types demonstrated significantly lower robustness coefficients compared to the random scenario, with strong negative effect sizes (Fig. 5a–e). In the montane regions, simulations targeting wetlands demonstrated the strongest negative effect sizes (Dunn test: p < 0.001, Cliff’s delta: r = −1.000, n = 2000, Fig. 5a). In the lowland regions, simulations targeting agroecosystems demonstrated the strongest negative effect sizes (Dunn test: p < 0.001, Cliff’s delta: r = −1.000, n = 2000, Fig. 5e).
Montane robustness coefficients were significantly lower than in the lowlands, with strong effect sizes for the scenarios targeting grassland (Dunn test: p < 0.001, Cliff’s delta = -0.989, n = 2000), wetland (p < 0.001, Cliff’s delta = 1.000, n = 2000), aquatic (p < 0.001, Cliff’s delta = -0.831, n = 2000), and forest (p < 0.001, Cliff’s delta = -0.997, n = 2000) species. In the random scenario, robustness coefficients did not differ significantly between montane and lowland regions (p = 0.031, Cliff’s delta = −0.048, negligible effect, n = 2000). Conversely, robustness coefficients for scenarios targeting the removal of agroecosystem-associated species were significantly higher in the montanes than in the lowlands, with a strong effect size (p < 0.001, Cliff’s delta = 0.615, n = 2000).
Simulations of species extinction scenarios by abundance
Compared to the random extinction scenario, removing rare species first resulted in a slower increase in cumulative secondary extinctions in both the montane and the lowland regions (Fig. 6). Moreover, the removal of rare species first resulted in later network collapse than the random scenario in both the montane (after 1577 ± 4, n = 1000) and 1538 ± 15 (n = 1000) mean primary extinctions, respectively) and the lowland regions (after 3867 ± 12, n = 1000 and 771 ± 35, n = 1000, mean primary extinctions, respectively). Contrastingly, the loss of common species resulted in a steeper increase in cumulative secondary extinctions (Fig. 6) and earlier network collapse (after 1429 ± 5, n = 1000) primary extinctions in the montanes and after a mean of 3678 ± 32 (n = 1000) primary extinctions in the lowlands).
The left panels represent the number of cumulative secondary extinctions as a function of the number of primary extinctions for targeted extinctions of rare species (dark pink), common species (mint green) and randomly (black). On these panels, curves represent the median of 1000 simulations, the darker ribbons represent the interquartile range, and the lighter ribbons the 95th percentile confidence interval for montanes (above) and lowlands (below) in the Northern Alps biogeographic region. On the right are boxplots of the robustness coefficient for each extinction scenario and the adjusted p-values for the pairwise Dunn tests and Cliff’s delta effect sizes. The violin plots visualise the distributions of the data points.
Simulations removing rare species first showed significantly higher robustness coefficients with strong effect sizes than the random scenario, both in the montane (p < 0.001, Cliff’s delta = -1, n = 2000, Fig. 6) and lowland (p < 0.001, Cliff’s delta = -1, n = 2000, Fig. 6) regions. For simulations removing rare species first, robustness coefficients were significantly higher in the montane regions in comparison to the lowlands, with a moderate effect size (p < 0.001, Cliff’s delta = 0.437, n = 2000). Conversely, for simulations removing common species first, robustness coefficients were significantly lower in the lowland regions in comparison to the montanes, with a strong effect size (p < 0.001, Cliff’s delta = −1.000, n = 2000). The robustness coefficients for the random scenario showed no significant difference between the montane and lowland regions (p = 0.245, Cliff’s delta = 0.018, n = 2000).
Discussion
By simulating the extinction of species in regional food webs according to different ecological extinction scenarios, we aimed to better understand how habitat degradation and resulting species loss affect biodiversity through cascading effects in food webs across space. We demonstrate that the connectance of regional multi-habitat food webs is driven by different relative proportional covers of habitat types at montane and lowland elevations. Across multiple extinction scenarios, measures of robustness were generally lower in the montane regions than in the lowlands. We also show that—across biogeographic regions and elevation classes—regional multi-habitat food webs are less robust to the scenarios in which species in aquatic, agroecosystem, forest, grassland and wetland habitat types were more likely to become extinct than when species were removed randomly. This implies that in regional food webs encompassing multiple habitats, the effects of removing species associated with certain habitat types can cascade across the whole region through species’ dependencies to multiple habitat types. We also demonstrate that the loss of rarer species is less detrimental to network robustness than the loss of common species. Thus, while rare species can be functionally unique36, common species contributed significantly more to maintaining the robustness of food webs against sustained extinctions. As global change drives non-random species losses across local and regional scales, biodiversity conservation must consider these complex responses driven by between-habitat associations and species abundances.
