Abstract
Determining whether organisms can undergo adaptive evolution at a pace commensurate with contemporary climate change is critical to understanding and predicting the consequences of such change. Hybrid introgression is a mechanism of rapid evolution by which species may adapt to climatic shifts. Here, we examine variation in growth and survival in a long-term common garden experiment with a foundation tree species to determine if introgression is enhancing climate change resilience. Two naturally hybridizing tree species, low elevation Populus fremontii and high elevation Populus angustifolia, and hybrid and backcross genotypes were planted in a low elevation, warm common garden. We show that P. angustifolia and backcross trees are vulnerable to warming, and their survival is related to climate and transfer distance (proxies for climate change). Increased odds of survival are associated with genetic introgression, as indicated by RFLP genetic markers. Thus, for these long-lived foundation trees, hybrid introgression is associated with increased resistance to selection pressures in warmer, drier climates. These data highlight the importance of evolutionary patterns and processes in shaping ecosystem responses to climate change. If adaptive introgression through hybrid zones is common, hybrid-specific conservation policies and restoration should be reconsidered in the context of global change.
Similar content being viewed by others
Introduction
Identifying the ecological and evolutionary factors underlying ecosystem-level responses to climate change is an urgent research frontier. Climate change often acts as a selective force, shifting plant functional traits. The impacts of these shifts, especially when they occur in foundation species, extend to ecosystem function and associated communities (e.g., soil microbes, herbivores)1,2,3,4,5,6,7,8. Evolutionary changes in functional traits can occur on ecological timescales (i.e., rapid evolution)9,10,11,12. Investigations into the potential for rapid evolution and adaptation to warmer and drier conditions via interspecific gene flow (i.e., genetic introgression between hybridizing species) have demonstrated that for diverse taxa, ancient introgression has been an important mechanism of adaptation to historical climate change13,14,15. Evidence for the mitigation of some of the negative impacts of contemporary climate change via enriched genetic diversity and adaptive potential resulting from introgression has also been shown across levels of biological organization, from populations to ecosystems16,17,18,19. However, we lack data on genotype-level selection over multiple decades in concert with measurements of key organismal traits and information about the intraspecific genetic variation underlying phenotypic changes.
Variation in survival and growth under changing climates among foundation plant species represents a mechanism through which climate change-driven selection can affect ecosystem functions (e.g., carbon sequestration)6. This has been explored with naturally hybridizing foundation tree species in the Populus genus. Natural hybrid populations act as selective filters for introgressed genes from the low elevation, warm-adapted Populus fremontii moving into relatively high elevation, cool-adapted Populus angustifolia populations20. In a common garden, the presence of these introgressed P. fremontii genetic markers has been associated with unique arthropod communities and greater asexual reproduction21. Further, genotype-level variation has been demonstrated to influence ecosystem processes in this system. For example, genetically-based differences in herbivore susceptibility among these species and their hybrids influence aquatic leaf litter decomposition rates and carbon cycling16 and ecosystem-level carbon budgets both in the field and in common gardens vary with tree cross type22,23. These studies demonstrate that hybrid introgression between foundation tree species can result in changes to key fitness traits and shape organismal and ecosystem-level responses to climate change.
Here, we use a 31-year-old common garden experiment to understand the consequences of introgression for species’ growth and survival in a climate change context to explore the potential ecosystem-level impacts of hybridization24. A change in climate was imposed on Populus fremontii and Populus angustifolia genotypes, as well as hybrid and backcross trees originating from natural populations along a temperature and elevational gradient, by planting them in a low-elevation, warm common garden. Over three decades, nearly half of all trees planted were lost to selective pressures (i.e., mortality in the climate change conditions in the warm, dry common garden) after reaching reproductive maturity. We hypothesized that introgression influences tree fitness and tested this by comparing measures of growth and selection over time. We tested three predictions: (1) In the common garden, P. fremontii trees will have higher survival and greater biomass accumulation (i.e., carbon) than P. angustifolia trees, and survival and growth for hybrid and backcross genotypes will fall between the parental species (i.e., will be additive), (2) among the more vulnerable P. angustifolia and backcross genotypes growing in warmer conditions, tree mortality will increase as a function of the magnitude of climate change (proxied by geographic and climatic transfer distance), and (3) among P. angustifolia and backcross trees the presence of introgressed genetic markers will be associated with increased survival and biomass accumulation. Testing these predictions is important because, if supported, they suggest a potentially common hybrid pathway by which a foundation tree species can adapt to a rapidly changing environment.
Results
Parental species and hybrid patterns of growth and survival
After 31 years in the common garden there were significant differences in survival among the four cross types that varied in their climatic origin (i.e., parental species P. fremontii and P. angustifolia, F1 hybrids, and F1 × P. angustifolia backcross hybrids). In the low elevation common garden approximately 90% of the low-elevation-adapted P. fremontii and 100% of F1 hybrid cottonwood genotypes survived, while among the high-elevation-adapted backcross hybrid and P. angustifolia genotypes, only approximately 30% and 25% survived, respectively (Table S1, Fig. 1A). We also found significant differences in accumulated biomass among the four cross types, the patterns of which were consistent with previous studies (Table S2); F1 hybrid genotypes had the highest biomass, followed by the P. Fremontii genotypes. Mean biomass for the backcross and P. angustifolia genotypes was approximately 37% lower than for P. fremontii genotypes (Fig. 1B). Tukey post-hoc analyses showed that mean biomass for P. angustifolia and backcross genotypes were significantly different from P. fremontii genotypes, while F1 genotypes were not significantly different from either parental species or the backcrosses.
