Abstract
Resistance to poly(ADP‒ribose) polymerase inhibitors (PARPis) remains a significant challenge in ovarian cancer (OC) treatment. TTK protein kinase (TTK) has been implicated in cisplatin resistance in OC, but its role in PARPi resistance remains unclear. In this research, we found that TTK inhibition overcome olaparib resistance in HR-proficient OC cells, whereas TTK promotes olaparib resistance in HR-deficient OC cells. Mechanistically, TTK directly interacts with RPA2, facilitating phosphorylation of its S33 residue to activate the ATR signaling pathway. Knocking down RPA2 increased olaparib sensitivity in OC cells. Additionally, TTK-mediated resistance to olaparib through the RPA2/ATR signaling pathway was confirmed via both in vitro and in vivo models. In conclusion, TTK inhibition overcomes olaparib resistance in HR-proficient OC cells, in part by suppressing RPA2-S33 phosphorylation and attenuating ATR signaling. TTK inhibitors offer a promising strategy to increase the therapeutic efficacy of PARPis in OC patients.
Similar content being viewed by others
Introduction
Ovarian cancer (OC), with a 5-year overall survival rate of ~30%, represents the gynecologic cancer subtype with the highest mortality1,2. Poly (ADP‒ribose) polymerase (PARP) inhibitors (PARPis), particularly olaparib, have significantly improve progression-free survival (PFS) in OC patients and have been recommended as maintenance therapies for patients who achieve a sustained complete response (CR) or partial response (PR) following platinum-based chemotherapy3. However, PARPis resistance has been reported, and several mechanisms underlying this resistance have been identified. These include decreased PARP trapping, restoration of homologous recombination (HR), protection of stalled replication forks, reversion of mutations in BRCA genes, restoration of ADP-ribosylation (PARylation), and loss of 53BP14,5,6,7. Despite these findings, the exact mechanisms of PARPi resistance remain unclear. Therefore, further investigations of these resistance mechanisms and the development of new strategies or drug combinations to overcome PARPi resistance are crucial.
TTK protein kinase (TTK), also known as monopolar spindle 1 (Mps1), is a dual-specificity protein kinase capable of phosphorylating tyrosine, serine, and threonine residues. It plays a critical role as a mitotic checkpoint protein, regulating cell proliferation, the cell cycle, and the DNA damage response8,9,10. Additionally, TTK has been implicated in promoting the tumorigenesis of various cancers11,12,13,14,15,16,17,18. As a result, several TTK-specific inhibitors have been developed and evaluated in clinical trials, including CFI-402257 (NCT03568422), BAY1161909 (NCT02138812), BAY1217389 (NCT02366949), and BOS172722 (NCT03328494)19,20,21,22. TTK inhibition has also been shown to enhance the effectiveness of chemotherapy and radiotherapy in different cancer types14,23,24,25,26. Our previous study demonstrated that TTK is highly expressed in OC and that its upregulation contributes to cisplatin resistance in OC27. However, the role of TTK in resistance to PARPis in OC remains unclear.
Replication protein A2 (RPA2) is a subunit of the heterotrimeric replication protein A (RPA) complex, which is composed of three subunits: RPA1, RPA2, and RPA328. RPA plays a vital role in DNA metabolism, including DNA replication, recombination, and the DNA damage response29. In response to DNA damage, RPA2 undergoes phosphorylation of several amino acid residues within its N-terminal domain, which is referred to as RPA2 hyperphosphorylation30,31,32,33,34. Nine potential phosphorylation sites have been identified, with threonine 21, serine 4, 8, and 33, along with at least one phosphoserine at residues 11–13, being involved in the DNA damage response.
In response to the DNA damage response, especially HR (homologous recombination)-mediated repair, RPA2 activates ataxia telangiectasia and Rad3-related protein (ATR) kinase. This activation facilitates the recruitment of RAD51, or RPA2 directly interacts with RAD51 to enhance repair processes35. RPA has also been linked to cisplatin resistance in OC36. Cisplatin induces the phosphorylation of RPA2 in head and neck squamous cell carcinoma cells, and higher levels of RPA2 phosphorylation correlate with reduced cisplatin sensitivity37. However, the role of RPA2 in regulating resistance to PARPis remains unclear and warrants further investigation to understand whether and how RPA2 contributes to this form of resistance in OC.
In this study, we found that TTK inhibition overcomes olaparib resistance in HR-proficient OC cells, in part by suppressing RPA2-S33 phosphorylation and attenuating ATR signaling. TTK inhibitors offer a promising strategy to increase the therapeutic efficacy of PARPis in OC patients.