Differences in proportions of different habitat types can influence the structure of regional food webs. Almost all regions contained relatively higher proportions of grassland and forest cover in comparison to other habitat types (except the montane Eastern Central Alps, Fig. 3b), and we also found more species associated with forest and grassland habitat types (Fig. 3c). This is understandable, as certain types of grasslands (e.g., semi-natural grasslands72) are known hotspots of diversity in central Europe73. Moreover, although the majority of forests in Switzerland have long been heavily managed, they serve as habitat for 60% of recorded animals, plants and fungi74. Indeed, even managed forests serve as habitat for many species, underlining their overall importance for biodiversity75. Connectance was significantly higher in montane than in lowland regions. Additionally, connectance decreased along the second principal component, which captured increasing proportions of less productive (barren) or intensively used (agricultural and urban) habitat types. This suggests that both elevation and land use influence food web structure. The relationship between connectance and elevation remains unresolved across empirical studies, with reported patterns varying in direction and significance76, though connectance generally decreases with increasing species richness24,76,77. Additionally, regions with high human footprint (often strongly negatively correlated with regions with high elevation), demonstrate lower connectance78, likely reflecting differences in species richness, interaction structure, and local environmental filtering76.
Wetlands, grasslands, forests, and even some agroecosystems may often serve as corridors and play important roles in maintaining landscape-level biodiversity and spatial connectivity, which may drive their critical role in maintaining network robustness79. Robustness coefficients of scenarios targeting wetland species were significantly lower than the others in the montane regions, while in the lowlands, this was the case for scenarios targeting agroecosystem species. Lower robustness coefficients are evidence of the fragmentation of the network into larger isolated fragments, rather than the ability to maintain a giant component31. This may point towards a higher probability that species associated with montane wetlands are responsible for maintaining the function of the multi-habitat food web. Small, isolated habitat fragments may disproportionately contribute to regional network robustness by functioning as sources of unique species and interactions80. The removal of all wetland-associated species from the metaweb (Supplementary Fig. 15) results in a notable reduction in the species richness of EPT (Ephemeroptera, Plecoptera and Trichoptera) and vertebrates. These groups are particularly well-known cross-habitat connectors: EPT taxa are aquatic insects with terrestrial adult stages, and vertebrates frequently have large spatial ranges and generalist diets, linking otherwise disparate parts of a region81,82. Most wetlands in Switzerland have long been highly fragmented12, yet even small alpine wetlands in a region can support diverse plant communities and lead to an increase in both local and regional species richness83. Such wetlands in landscapes can play an important role as vegetation refugia for wetland specialists as well as many other upland species83. Although wetland species accounted for 31% of all taxa, their removal led to the loss of 69% of all recorded interactions, indicating that they are involved in a disproportionately high number of trophic links (Supplementary Fig. 15). Therefore, conservation actions prioritising such fragments of wetland habitat may enhance regional network robustness. Our simulations are conducted at a coarser grain size, thus masking some of the fine-scale processes occurring in these smaller habitats. Nevertheless, they provide preliminary insights into broader patterns of biodiversity. In the lowlands, primary extinctions of agroecosystem-associated species led to a faster rate of secondary extinctions. Disturbed landscapes, such as lowland agroecosystems, generally contain fewer plant species and more generalist animals84; both are characteristics of species whose loss would be more likely to trigger secondary extinctions. Thus, the critical impact is likely not due to the loss of agroecosystem specialists, but rather the loss of generalist species, whose disappearance reverberates across the entire regional multi-habitat food web. Robustness coefficients of scenarios targeting aquatic species were significantly higher than the others, providing evidence that the food web retains a larger giant component as primary extinctions continue. Aquatic food webs have been demonstrated to be more highly connected than terrestrial ecosystems47,85,86 and exhibit higher link density, pointing towards more generalised feeding behaviours86, which may result in greater robustness to primary species extinctions. Additionally, while aquatic-terrestrial flows of species and energy are documented87, they are likely to be less influential than flows between different terrestrial habitat types. For example, spillover of species to and from agroecosystems to other habitat types has been documented and demonstrated to be both beneficial to the pollination of crops and detrimental to the pollination of wild plants88. Alternatively, as aquatic and terrestrial food webs have historically been studied separately89, these results may be an artefact due to a lack in existing data on aquatic-terrestrial flows. Nonetheless, we argue that not only do habitat associations play an important role in the robustness of regional food webs to species extinctions, the variation in habitat-type associations as well as the types of potential spillovers between habitat types may drive the decrease in robustness.
Rare species—while remaining functionally crucial36,90 within habitats—may serve a less prominent role in maintaining regional multi-habitat food web robustness due to their low population size, restricted geographic ranges and specific habitat requirements. This is evidenced by our findings that the preferential loss of rare species results in fewer secondary extinctions and higher robustness coefficients than the scenario targeting common species and the random scenario. Common species remain crucial for many ecosystem processes and functions due to their high abundance and potential to be habitat generalists with large geographic ranges39,40. Thus, the loss of common species may result in the faster loss of links between habitats and increased network fragmentation. We demonstrate that the regional food webs are least robust against initial primary extinctions of common species. A moderate positive correlation exists between a species’ relative abundance and the number of recorded interactions in which it partakes (see Supplementary Fig. 13). We additionally assessed how common and rare species contribute to food web structure by examining their influence on nestedness (Supplementary Methods Note 2). Removing the most common species led to a consistent reduction in nestedness across regional food webs, suggesting that these species form a structural core around which rarer species’ interactions are organised (see Supplementary Fig. 14). In the literature, abundant generalist pollinators have been shown to maintain network robustness in agricultural pollination networks under pressure91, and generalist plant species were responsible for the maintenance of network robustness under random extinction scenarios92. Therefore, as our results also confirm, natural food webs may be robust against the loss of rare, often more specialised (see Supplementary Fig. 13) species. However, such realistic extinction sequences may generate food webs where many species are dependent on a few core generalist species93,94. Biotic homogenisation also results in lower β-diversity among local communities through decreased spatial asynchrony and consequent destabilisation of ecosystem functioning95. In homogenised food webs, the loss of these common species would pose a significant threat to food web robustness, as there is a lack of functional redundancy, and all remaining species in the food web must play crucial roles in maintaining its structure and function. While conservation efforts often focus on rare species, anthropogenic stressors have negatively impacted the populations of common species for centuries43. The loss of common species can disrupt community dynamics and lead to negative impacts on ecosystem functions and processes96. Thus, not only rare but also common species with declining populations require conservation intervention and reduction of exposure to anthropogenic stressors. Further studies considering spatially explicit local communities, population declines and the turnover of interactions may shed light on the stability of homogenised networks in space in the face of continuous species loss.