Survival (A) and growth (B) for two hybridizing species Populus fremontii and P. angustifolia, F1 hybrids and backcross genotypes differ after 31 years of growth in a common garden. Leaf icons correspond to cross types, letters correspond to significantly different groups identified with a Tukey HSD post hoc test.
Populus angustifolia and backcross survival is related to climate change
Consistent with the second hypothesis that warming and drying would select against high elevation populations from cooler and wetter sites, survival among P. angustifolia and backcross trees varied significantly, with both geographic and climatic transfer distance, after 31 years in the common garden. Populus angustifolia and backcross trees from populations closer and more climatically similar to the common garden were more likely to survive than those originating from more distant and dissimilar populations (Fig. 2A; Table S3). Overall, we found that for each 1 °C increase in temperature, the odds of survival decreased by 7.5%, and that there was greater than 90% mortality among P. angustifolia and backcross trees when mean annual temperature (MAT) (MAT, here, a proxy for a number of temperature and precipitation variables, see Fig. S1) differed more than 4 °C between source populations and the common garden.
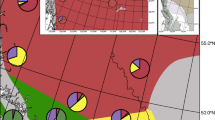
Mortality of Populus angustifolia and backcross trees (i.e., the percentage of trees from each population that died between 1991 and 2022) increased as both A geographic distance and B climatic distance (i.e., the absolute value of the difference in mean annual temperature (MAT) between the common garden and source population) increased. Individual data points have been jittered vertically to increase visibility. C A map of showing the location of the common garden (green star) and the populations from which the trees were collected. Orange dots indicate populations from which all four cross types were collected, while blue dots indicate populations from which only P. angustifolia and backcross trees were collected. Map in C was created using the Open Source Geospatial Foundation Project through QGIS Geographic Information System (2024).
Marker-trait associations among surviving backcross and P. angustifolia genotypes
While high elevation backcross hybrid and P. angustifolia genotypes had the highest mortality overall, survival among these cross types was associated with the presence of a genetic marker introgressed from P. fremontii. We found that the presence of markers RFLP-755, RFLP-754, and RFLP-1286 were the most important predictors of survival for P. angustifolia and backcross genotypes, and that there was a significant difference in survival among individuals with RFLP-1286 compared to those without this marker (Table S4, single factor ANOVA results in Table S5). Overall, backcross hybrid and P. angustifolia trees that carried P. fremontii marker RFLP-1286 had approximately 75% greater survival after 31 years in the common garden relative to the trees that did not have the marker (Fig. 3). Further, all backcross individuals with this marker remained alive in 2022. This pattern had not yet emerged twelve years after planting; we found no significant difference in mortality based on marker presence for RFLP-1286 20 years ago, indicating that warming conditions over the last two decades have likely been important selective agents. We further investigated the effects of RFLP-1286 as a predictor for tree traits but found no significant effect (Table S6).
Discussion
Long-term common garden experiments are rare, especially for large, long-lived species, but provide valuable insight into how interspecific interactions, such as hybridization and introgression, can influence ecosystem persistence (or change) under contemporary climate change. Our results shed light on the broad consequences of natural hybridization. We found strong differences in selection among two species and their hybrids, and that selection and growth differences were strongly associated with genetic factors. Specifically, the lower elevation P. fremontii had greater survival in the low elevation common garden than the higher elevation P. angustifolia (Fig. 1). F1 hybrids were more similar in growth to P. fremontii, suggesting that P. fremontii traits in these hybrids are dominant over P. angustifolia traits. In another study examining P. fremontii and P. angustifolia survival after a record drought, Hultine et al.25 found that both parental species suffered 3-4 times greater mortality in the wild relative to naturally occurring F1 hybrids. This suggests that hybrid vigor, or heterosis, is also a potential outcome of hybridization in this system (also see refs. 23,24). Backcross trees had similar survival and biomass to P. angustifolia genotypes. F1 hybrids did not significantly differ from either parental species in biomass or survival, suggesting that hybridization increases phenotypic variability in these individuals. In natural settings, these patterns are likely to have consequences for both individual species distributions and the composition and abundance of parental species and crosses in hybrid zones. Our results also show that the probability of survival decreased for trees as the distance and MAT between the common garden and source population increased. Thus, the deleterious impacts of climate change on P. angustifolia are likely to increase due to stronger selection pressures as the magnitude of climate change increases.
Although we saw high overall mortality among the P. angustifolia and backcross genotypes in the common garden, our study also provides evidence that hybrid introgression can be adaptive and has the potential to mitigate the negative impacts of climate change by enriching species’ genetic variation. Greater genetic variation increases adaptive capacity, and therefore also increases the probability of persistence under climate change. Our results demonstrated this effect, as we show that a P. fremontii genetic marker is associated with greater survival among the surviving P. angustifolia and backcross trees. These findings provide evidence that introgression can be a means of surviving the strong selective forces of climate change. At larger scales, climate change effects are likely to alter the genetic and trait-based landscape of western riparian forests, and climate-driven shifts in genetic variation may result in altered ecosystem processes.
Rapid evolution in response to climate change
Determining whether species can undergo adaptive evolution at a pace commensurate with contemporary climate change (i.e., rapid evolution) is a critical step in predicting the long- and short-term consequences of changes in abiotic conditions26,27,28,29. A growing body of evidence shows that rapid evolution over the course of a few years to decades is possible. For example, a resurrection study comparing Brassica rapa plants grown from four cohorts of seeds collected before and after major drought events found that evolutionary change had occurred within 18 generations30. Post-drought plants diverged in traits from pre-drought plants, overall showing both earlier phenology (here, flowering time), smaller stem diameters, and lower water use efficiency relative to pre-drought plants30. Interestingly, B. rapa flowering time shifts tracked oscillations between wet and dry periods. Seeds from the youngest cohort flowered earlier than the oldest cohort, but the intervening cohorts showed a shift back to later flowering time for seeds produced after a two-year wet period. After this intermediary wet period, the drought returned, and flowering time shifted back to the earlier timeframe. Rapid evolution has also recently been demonstrated for the marsh sedge Schoenoplectus americanus; Vahsen et al.6 used a resurrection approach to compare plants grown from seeds collected from an age-stratified seed bank. They found differences in the distribution and magnitude of belowground biomass and root:shoot ratios (~50–70% of which was explained by heritable variation) both between geographic provenances and between seed age cohorts. Importantly, these differences resulted in projected shifts in ecosystem-level processes related to the capacity of marsh resilience to sea level rise expected under climate change (i.e., soil surface accretion and carbon accumulation)6. Given this evidence demonstrating that rapid evolution in plants is possible, a current frontier in climate change adaptation studies is to determine the mechanisms by which the adaptive traits and underlying genetic variation associated with evolutionary change are obtained in populations.
Hybridization as a mechanism for rapid evolution
Hybrid introgression is a mechanism for rapid adaptive evolution that has gained attention for its potential to increase organismal and ecosystem resilience to climate change17. It is particularly interesting to consider for plant species, among which hybridization is extremely common; about 25% of angiosperms can hybridize in natural settings, compared to ~10% of animal species31. Hybridization can have various consequences for parental species, ranging from the initiation of speciation (reduplication events associated with hybridization are thought to be a major speciation mechanism for vascular plants) to an overall loss of genetic diversity32,33, which can lead to extinction34. As early as the mid-20th century, researchers speculated that one of the most common and important outcomes of hybridization could be introgression, which is an intermediate to these extreme outcomes35. Alleles obtained through introgression can become fixed through neutral processes (i.e., genetic drift), but can also be acted upon by natural selection when they provide a fitness advantage. This process is designated as adaptive introgression, which is a specific case of hybrid introgression (the broader term includes the transmission of any allele, whether it be beneficial, neutral, or deleterious, between species). In fact, introgression is a well-established mechanism for adaptation to changing environmental conditions. Two examples of this are (1) among Iris species, in which the introgression of alleles from a flood tolerant species to a dry-adapted species resulted in increased flooding tolerance in the latter in a common garden setting36,37, and (2) in Helianthus species, in which introgression is associated with altered herbivory and reproductive output, as well as with increased drought tolerance38. Hybrid introgression has also been leveraged as a tool for increasing genomic diversity and tolerance to environmental stressors extensively in agricultural settings through introgression breeding or selective introgression. For example, in agricultural crops, introgression breeding has been used to increase disease resistance39; it is also frequently used in wheat crops to transfer beneficial alleles from wild to cultivated species (reviewed in ref. 40,41). Similarly, the potential use of introgression as a tool for restoration both via genetic engineering (e.g., to increase blight tolerance in the American Chestnut42) and through assisted gene flow has gained interest41. The frequency, magnitude, direction, and adaptive value of introgressed alleles in natural settings is not well understood, particularly at large spatial scales. However, as the evidence of its adaptive potential continues to grow, more researchers are identifying the genes and traits being transmitted by this mechanism.
Hybrid introgression has been explored in the Populus genus, and some progress has been made towards identifying both its fitness consequences and the specific genes transmitted by this mechanism. The gene regions associated with the RFLP markers we use here have not and cannot be identified43, nevertheless, previous studies provide some insight into the types of genes and traits that have moved between Populus species. For example, previous studies show evidence that hybrid introgression influences the range dynamics of Populus species. In a 2018 study, Suarez-Gonzales et al.44 showed that introgression from the lower elevation species P. trichocarpa (black cottonwood) to the higher elevation species P. balsamifera (balsam poplar) facilitates northern range expansion. Further, they identified the nitrate transporter AtNRT2 as a candidate gene for traits related to phenology (e.g., leaf out date) and biomass (height) that was acquired through introgression. Nitrate regulation is an important component of physiological changes related to dormancy cycling45. Chhatre et al.15 characterized introgression in a tri-species hybrid complex at the range margins of P. angustifolia (narrowleaf cottonwood), P. balsamifera (balsam poplar), and P. trichocarpa (black cottonwood) and found introgression occurring among all three species, although it was most commonly observed as a transfer from the narrowleaf cottonwood to the balsam poplar. They identified introgression outliers (i.e., introgressed SNPs that were present more frequently than expected under a neutral demographic model and are thus likely under positive selection) and found that the gene regions flanking these outliers were related to photoperiodic regulation and the synthesis of cell wall components important for dormancy. Relative to the balsam poplar, the narrowleaf cottonwood is adapted to warmer climates with longer photoperiods in the growing season thus, it seems that the introgression of narrowleaf alleles related to photoperiodic regulation facilitates balsam poplar adaptation at its southern range margin. In this scenario, Populus angustifolia is analogous to P. fremontii in study system utilized here. Although we expect that many introgressed alleles will be neutral in their fitness effects, these examples demonstrate that adaptive introgression is likely common among Populus species and that it is likely possible to identify specific introgressed alleles associated with phenotypic traits that confer fitness advantages.