The simulations were built on an empirically based trophic metaweb and a broad habitat-association data set. Biases in the existing data have been carried into this analysis, such as the bias towards higher availability of empirical data for plant-pollinator, bird-plant, and plant-parasite interactions, as well as biases between different types of interactions (e.g., insect predation vs pollination)48. However, the usage of the metaweb approach aims to standardise—if not minimise—these biases, as the biases are equally spread across the inferred regional food webs, making them comparable. A simple geographic model had been utilised to further infer trophic interactions for generalists based on low-resolution interaction data to partly fill these gaps48. For specific taxa, further modelling based on functional or phylogenetic matching for suitable taxa may improve data coverage in the future97,98,99.
A metaweb contains all possible interactions, including those that may not occur as the species may not have co-occurred and have the opportunity to interact despite matching in other traits98. While some of these impossible links have been removed by the inferences of the regional food webs, these regional food webs still represent geographically large regions. Many species within our twelve regional food webs are still unlikely to interact at the local scale, overestimating the connectivity of the networks and the diet breadth of the species. This is particularly true for some inferred interactions in the trophiCH metaweb, which are based on expert knowledge based on habitat associations (e.g., bird-plant interactions)53. Therefore, the results presented here may be an underestimation of the real vulnerability of these regions to sequential extinctions or, alternatively, an overestimation of vulnerability due to the overrepresentation of links that are not locally plausible, leading to increased connectivity.
In contrast to the simulations conducted in this study, trophic interactions within food webs are not static. Firstly, novel interactions may arise as new species co-occur due to climate-induced species range shifts100,101,102 and introductions103,104. Particularly, when generalist non-native species are introduced into a system, they have a tendency to enhance network redundancy and occupy central nodes105. While such species are less likely to engage in novel interactions with specialists, ecosystem functions provided by the interactions may be retained, even though taxonomic diversity and functionally unique interactions are lost99,105. In Switzerland, 1305 species have been classified as non-native, of which 737 species were included in this study (9.43% of the included taxa). Of these, 128 species have been classified as invasive or potentially invasive106. A spatially explicit study with more localised food webs would be necessary to adequately address the impact of novel interactions on food webs in space, as the degree to which non-native species act as disruptors and contributors to ecosystem functionality depends on the spatial resolution and dispersal capacity of the species107. Future studies may benefit from inferring future food webs through co-occurrence based future species distribution models101 and the explicit addition of non-native species as a proxy for novel interactions99. Secondly, declines in prey populations may drive species to switch to other potential resources, especially for consumers with generalised feeding behaviours108. The trophiCH database inferred many interactions for generalists based on a taxonomic approach53 and thus considers many potential and locally unrealised interactions85,98. It has been established that emerging interactions are often limited to within well-defined taxonomic levels, mostly at the genus level3. Thus, our approach implicitly allows for the potential rewiring to resources that the species is known to feed on in other locations. Nevertheless, the potential is limited by our existing knowledge of interactions, and the approach does not account for species switching their diets towards resource taxa with functional—rather than taxonomic—similarities. Over longer time scales, many species may adapt to new modes of feeding completely, rather than immediately becoming extinct109,110. Accounting for adaptation in food webs is challenging due to the complexity and variability of adaptive processes, which can occur through shifts in species’ behaviour, physiology, or evolution over different time scales109. Adaptation depends on local factors such as competition, resource availability, and habitat conditions, and often lags behind rapid environmental changes like habitat loss or climate shifts111,112. Future studies could consider incorporating higher temporal resolution, species-specific rates of adaptation, and local ecological dynamics. However, they may be hindered by data gaps at large spatial scales for regional food webs, which often contain thousands of species. Alternatively, model food webs based on theories such as allometric optimal foraging theory could be employed to explore potential adaptive responses113.
This present study focused on food web topology but does not account for interaction strengths or shifts in species abundances, which are critical for understanding food web dynamics. Species do not disappear from an ecosystem immediately due to disturbance, instead, they experience reductions in population size. While our static approach emphasises bottom-up effects of food web dependencies as species are fully lost, top-down effects are also well-established114, where predator populations regulate entire ecosystems by controlling the prey populations. Spatial fluctuations in initial predator populations may also drive different responses to disturbances115. Incorporating these factors at large spatial scales are often infeasible; however, future studies could benefit from integrating interaction strengths and species abundance data, or modelling, to provide a more comprehensive view of food web dynamics across large regions. Moreover, recent work has demonstrated that while regional context is important for assessing the impact of secondary extinctions of regional networks, results of simulations can differ significantly when species are removed from the whole region or from some local sites within the region116. Further work, including networks resolved at a finer resolution based on information on real-world communities, which also account for regional context, may further increase our understanding of future outcomes of species loss.