Previous work in this 31-year-old common garden also provides clues as to what kinds of genes and traits may be involved in adaptive introgression between P. fremontii and P. angustifolia. For example, marker RFLP-755 has been identified as being associated with increased cloning, which may be a particularly important as the Western US continues to undergo aridification21. Martinsen et al.20 demonstrated the adaptive value of introgression by showing that hybrids act as a selective filter (i.e., introgressed fragments of different sizes move into P. angustifolia populations at different rates, and fragments which introgress further into the P. angustifolia zone are candidates for adaptative introgression). Our marker-trait associations support this idea, as individuals with RFLP-1286 had increased odds of survival relative to individuals without the marker. Although our study did not directly test the effects of introgression on range dynamics, we show that mortality increases with the magnitude of climate change based on transfer distance. Recent ecophysiological studies with P. fremontii have not yet attempted to tie functional traits to specific markers; however, they have identified traits that have clear ties to greater tolerance to increased temperatures associated with climate change that could potentially benefit P. angustifolia populations via adaptive introgression. In common garden studies, Blasini et al.46,47 showed adaptive trait syndromes of phenology, wood density, xylem vessel size, and leaf economic spectrum traits of leaf area, stomatal densities, and stomatal conductance genetically varied among populations and allowed trees to tolerate temperatures exceeding 41 °C. Similarly, in a 2017 study Fischer et al.48 found strong differentiation among P. fremontii genotypes in phenology and productivity in a southern Arizona common garden. In combination, these studies imply that local adaptation in P. angustifolia may be more likely to occur in warm edge, or low elevation populations that are more likely to overlap with P. fremontii populations.
Introgression between hybridizing species can alter allele frequencies in focal species’ populations through positive selection on beneficial alleles or the integration of neutral alleles into the genome. These shifts can potentially impact larger scales of biological organization, depending on the trait these alleles are related to. Here, we show that climate-driven selection is favoring P. angustifolia backcrosses with genetic material obtained via introgression from P. fremontii. The strong selective pressures associated with climate change (e.g., increased temperatures, altered precipitation regimes) will continue to act on these markers, likely making these more productive phenotypes dominant under global change. Under climate change, P. fremontii and F1 hybrid genotypes may become more dominant in regions where the range of the former overlaps with P. angustifolia due to greater heat tolerance. Such shifts may have implications for C sequestration, as we have shown P. fremontii trees are significantly more productive, and also cycle C faster due to enhanced tissue decomposition and belowground respiration22,23. Either of these scenarios, if occurring in natural populations, could represent examples of ecosystem evolution (i.e., a genetically-based change to an ecosystem process) occurring via genetic change to a foundation species but further investigation is required to characterize these impacts at large scales and across different climatic gradients.
Hybridization and conservation policy
In the United States, the “hybrid policy” advocated by O’Brien and Mayr49 is to “discourage hybridization between species” because hybridization may “disintegrate the genetic organization of the species in contact.” However, this view, especially with regard to plants, has long been challenged as there are a number of significant counter arguments (see refs. 50,51). In addition to hybrid zones being hot spots of biodiversity and critical habitat for unique species52, hybridization in plants has been documented as a common pathway in plant evolution and speciation32,33,53,54,55,56. In a 30-year review of the Endangered Species Act, Haig and Allendorf57 note that hybrids are not included in the definition of species and thus not protected. Nevertheless, some hybrid species are assessed on a case-by-case basis. For example, specific hybrid palms and cliffroses are now officially protected and explicitly identified in recovery plans50. In the latter case, two species of cliffrose (Purshia subintegra and P. stansburiana) hybridize naturally, and although these hybrids are beyond the definition of a species, the U.S. Fish and Wildlife Services’ recovery plan recognizes that their hybrid swarms “illustrate the migratory and dynamic nature of evolving plant populations. Plants in the hybrid swarms are genetically and phenotypically variable, represent a piece of the evolutionary history of Purshia, and may provide the key to the future of the genus and species,” thus justifying the need for conservation of these specific hybrid populations58. The present study adds to this argument by presenting an example of adaptive introgression that could be important for promoting evolutionary responses to global warming that are occurring naturally in native hybridizing species.
Long-term common garden experiments can provide a unique insight into the potential for natural hybridization to be utilized in conservation and management practices. It has been suggested that interspecific gene flow in the form of facilitated introgression is an underutilized conservation tool41. This tool has been explored as an option in the restoration of the American Chestnut (Castenea dentata), which has been driven to near extinction by the fungal pathogen Cryphonectria parasitica42. The intentional introgression of blight resistance genes from Asian chestnut species using genetic engineering is suggested as a potential avenue for reintroducing the American chestnut into natural settings. Our study shows that hybrid introgression has the potential to increase climate change resilience, and if the specific genes underlying resilience-related traits can be identified, intentional introgression could be more realistically considered for conserving this rare but important riparian forest ecosystem type.
While there has been interest in hybrid introgression as a form of gene flow and a mechanism for adaptive evolution, its prevalence in natural populations has long been unclear. Our results provide compelling evidence that hybrid introgression may be an under-recognized mechanism of rapid evolution and adaptation in response to climate change. Further, we show that hybridization, broadly, and introgression, specifically, can lead to shifts in traits, the consequences of which may extend beyond the species’ level to alter ecosystem function. Elucidating the extent, distribution, and fitness effects of introgression at larger spatial scales and identifying and characterizing the effects of candidate genes for these introgressed regions of the genome on populations, communities, and ecosystems will allow us to better understand and predict the fate of species under climate change.
Foundation species, like P. angustifolia and P. fremontii, have a disproportionately large impact on ecosystem function and modulate a number of ecosystem processes59. Because these species dominate the landscapes they inhabit and influence their associated communities through largely non-trophic and mutualistic effects (e.g., shaping microclimates, soil properties, providing habitat, etc.) changes in intraspecific genetic variation in traits related to those interactions and processes (i.e., shifts in the mean or variance of phenotypes within and/or among populations) can have cascading effects to influence higher levels of organization59,61,62,62. For example, it has been shown that declines in populations of a number of large foundation tree species, including eastern hemlocks (Tsuga canadensis) and whitebark pine (Pinus albicaulis), alters ecosystem function by decreasing net primary productivity60. Our results suggest that introgression and hybridization result in higher biomass phenotypes in populations of the dominant, foundation Populus species. Populus angustifolia co-occurs and hybridizes with P. fremontii in numerous populations across the Intermountain West; if introgression impacting biomass accumulation in this manner is widespread, it is likely to shift aspects of C cycling and soil properties (e.g., through leaf litter decomposition, moisture), and microclimates (through changes in canopy density associated with increased growth) across this geographic region. Characterizing how introgression and hybridization dynamics vary throughout the species’ range in future studies will give more insight into the potential ecosystem-level impacts of trait change driven by these processes.
Methods
Common garden
Many riparian forests across the western US are dominated by Populus species. High elevation localities are frequently occupied by P. angustifolia (narrowleaf cottonwoods), while P. fremontii (Fremont cottonwoods) occupy lower elevation sites. Hybridization is common where the distributions of these species overlap, and introgression has been shown to occur in a unidirectional pattern with P. fremontii genes moving into P. angustifolia populations, but not vice versa63,64. Four cross types were planted in the common garden we used for this study: “pure” P. fremontii and P. angustifolia trees, F1 hybrids, and backcross hybrids (F1 hybrid x P. angustifolia crosses). Genotypes were collected from multiple natural populations along the Weber River, Utah, which has a 13 km hybrid zone20,63. P. angustifolia populations were collected at elevations between 1500–2350 m, hybrid and backcross hybrid populations between 1330–1500 m, and P. fremontii populations between 1280–1330 m. In 1990 and 1991, 199 clones, representing 63 naturally occurring genotypes of both parental species, F1, and backcross hybrids, were randomly planted on 4 m centers in a common garden in Ogden, Utah, at the Ogden Nature Center (1300 m). Geographic transfer distance (the distance between the common garden and the population of origin) and climatic transfer distance (the absolute value of the difference between the MAT of the common garden and the population of origin) were calculated for each source population. The site receives approximately 459 mm of precipitation annually (MAP) and MAT is 10.3 °C. MAT among source populations ranges from 1.6 °C to 8 °C, while MAP ranges from 491 to 663 mm. The soil at the common garden is in the Entisol USDA Soil Taxonomic order and is composed of ~60% sand, ~30% silt, and ~10% clay22,65,66. The cross type status (i.e., P. fremontii, P. angustifolia, F1 hybrid, or backcross hybrid) for each tree was determined in earlier studies20,63 based on the presence of P. fremontii-specific RFLP molecular markers (Fig. S2)67. Since RFLPs are codominant, it is possible to identify diagnostic species-specific markers (i.e., stable interspecific polymorphisms), which can then be used to assign hybrid classes based on the frequency of these loci in individuals20. Keim et al. identified a set of 35 RFLP markers (originally developed by Bradshaw et al.67) that represented stable polymorphisms between P. fremontii and P. angustifolia. The genotypes used in this study were a randomly selected subset of the 550 individuals genotyped by Martinsen et al.20,23. Additionally, the parental species and their hybrids can be distinguished from one another based on morphological differences; P. fremontii has cordate leaves with wide-toothed margins, while P. angustifolia has lanceolate leaves with fine-toothed margins. Hybrids have an intermediate morphology, with ovate leaves with intermediately toothed margins (see leaf icons in Fig. 1). These morphological differences have been verified to reliably correspond with the cross types63. The combination of morphological differences and RFLP marker data thus provides a robust system for classifying trees by cross type.
In 2003, the 199 trees were surveyed for diameter at 1.4 m above the ground surface (diameter at breast height; hereafter DBH;) and survival23. In September 2022, nearly 20 years later, the remaining living trees in the common garden were resurveyed (n = 99). DBH data from both surveys were used to estimate aboveground biomass carbon accumulation based on locally-developed allometric equations relating tree dry biomass to DBH for both species and hybrids between P. angustifolia and P. fremontii (where ~50% of biomass represents carbon)22,23.