We demonstrate clear differences between elevational groups in the response of regional multi-habitat food webs to primary extinctions. However, elevation is correlated with land-use intensity; montane ecosystems are generally less intensively influenced by humans than lowland ecosystems117. Thus, it is challenging to disentangle whether the observed effects between elevational groups are primarily driven by inherent abiotic factors associated with elevation or by differences in land-use intensity.
This study employs a country-based approach, which is relevant for biodiversity conservation conducted at policy-relevant spatial scales118,119,120,121,122. Socio-political boundaries influence biodiversity and efforts to conserve it due to divided governance and management123, yet ecological processes often extend beyond these borders. For example, the ability of a species to move between suitable habitats is not limited by open political boundaries. However, this present study accounts for habitat loss implicitly, through the use of species’ habitat associations and trophic dependencies across habitat types. Spatially explicit explorations of habitat loss, fragmentation and degradation on food web structure would require a consideration of species’ habitat connectivity and resource availability across national borders. Moreover, species may exhibit biotic niche truncation, as species without feeding resources within a biogeographic region in Switzerland may be able to find suitable sources by moving outside the national borders122. We used an empirical metaweb based on documented interactions in and around Switzerland, thus, many interactions for species along the border are already accounted for, even if those interactions have not been documented directly in Switzerland. In addition, our regional food webs represent potential constructs that consider biogeographic regions with large spatial extents. This likely overestimation of the potential interactions occurring at any one location within these regions functions as a way to counteract the potential underestimation of feeding resources due to niche truncation along political boundaries. Future studies should aim to address these border effects explicitly by integrating cross-border data as far as available, to provide a more comprehensive understanding of the effects of habitat fragmentation and species connectivity on food web structure.
Drivers of global change place unequal pressures upon various habitat types across different regions in increasingly fragmented landscapes. Our results underscore the cascading effects of species loss across regional networks containing a legion of species moving through multiple habitat types, emphasising the need for urgent, integrated conservation strategies. The national scale of this study offers a unique opportunity to inform conservation approaches that align efforts across regions, ensuring policies are consistent and coordinated to address habitat connectivity and species interdependencies across diverse ecosystems. Both habitat-specific interventions and broader landscape considerations are necessary to effectively confront the sustained and escalating threats that biodiversity faces today.
Data availability
Raw occurrence data from the Swiss Species Information Centre InfoSpecies (www.infospecies.ch) is available upon request to the organisation. Data produced from this study are available through the Envidat repository124. All other data sets used in this study are publicly available and have been cited within the methods.
Code availability
All code and data needed to run the simulations and statistical analyses are available here on Envidat124. The most up-to-date code reproducing the simulations is available here: https://github.com/mrejichacko/casCHades.
References
Kagata, H. & Ohgushi, T. Bottom-up trophic cascades and material transfer in terrestrial food webs. Ecol. Res. 21, 26–34 (2006).
Kehoe, R., Frago, E. & Sanders, D. Cascading extinctions as a hidden driver of insect decline. Ecol. Entomol. 46, 743–756 (2021).
Pearse, I. S. & Altermatt, F. Extinction cascades partially estimate herbivore losses in a complete Lepidoptera-plant food web. Ecology 94, 1785–1794 (2013).
Strona, G. & Bradshaw, C. J. A. Coextinctions dominate future vertebrate losses from climate and land use change. Sci. Adv. 8, eabn4345 (2022).
Scherber, C. et al. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature 468, 553–556 (2010).
Barnes, A. D. et al. Energy Flux: The link between multitrophic biodiversity and ecosystem functioning. Trends Ecol. Evol. 33, 186–197 (2018).
Thompson, R. M. et al. Food webs: reconciling the structure and function of biodiversity. Trends Ecol. Evol. 27, 689–697 (2012).
Jaureguiberry, P. et al. The direct drivers of recent global anthropogenic biodiversity loss. Sci. Adv. 8, eabm9982 (2022).
Dudgeon, D. et al. Freshwater biodiversity: importance, threats, status and conservation challenges. Biol. Rev. 81, 163–182 (2006).
Gozlan, R. E., Karimov, B. K., Zadereev, E., Kuznetsova, D. & Brucet, S. Status, trends, and future dynamics of freshwater ecosystems in Europe and Central Asia. Inland Waters 9, 78–94 (2019).
Markovic, D. et al. Europe’s freshwater biodiversity under climate change: distribution shifts and conservation needs. Divers. Distrib. 20, 1097–1107 (2014).
Gimmi, U., Lachat, T. & Bürgi, M. Reconstructing the collapse of wetland networks in the Swiss lowlands 1850-2000. Landsc. Ecol. 26, 1071–1083 (2011).
Sandor, M. E., Elphick, C. S. & Tingley, M. W. Extinction of biotic interactions due to habitat loss could accelerate the current biodiversity crisis. Ecol. Appl. 32, e2608 (2022).
Zhang, H. et al. A spatial fingerprint of land-water linkage of biodiversity uncovered by remote sensing and environmental DNA. Sci. Total Environ. 867, 161365 (2023).