Statistics & reproducibility
Species and cross type patterns of growth and survival
To test our first prediction, that there would be differences in growth among the two species and their crosses, we used a linear mixed effects model (“lme4” and “car” in R68,69). Biomass was the response variable, cross type was the fixed effect, and genotype was included as a random effect. Biomass data were transformed to meet model assumptions. Ten trees were excluded from analyses due to missing data (source population and genotype, n = 185). Similarly, to test whether there were differences in cross type survival, we used a generalized linear mixed effects model with a binomial distribution where mortality (live or dead) was included as the response variable, species cross type as the fixed effect, and genotype as a random effect (“lme4” and “car” packages in R68,69). P-values were adjusted using a sequential Bonferroni correction to account for multiple testing. For both models, we used a Tukey’s Honest Significant Difference test to identify differences among groups using the “multcomp” package in R70.
Populus angustifolia and backcross survival is related to climate change
To test our second prediction, that mortality among P. angustifolia and backcross trees would be related to climatic differences between source populations and the common garden, we used generalized linear models with geographic transfer distance (km) or climatic transfer distance (°C) as proxies for climate change for each source population that backcross and/or P. angustifolia trees were collected from as fixed effects and mortality (binary variable of live/dead) as the response variable and included genotype as a random effect68. P-values were adjusted using a sequential Bonferroni correction to account for multiple testing. These analyses utilized the same R packages described above. Transfer distance for P. fremontii and F1 hybrids was not analyzed as we expected them to be acclimated to the climatic conditions in the common garden due to their source populations being nearby. We extracted MAT for each source population from WorldClim71. Climatic transfer distance for each source population was calculated by taking the absolute value of the difference between the MAT for each population and the MAT for the common garden. MAT is correlated with numerous temperature and precipitation-related bioclimatic variables (Fig.S1) and, therefore, is employed here as a proxy for broad-scale climatic differences among source populations and the common garden. Additionally, previous ecological niche modelling studies have shown temperature and precipitation variables to be the primary environmental drivers of P. angustifolia occurrence patterns72. Both the geographic and climatic transfer distance response variables were transformed to meet model assumptions.
Marker-trait associations among surviving backcross and P. angustifolia trees
To test our third prediction, that hybrid introgression would increase fitness in the common garden, we first identified associations between the RFLP markers and patterns of survival among backcross and P. angustifolia trees. To do this, we used a model selection approach based on Aikake’s information criterion to identify a subset of the RFLP markers to include in statistical analyses (“MASS” package in R73). Prior to model selection, we first took a subset of ten of the of 35 P. fremontii-specific RFLP by removing highly correlated markers (r > 0.7) so that final set of ten candidate markers could be used in analyses. We further investigated the impact of markers identified by this process on tree biomass using an LMM with markers of interest as fixed effects and genotypes as random effects. The p-values for all analyses were corrected using a sequential Bonferroni to account for multiple testing.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data available in supplementary file.
Code availability
Code available at https://github.com/ashlynnhord/ONC.
References
Loreau, M., Mazancourt, C. D. & Holt, R. D. Evolutionary Conservation Biology. 327–343 (Cambridge University Press, 2004)
Bayliss, S. L. J., Mueller, L. O., Ware, I. M., Schweitzer, J. A. & Bailey, J. K. Plant genetic variation drives geographic differences in atmosphere–plant–ecosystem feedbacks. Plant Environ. Interact. 1, 166–180 (2020).
Ware, I. M. et al. Climate-driven divergence in plant-microbiome interactions generates range-wide variation in bud break phenology. Commun. Biol. 4, 748 (2021).
Nuland, M. E. V. et al. Natural soil microbiome variation affects spring foliar phenology with consequences for plant productivity and climate-driven range shifts. N. Phytol. 232, 762–775 (2021).
Love, S. J., Schweitzer, J. A. & Bailey, J. K. Climate-driven convergent evolution in riparian ecosystems on sky islands. Sci. Rep.-UK 13, 2817 (2023).
Vahsen, M. L. et al. Rapid plant trait evolution can alter coastal wetland resilience to sea level rise. Science 379, 393–398 (2023).
Bailey, J. K. et al. Indirect genetic effects: an evolutionary mechanism linking feedbacks, genotypic diversity and coadaptation in a climate change context. Funct. Ecol. 28, 87–95 (2014).
Harmon, L. J. et al. Evolutionary diversification in stickleback affects ecosystem functioning. Nature 458, 1167–1170 (2009).
Franks, S. J., Sim, S. & Weis, A. E. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. USA 104, 1278–1282 (2007).
Franks, S. J. & Weis, A. E. A change in climate causes rapid evolution of multiple life-history traits and their interactions in an annual plant. J. Evol. Biol. 21, 1321–1334 (2008).
Smith, D. S. et al. Rapid plant evolution in the presence of an introduced species alters community composition. Oecologia 179, 563–572 (2015).
Gehring, C. A., Sthultz, C. M., Flores-Rentería, L., Whipple, A. V. & Whitham, T. G. Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. USA 114, 11169–11174 (2017).
Lan, T. et al. Insights into bear evolution from a Pleistocene polar bear genome. Proc. Natl. Acad. Sci. USA 119, e2200016119 (2022).