Devoto, M., Bailey, S. & Memmott, J. Ecological meta-networks integrate spatial and temporal dynamics of plant–bumble bee interactions. Oikos 123, 714–720 (2014).
Schneider, G., Krauss, J., Boetzl, F. A., Fritze, M.-A. & Steffan-Dewenter, I. Spillover from adjacent crop and forest habitats shapes carabid beetle assemblages in fragmented semi-natural grasslands. Oecologia 182, 1141–1150 (2016).
Frost, C. M., Didham, R. K., Rand, T. A., Peralta, G. & Tylianakis, J. M. Community-level net spillover of natural enemies from managed to natural forest. Ecology 96, 193–202 (2015).
Evans, D. M., Pocock, M. J. O. & Memmott, J. The robustness of a network of ecological networks to habitat loss. Ecol. Lett. 16, 844–852 (2013).
Isbell, F. et al. Linking the influence and dependence of people on biodiversity across scales. Nature 546, 65–72 (2017).
Breviglieri, C. P. B. & Romero, G. Q. Terrestrial vertebrate predators drive the structure and functioning of aquatic food webs. Ecology 98, 2069–2080 (2017).
Pace, M. L., Cole, J. J., Carpenter, S. R. & Kitchell, J. F. Trophic cascades revealed in diverse ecosystems. Trends Ecol. Evol. 14, 483–488 (1999).
Sahasrabudhe, S. & Motter, A. E. Rescuing ecosystems from extinction cascades through compensatory perturbations. Nat. Commun. 2, 170 (2011).
Dunne, J. A., Williams, R. J. & Martinez, N. D. Network structure and biodiversity loss in food webs: Robustness increases with connectance. Ecol. Lett. 5, 558–567 (2002).
Dunne, J. A., Williams, R. J. & Martinez, N. D. Food-web structure and network theory: The role of connectance and size. Proc. Natl. Acad. Sci. 99, 12917–12922 (2002).
Holme, P., Kim, B. J., Yoon, C. N. & Han, S. K. Attack vulnerability of complex networks. Phys. Rev. E 65, 056109 (2002).
Landi, P., Minoarivelo, H. O., Brännström, Å, Hui, C. & Dieckmann, U. Complexity and stability of ecological networks: a review of the theory. Popul. Ecol. 60, 319–345 (2018).
Gilarranz, L. J., Rayfield, B., Liñán-Cembrano, G., Bascompte, J. & Gonzalez, A. Effects of network modularity on the spread of perturbation impact in experimental metapopulations. Science 357, (2017).
Gaichas, S. K. & Francis, R. C. Network models for ecosystem-based fishery analysis: a review of concepts and application to the Gulf of Alaska marine food web. Can. J. Fish. Aquat. Sci. 65, 1965–1982 (2008).
Solé, R. V. & Montoya, M. Complexity and fragility in ecological networks. Proc. R. Soc. Lond. B Biol. Sci. 268, 2039–2045 (2001).
Rodriguez, I. D. & Saravia, L. A. Potter Cove’s Heavyweights: Estimation of Species’ Interaction Strength of an Antarctic Food Web. Ecol. Evol. 14, e70389 (2024).
Piraveenan, M., Thedchanamoorthy, G., Uddin, S. & Chung, K. S. K. Quantifying topological robustness of networks under sustained targeted attacks. Soc. Netw. Anal. Min. 3, 939–952 (2013).
Johnson, C. N. Species extinction and the relationship between distribution and abundance. Nature 394, 272–274 (1998).
Purvis, A., Gittleman, J. L., Cowlishaw, G. & Mace, G. M. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B Biol. Sci. 267, 1947–1952 (2000).
IŞIK, K. Rare and endemic species: why are they prone to extinction?. Turk. J. Bot. 35, 411–417 (2011).
Leitão, R. P. et al. Rare species contribute disproportionately to the functional structure of species assemblages. Proc. R. Soc. B Biol. Sci. 283, 20160084 (2016).
Basile, M. Rare species disproportionally contribute to functional diversity in managed forests. Sci. Rep. 12, 5897 (2022).
Ho, H.-C. & Altermatt, F. Associating the structure of Lepidoptera-plant interaction networks across clades and life stages to environmental gradients. J. Biogeogr. 51, 725–738 (2024).
Burner, R. C. et al. Functional structure of European forest beetle communities is enhanced by rare species. Biol. Conserv. 267, 109491 (2022).
Winfree, R. et al. Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. 18, 626–635 (2015).
Chapman, A. S. A., Tunnicliffe, V. & Bates, A. E. Both rare and common species make unique contributions to functional diversity in an ecosystem unaffected by human activities. Divers. Distrib. 24, 568–578 (2018).
Bellingeri, M., Cassi, D. & Vincenzi, S. Increasing the extinction risk of highly connected species causes a sharp robust-to-fragile transition in empirical food webs. Ecol. Model. 251, 1–8 (2013).
McGill, B. J. et al. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol. Lett. 10, 995–1015 (2007).
Gaston, K. J. & Fuller, R. A. Biodiversity and extinction: losing the common and the widespread. Prog. Phys. Geogr. Earth Environ. 31, 213–225 (2007).
Dunne, J. A. The network structure of food webs. in Ecological Networks: Linking Structure to Dynamics in Food Webs (eds Pascual, M. & Dunne, J.) 25–76 (Oxford University Press, Oxford, 2006).
Saravia, L. A., Marina, T. I., Kristensen, N. P., De Troch, M. & Momo, F. R. Ecological network assembly: How the regional metaweb influences local food webs. J. Anim. Ecol. 91, 630–642 (2022).
O’Connor, L. M. J. et al. Unveiling the food webs of tetrapods across Europe through the prism of the Eltonian niche. J. Biogeogr. 47, 181–192 (2020).
Albouy, C. et al. The marine fish food web is globally connected. Nat. Ecol. Evol. 3, 1153–1161 (2019).
Reji Chacko, M. et al. trophiCH - a food web for Switzerland. EnviDat https://doi.org/10.16904/ENVIDAT.467 (2024).
Statistisches Jahrbuch der Schweiz 1997. (NZZ Libro (NZZ_BUCHV), 1996).
Gonseth, Y., Klingl, T. & Nöthiger-Koch, U. Die Biogeographischen Regionen Der Schweiz: Erläuterungen Und Einteilungsstandard = Les Régions Biogéographiques de La Suisse: Explications et Division Standard. (Bezugsquelle: BUWAL, Dokumentation, Bern, 2001).
BAFU. Artenvielfalt in der Schweiz. Bundesamt für Umwelt BAFU https://www.bafu.admin.ch/bafu/de/home/themen/biodiversitaet/zustand-der-biodiversitaet-in-der-schweiz/zustand-der-artenvielfalt-in-der-schweiz.html (2023).
CSCF. info fauna | Data Server. info fauna Nationales Daten- und Informationszentrum der Schweizer Fauna https://lepus.infofauna.ch/tab/ (2017).
Reji Chacko, M. et al. A species-level multi-trophic metaweb for Switzerland. Sci. Data https://doi.org/10.1038/s41597-025-05487-7 (2025).
Maiorano, L., Montemaggiori, A., Ficetola, G. F., O’Connor, L. & Thuiller, W. TETRA-EU 1.0: A species-level trophic metaweb of European tetrapods. Glob. Ecol. Biogeogr. 29, 1452–1457 (2020).
Morales-Castilla, I., Matias, M. G., Gravel, D. & Araújo, M. B. Inferring biotic interactions from proxies. Trends Ecol. Evol. 30, 347–356 (2015).
Landolt, E. et al. Flora indicativa - Ökologische Zeigerwerte und biologische Kennzeichen zur Flora der Schweiz und der Alpen. Haupt Bern. 7, 378 (2010).
BAFU. Biogeographical regions of Switzerland (CH). (2022).
Chauvier, Y. et al. Resolution in species distribution models shapes spatial patterns of plant multifaceted diversity. EnviDat https://doi.org/10.16904/envidat.334 (2022).
Külling, N. & Adde, A. SWECO25: Topographic (topo). Zenodo https://doi.org/10.5281/zenodo.7973960 (2023).
Delarze, R., Gonseth, Y., Eggenberg, S. & Vust, M. Lebensräume Der Schweiz: Ökologie - Gefährdung - Kennarten. (Ott-Verlag, Thun, 2015).
Price, B., Huber, N., Ginzler, C., Pazúr, R. & Rüetschi, M. The Habitat Map of Switzerland v1. Envidat https://doi.org/10.16904/envidat.262 (2021).
Gillies, S. Rasterio: geospatial raster I/O for Python programmers. Mapbox (2013).
Jordahl, K. et al. geopandas/geopandas: v0.8.1. Zenodo https://doi.org/10.5281/zenodo.3946761 (2020).
Csárdi, G. et al. igraph: Network Analysis and Visualization in R. https://doi.org/10.5281/zenodo.7682609 (2024).
De Vos, J. M., Joppa, L. N., Gittleman, J. L., Stephens, P. R. & Pimm, S. L. Estimating the normal background rate of species extinction. Conserv. Biol. 29, 452–462 (2015).
Harris et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Hagberg, A., Swart, P. & S Chult, D. Exploring Network Structure, Dynamics, and Function Using NetworkX. (2008).
VanRossum, G., Drake, F. L. Python 3 Reference Manual. CreateSpace: Scotts Valley, CA, 2009.
RStudio Team. RStudio: Integrated Development Environment for R. RStudio, PBC. (2020).
Dinno, A. dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. (2017).
Torchiano, M. effsize: Efficient Effect Size Computation. https://doi.org/10.5281/zenodo.1480624 (2020).
Shipley, J. R. et al. Agricultural practices and biodiversity: Conservation policies for semi-natural grasslands in Europe. Curr. Biol. 34, R753–R761 (2024).
Habel, J. C. et al. European grassland ecosystems: threatened hotspots of biodiversity. Biodivers. Conserv. 22, 2131–2138 (2013).
Brändli, U. B., Abegg, M. & Düggelin, C. Biologische Vielfalt. in Schweizerisches Landesforstinventar. Ergebnisse der vierten Erhebung 2009–2017. 189–237 (Swiss Federal Institute for Forest, Snow and Landscape Research, Federal Office for the Environment, Birmensdorf, (2020).
Shipley, J. R., Gossner, M. M., Rigling, A. & Krumm, F. Conserving forest insect biodiversity requires the protection of key habitat features. Trends Ecol. Evol. 38, 788–791 (2023).