Mao, Y., Economo, E. P. & Satoh, N. The roles of introgression and climate change in the rise to dominance of acropora corals. Curr. Biol. 28, 3373–3382.e5 (2018).
Chhatre, V. E., Evans, L. M., DiFazio, S. P. & Keller, S. R. Adaptive introgression and maintenance of a trispecies hybrid complex in range-edge populations of Populus. Mol. Ecol. 27, 4820–4838 (2018).
LeRoy, C. J., Fischer, D., Schweitzer, J. A. & Bailey, J. K. Aphid gall interactions with forest tree genotypes influence leaf litter decomposition in streams. Forests 11, 182 (2020).
Turbek, S. P. & Taylor, S. A. Hybridization provides climate resilience. Nat. Clim. Change 13, 212–213 (2023).
Brauer, C. J. et al. Natural hybridization reduces vulnerability to climate change. Nat. Clim. Change 13, 282–289 (2023).
Rocha, J. L. et al. North African fox genomes show signatures of repeated introgression and adaptation to life in deserts. Nat. Ecol. Evol. 7, 1267–1286 (2023).
Martinsen, G. D., Whitham, T. G., Turek, R. J. & Keim, P. Hybrid populations selectively filter gene introgression between species. Evolution 55, 1325–1335 (2001).
Bailey, J. K. et al. From genes to ecosystems: an emerging synthesis of eco-evolutionary dynamics. In The Ecology of Plant Secondary Metabolites vol. 184 746–749 (Cambridge University Press, 2012).
Fischer, D. G., Hart, S. C., LeRoy, C. J. & Whitham, T. G. Variation in below-ground carbon fluxes along a Populus hybridization gradient. N. Phytol. 176, 415–425 (2007).
Lojewski, N. R. et al. Genetic basis of aboveground productivity in two native Populus species and their hybrids. Tree Physiol. 29, 1133–1142 (2009).
Whitham, T. G. et al. A framework for community and ecosystem genetics: from genes to ecosystems. Nat. Rev. Genet. 7, 510–523 (2006).
Hultine, K. R. et al. Adaptive capacity in the foundation tree species Populus fremontii: implications for resilience to climate change and non-native species invasion in the American Southwest. Conserv. Physiol. 8, coaa061 (2020).
Thompson, J. N. Rapid evolution as an ecological process. Trends Ecol. Evol. 13, 329–332 (1998).
Ellner, S. P., Geber, M. A. & Hairston, N. G. Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol. Lett. 14, 603–614 (2011).
Hendry, A. P. & Kinnison, M. T. Perspective: the pace of modern life: measuring rates of contemporary microevolution. Evolution 53, 1637–1653 (1999).
Hoffmann, A. A. & Sgrò, C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011).
Hamann, E., Weis, A. E. & Franks, S. J. Two decades of evolutionary changes in Brassica rapa in response to fluctuations in precipitation and severe drought. Evolution 72, 2682–2696 (2018).
Mallet, J., Besansky, N. & Hahn, M. W. How reticulated are species? Bioessays 38, 140–149 (2016).
Soltis, P. S. & Soltis, D. E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 60, 561–588 (2009).
Abbott, R. et al. Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013).
Rhymer, J. M. & Simberloff, D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27, 83–109 (1996).
Anderson, E. & Stebbins, J. L. Hybridization as an evolutionary stimulus. Evolution 4, 377–378 (1954).
Martin, N. H., Bouck, A. C. & Arnold, M. L. Detecting Adaptive Trait Introgression Between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics 172, 2481–2489 (2006).
Bennett, B. D. & Grace, J. B. Shade tolerance and its effect on the segregation of two species of Louisiana Iris and their hybrids. Am. J. Bot. 77, 100–107 (1990).
Whitney, K. D., Randell, R. A. & Rieseberg, L. H. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. N. Phytol. 187, 230–239 (2010).
Burgarella, C. et al. Adaptive introgression: an untapped evolutionary mechanism for crop adaptation. Front. Plant Sci. 10, 4 (2019).
Hao, M. et al. The resurgence of introgression breeding, as exemplified in wheat improvement. Front. Plant Sci. 11, 252 (2020).
Hamilton, J. A. & Miller, J. M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 30, 33–41 (2016).
Newhouse, A. E. & Powell, W. A. Intentional introgression of a blight tolerance transgene to rescue the remnant population of American chestnut. Conserv. Sci. Pract. 3, e348 (2021).
Marris, E. In the name of nature. Nature 443, 498–501 (2006).
Suarez-Gonzalez, A., Hefer, C. A., Lexer, C., Douglas, C. J. & Cronk, Q. C. B. Introgression from Populus balsamifera underlies adaptively significant variation and range boundaries in P. trichocarpa. N. Phytol. 217, 416–427 (2018).
Larisch, C. et al. Poplar wood rays are involved in seasonal remodeling of tree physiology. Plant Physiol. 160, 1515–1529 (2012).
Blasini, D. E. et al. Adaptive trait syndromes along multiple economic spectra define cold and warm adapted ecotypes in a widely distributed foundation tree species. J. Ecol. 109, 1298–1318 (2021).
Blasini, D. E. et al. Tradeoffs between leaf cooling and hydraulic safety in a dominant arid land riparian tree species. Plant Cell Environ. 45, 1664–1681 (2022).