Mestre, F. et al. Disentangling food-web environment relationships: A review with guidelines. Basic Appl. Ecol. 61, 102–115 (2022).
May, R. M. Will a Large Complex System be Stable?. Nature 238, 413–414 (1972).
Braga, J. et al. Spatial analyses of multi-trophic terrestrial vertebrate assemblages in Europe. Glob. Ecol. Biogeogr. 28, 1636–1648 (2019).
Shi, F. et al. Spatio-temporal dynamics of landscape connectivity and ecological network construction in Long Yangxia Basin at the Upper Yellow River. Land 9, 265 (2020).
Santos, M. et al. Robustness of a meta-network to alternative habitat loss scenarios. Oikos 130, 133–142 (2021).
Anderson, K. E. & Fahimipour, A. K. Body size dependent dispersal influences stability in heterogeneous metacommunities. Sci. Rep. 11, 17410 (2021).
Schofield, K. A. et al. Biota connect aquatic habitats throughout freshwater ecosystem mosaics. JAWRA J. Am. Water Resour. Assoc. 54, 372–399 (2018).
Flinn, K. M., Lechowicz, M. J. & Waterway, M. J. Plant species diversity and composition of wetlands within an upland forest. Am. J. Bot. 95, 1216–1224 (2008).
Devictor, V., Julliard, R. & Jiguet, F. Distribution of specialist and generalist species along spatial gradients of habitat disturbance and fragmentation. Oikos 117, 507–514 (2008).
Ho, H.-C. et al. Blue and green food webs respond differently to elevation and land use. Nat. Commun. 13, 6415 (2022).
Vitekere, K., Hua, Y. & Jiang, G. Complexity, connectance and link density in continental food webs: dissimilarities in aquatic and terrestrial food webs and their habitats. Appl. Ecol. Environ. Res. 19, 817–831 (2021).
Shipley, J. et al. Consumer biodiversity increases organic nutrient availability across aquatic and terrestrial ecosystems. Science 386, 335–340 (2024).
Magrach, A., González-Varo, J. P., Boiffier, M., Vilà, M. & Bartomeus, I. Honeybee spillover reshuffles pollinator diets and affects plant reproductive success. Nat. Ecol. Evol. 1, 1299–1307 (2017).
McFadden, I. R. et al. Linking human impacts to community processes in terrestrial and freshwater ecosystems. Ecol. Lett. 26, 203–218 (2023).
Säterberg, T., Jonsson, T., Yearsley, J., Berg, S. & Ebenman, B. A potential role for rare species in ecosystem dynamics. Sci. Rep. 9, 11107 (2019).
Maurer, C., Sutter, L., Martínez-Núñez, C., Pellissier, L. & Albrecht, M. Different types of semi-natural habitat are required to sustain diverse wild bee communities across agricultural landscapes. J. Appl. Ecol. 59, 2604–2615 (2022).
Albrecht, M., Padrón, B., Bartomeus, I. & Traveset, A. Consequences of plant invasions on compartmentalization and species’ roles in plant–pollinator networks. Proc. R. Soc. B Biol. Sci. 281, 20140773 (2014).
Heleno, R. H., Ripple, W. J. & Traveset, A. Scientists’ warning on endangered food webs. Web Ecol. 20, 1–10 (2020).
Gossner, M. M. et al. Land-use intensification causes multitrophic homogenization of grassland communities. Nature 540, 266–269 (2016).
Wang, S. et al. Biotic homogenization destabilizes ecosystem functioning by decreasing spatial asynchrony. Ecology 102, e03332 (2021).
Ellison, A. M. et al. Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front. Ecol. Environ. 3, 479–486 (2005).
Rodríguez-Castañeda, G. et al. Tropical forests are not flat: how mountains affect herbivore diversity. Ecol. Lett. 13, 1348–1357 (2010).
Adhurya, S., Agasti, N. & Park, Y. Metaweb and its applications in understanding ecological interactions. Preprint at https://doi.org/10.32942/X2SW3V (2023).
Fricke, E. C., Ordonez, A., Rogers, H. S. & Svenning, J.-C. The effects of defaunation on plants’ capacity to track climate change. Science 375, 210–214 (2022).
Chen, I.-C., Hill, J. K., Ohlemüller, R., Roy, D. B. & Thomas, C. D. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011).
Grünig, M., Mazzi, D., Calanca, P., Karger, D. N. & Pellissier, L. Crop and forest pest metawebs shift towards increased linkage and suitability overlap under climate change. Commun. Biol. 3, (2020).
Pecuchet, L. et al. Novel feeding interactions amplify the impact of species redistribution on an Arctic food web. Glob. Change Biol. 26, 4894–4906 (2020).
Seebens, H. et al. No saturation in the accumulation of alien species worldwide. Nat. Commun. 8, 14435 (2017).
Seebens, H. et al. Global rise in emerging alien species results from increased accessibility of new source pools. Proc. Natl. Acad. Sci. 115, E2264–E2273 (2018).
Aslan, C. E. Implications of non-native species for mutualistic network resistance and resilience. PLOS ONE 14, e0217498 (2019).
InfoSpecies. Artinformationen, Verbreitungsdaten. InfoSpecies https://www.infospecies.ch/de/neobiota/artinformationen-verbreitungsdaten.html (2021).
Bartley, T. J. et al. Food web rewiring in a changing world. Nat. Ecol. Evol. 3, 345–354 (2019).
Moorhouse-Gann, R. J., Kean, E. F., Parry, G., Valladares, S. & Chadwick, E. A. Dietary complexity and hidden costs of prey switching in a generalist top predator. Ecol. Evol. 10, 6395–6408 (2020).
Yacine, Y., Allhoff, K. T., Weinbach, A. & Loeuille, N. Collapse and rescue of evolutionary food webs under global warming. J. Anim. Ecol. 90, 710–722 (2021).
Tylianakis, J. M., Didham, R. K., Bascompte, J. & Wardle, D. A. Global change and species interactions in terrestrial ecosystems. Ecol. Lett. 11, 1351–1363 (2008).
Raubenheimer, D., Simpson, S. J. & Tait, A. H. Match and mismatch: conservation physiology, nutritional ecology and the timescales of biological adaptation. Philos. Trans. Biol. Sci. 367, 1628–1646 (2012).
Valdovinos, F. S., Ramos-Jiliberto, R., Garay-Narváez, L., Urbani, P. & Dunne, J. A. Consequences of adaptive behaviour for the structure and dynamics of food webs. Ecol. Lett. 13, 1546–1559 (2010).
Thierry, A. et al. Adaptive foraging and the rewiring of size-structured food webs following extinctions. Basic Appl. Ecol. 12, 562–570 (2011).
Ripple, W. J. & Beschta, R. L. Wolves and the Ecology of Fear: Can Predation Risk Structure Ecosystems?. BioScience 54, 755–766 (2004).
Paine, R. T. Food Web Complexity and Species Diversity. Am. Nat. 100, 65–75 (1966).
Ho, H.-C. & Altermatt, F. Predicted community consequences of spatially explicit global change-induced processes on plant–insect networks. Ecol. Evol. 14, e70272 (2024).
Körner, C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 22, 569–574 (2007).
Carroll, C., Noss, R. F., Dreiss, L. M., Hamilton, H. & Stein, B. A. Four challenges to an effective national nature assessment. Conserv. Biol. 37, e14075 (2023).
Külling, N. et al. Nature’s contributions to people and biodiversity mapping in Switzerland: spatial patterns and environmental drivers. Ecol. Indic. 163, 112079 (2024).
Jackson, S. T. et al. Toward a national, sustained US ecosystem assessment. Science 354, 838–839 (2016).
Vihervaara, P. et al. How essential biodiversity variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 10, 43–59 (2017).
Adde, A. et al. Projecting Untruncated Climate Change Effects on Species’ Climate Suitability: Insights From an Alpine Country. Glob. Change Biol. 30, e17557 (2024).
Dallimer, M. & Strange, N. Why socio-political borders and boundaries matter in conservation. Trends Ecol. Evol. 30, 132–139 (2015).
Reji Chacko, M. et al. Simulation data and analyses of regional food web robustness under habitat loss. EnviDat https://doi.org/10.16904/envidat.642 (2025).
Acknowledgements
We thank the Swiss Species Information Center InfoSpecies (www.infospecies.ch) for supplying species occurrences. We used Flaticon (https://www.flaticon.com/) for the animal and plant icons used in figures 1 and 2b, from creators amoghdesign, FreePik, max.icons, Park Jisun, Royyan Wijaya, Smashicons, and VectorPortal. The icons are curated within a collection (https://www.flaticon.com/collections/NTE0NjgxODY=). We are grateful to Théophile Sanchez for support with cluster computing and Bronwyn Price for technical support with utilising the Habitat Map of Switzerland. We thank the editor and four anonymous reviewers for their comments on previous versions of the manuscript. M.R. thanks A.T.Z. for support and inspiration. Funding for the trophiCH project was granted by the Action Plan of the Swiss Biodiversity Strategy by the Federal Office for the Environment (FOEN) to L.P. Funding to F.A. is by the Swiss National Science Foundation (Grant 31003A_1180513). Funding to H.H. is by MOE Taiwan (Grant No. MOE-112-YSFAG-0003-005-P1) and NSTC Taiwan (Grant No. NSTC 112-2621-B-002-006-MY3).
Author information
Authors and Affiliations
Contributions
F.A., N.F., M.R. and L.P. conceived the study. M.R., A.J., C.A., F.A., V.B., H.H., L.P., N.F. developed the methodology. M.R. and A.J. developed the simulation software. M.R. curated the data, conducted the investigation, validation and formal analyses, and wrote the original draft. F.A., L.P., M.G., M.B., F.N. provided resources. All co-authors reviewed and edited the manuscript. L.P. and F.A. supervised the project, with support from C.A. and H.H. L.P. acquired the funding for the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Micaela Santos, Eoin O’Gorman, and the other anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Michele Repetto. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Reji Chacko, M., Albouy, C., Altermatt, F. et al. Species loss in key habitats accelerates regional food web disruption. Commun Biol 8, 988 (2025). https://doi.org/10.1038/s42003-025-08396-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08396-y
This article is cited by
-
A species-level multi-trophic metaweb for Switzerland
Scientific Data (2025)
-
Integrative study on Meizotropis pellita (Hook. f. ex Prain): phytochemistry, antibacterial potential, molecular insights, biological applications, and conservation strategies of a critically endangered Himalayan endemic plant
Vegetos (2025)