Fischer, D. G. et al. Tree genetics strongly affect forest productivity, but intraspecific diversity–productivity relationships do not. Funct. Ecol. 31, 520–529 (2017).
O’Brien, S. J. & Mayr, E. Bureaucratic mischief: recognizing endangered species and subspecies. Science 251, 1187–1188 (1991).
Whitham, T. G. & Maschinski, J. Current Hybrid Policy and the importance of hybrid plants in conservation. In Southwestern Rare and Endangered Plants: Proceedings of the Second Conference (eds. Maschinski, J., Hammond, D. H. & Holter, L.) 103–112 (USDA Forest Service, 1996).
Whitham, T. G., Morrow, P. A. & Potts, B. M. Conservation of hybrid plants. Science 254, 779–779 (1991).
Evans, L. M., Allan, G. J. & Whitham, T. G. Populus hybrid hosts drive divergence in the herbivorous mite, Aceria parapopuli: implications for conservation of plant hybrid zones as essential habitat. Conserv. Genet. 13, 1601–1609 (2012).
McKinnon, G. Reticulate evolution in higher plants. In Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants (ed. Henry, R. J.) 81–96 (CAB International, 2005).
Baack, E. J. & Rieseberg, L. H. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518 (2007).
Stace, C. A. Hybridization and the plant species. In Differentiation patterns in higher plants (ed. Urbanska, K. M.) 115–127 (Academic Press, 1987).
Rieseberg, L. H., Sinervo, B., Linder, C. R., Ungerer, M. C. & Arias, D. M. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272, 741–745 (1996).
Haig, S. M. & Allendorf, F. W. Hybrids and Policy. In The Endangered Species Act at Thirty (eds. Scott, J. M., Goble, D. D. & Davis, F. W.) vol. 2 150–163 (Island Press, 2006).
USFWS. Arizona Cliffrose (Purshia Subintegra) Recovery Plan. (USFWS, 1995).
Ellison, A. M. Foundation species, non-trophic interactions, and the value of being common. iScience 13, 254–268 (2019).
Ellison, A. M. et al. Loss of foundation species: consequences for the structure and dynamics of forested ecosystems. Front. Ecol. Environ. 3, 479–486 (2005).
Roches, S. D., Pendleton, L. H., Shapiro, B. & Palkovacs, E. P. Conserving intraspecific variation for nature’s contributions to people. Nat. Ecol. Evol. 5, 574–582 (2021).
Stange, M., Barrett, R. D. H. & Hendry, A. P. The importance of genomic variation for biodiversity, ecosystems and people. Nat. Rev. Genet. 22, 89–105 (2021).
Keim, P., Paige, K. N., Whitham, T. G. & Lark, K. G. Genetic analysis of an interspecific hybrid swarm of Populus: occurrence of unidirectional introgression. Genetics 123, 557–565 (1989).
Hersch-Green, E. I., Allan, G. J. & Whitham, T. G. Genetic analysis of admixture and patterns of introgression in foundation cottonwood trees (Salicaceae) in southwestern Colorado, USA. Tree Genet. Genomes 10, 527–539 (2014).
Schweitzer, J. A., Martinsen, G. D. & Whitham, T. G. Cottonwood hybrids gain fitness traits of both parents: a mechanism for theirlong-term persistence? Am. J. Bot. 89, 981–990 (2002).
Fischer, D. G., Hart, S. C., Whitham, T. G., Martinsen, G. D. & Keim, P. Ecosystem implications of genetic variation in water-use of a dominant riparian tree. Oecologia 139, 288–297 (2004).
Bradshaw, H. D. et al. Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor. Appl Genet 89, 167–178 (1994).
Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 1, 1–48 (2015).
Fox, J. & Weisberg, S. An R Companion to Applied Regression. vol. 3 (Sage Publication, 2019).
Hothorn, T., Bretz, F. & Westfall, P. Simultaneous inference in general parametric models. Biom. J. 50, 346–363 (2008).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315 (2017).
Bothwell, H. M. et al. Genetic data improves niche model discrimination and alters the direction and magnitude of climate change forecasts. Ecol. Appl. 31, e02254 (2021).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. (Springer, 2002).
Acknowledgements
The authors would like to thank the Ogden Nature Center, and all of the past members of the Whitham lab group who worked to keep these trees alive for all of these years. Additionally, many research assistants, field technicians, and collaborating scientists contributed significantly to this work over more than three decades. We would like to especially thank K.C. Larson, M. Kearsley, G. Martinsen, P. Keim, S. Difazio, R. Lindroth, S. Hart, K. Gehring, A. Keith, N. Lojewski, and G. Wimp. This work was initially funded by US National Science Foundation for financial support through grants DEB-0078280 and DEB-0425908. A.H. is supported from the NSF Graduate Research Fellowship (2023348830).
Author information
Authors and Affiliations
Contributions
J.B. and D.F. collected data. J.B., D.F., and T.W. designed the experiment. A.H. performed data analyses. J.B., D.F., A.H., C.L., J.S., and T.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Shouli Li, Michele Repetto, Luke Grinham.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hord, A.M., Fischer, D.G., Schweitzer, J.A. et al. Hybrid introgression as a mechanism of rapid evolution and resilience to climate change in a riparian tree species. Commun Biol 8, 1173 (2025). https://doi.org/10.1038/s42003-025-08410-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08410-3