Results
TTK promotes olaparib resistance in OC cells
To investigate the role of TTK in the sensitivity of OC cells to olaparib, we utilized HR-proficient OC cell lines (OVCAR838,39 and OV9039,40), HR-deficient UWB1.289 cells41,42,43, and olaparib-resistant SKOV3 (SKOV3/OLA) cells44,45. The SKOV3/OLA cells were established by gradually exposing parental SKOV3 cells40 to increasing concentrations of olaparib in our laboratory44,45. Firstly, we transfected OVCAR8, OV90, and SK/OLA cells with TTK shRNA to knock down TTK expression (Fig. 1A and Supplementary Fig. 1A). Additionally, TTK was overexpressed in OVCAR8, OV90, and UWB1.289 cells (Fig. 1B and Supplementary Fig. 1B). TTK-knockdown OVCAR8, OV90, and SK/OLA cells were treated with gradient concentrations of olaparib. MTT assays revealed that TTK knockdown sensitized these cells to olaparib treatment (Fig. 1C). Colony survival assays confirmed these findings (Fig. 1D and Supplementary Fig. 1C, D). Conversely, cells overexpressing TTK presented increased resistance to olaparib (Fig. 1E, F and Supplementary Figs. 1E and 2).
A OVCAR8, OV90, and SK/OLA cells were stably transfected with PLKO.NC or TTK shRNA (shTTK1 and shTTK2). TTK protein levels were assessed by Western blot. B OVCAR8, OV90, and UWB1.289 cells were stably transfected with PCMV or PCMV-TTK plasmids. TTK protein levels were determined by Western blot. C, D OVCAR8, OV90, and SK/OLA cells stably transfected with PLKO.NC or TTK shRNA (shTTK1 and shTTK2) were treated with increasing concentrations of olaparib (0, 25, 50, and 100 μM for OVCAR8 and OV90 cells; 0, 25, 50, 100, 200, and 400 μM for SK/OLA cells). The effects of TTK on olaparib sensitivity were measured by MTT assay (C) and colony survival assay (D). Data are mean ± SD, n = 3, one-way ANOVA. a: p relative to TTK knockdown alone; b: p relative to olaparib treatment alone. E, F OVCAR8, OV90, and UWB1.289 cells stably transfected with PCMV or PCMV-TTK plasmids were treated with varying concentrations of olaparib. The effects of TTK on olaparib sensitivity were evaluated by MTT assay (E) and colony survival assay (F). Data are mean ± SD, n = 3, one-way ANOVA. a: p relative to TTK overexpression alone; b: p relative to olaparib treatment alone. (Data are mean ± SD, one-way ANOVA, *P < 0.05, **P < 0.01, n = 3).
In conclusion, these results reveal that inhibition of TTK enhances sensitivity to olaparib in HR-proficient OC cells, whereas TTK overexpression promotes olaparib resistance in HR-deficient OC cells.
TTK inhibitors enhance the effect of olaparib in OC cells
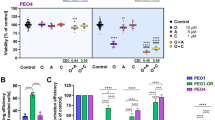
We further investigated the impact of TTK inhibitors on olaparib sensitivity in OC cells using two different TTK inhibitors: CFI402257 (referred to as CFI) and BAY1161909 (referred to as B909). The IC50 values for both inhibitors were first determined using the MTT assay. In OVCAR8 and OV90 cells, the IC50 values of CFI were 528.84 nM (Fig. 2A and Supplementary Fig. 3A) and 1011 nM27, respectively. The IC50 values of B909 were 217.31 nM and 358.34 nM in OVCAR8 and OV90 cells, respectively (Fig. 2B and Supplementary Fig. 3B). Both MTT and colony survival assays demonstrated that CFI and B909 enhanced the efficacy of olaparib in OC cells (Fig. 2C–F and Supplementary Fig. 3C, D).
A The IC50 of CFI402257 (CFI) in OVCAR8 cells (IC50 = 528.84 nM, 96 h). Data are mean ± SD, n = 3. B The IC50 of BAY1161909 (B909) in OVCAR8 cells (IC50 = 217.31 nM, 96 h) and OV90 cells (IC50 = 358.34 nM, 96 h). Data are mean ± SD, n = 3. C, D MTT, and colony survival assays were performed to assess the efficacy of the combined treatment of CFI and olaparib (OLA) in OVCAR8 and OV90 cells. Data are mean ± SD, n = 3, one-way ANOVA. a: p relative to CFI treatment alone; b: p relative to olaparib treatment alone. The effect of B909 on olaparib sensitivity was evaluated using the MTT assay (E) and colony survival assay (F). Data are mean ± SD, n = 3, one-way ANOVA. a: p relative to B909 treatment alone; b: p relative to olaparib treatment alone. (Data are mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, n = 3).
These results suggest that TTK inhibitors can potentiate the therapeutic effect of olaparib in OC cells.
TTK directly interacts with RPA2
To investigate the mechanism through which TTK promotes resistance to olaparib in OC, we performed MS analysis. The results revealed that RPA2 was significantly enriched among the pull-down proteins (Fig. 3A and Supplementary Data 1). To confirm the endogenous interaction between TTK and RPA2, Co-IP assays were conducted in OVCAR8 and OV90 cells using control IgG, anti-TTK, or anti-RPA2 antibodies. The results demonstrated that TTK interacts with RPA2 endogenously (Fig. 3B, C). Furthermore, immunofluorescence staining revealed that both TTK and RPA2 are localized in the nucleus of OC cells (Fig. 3D). A proximity ligation assay (PLA) further confirmed the interaction between TTK and RPA2, with the red dots indicating protein interactions in OVCAR8 and OV90 cells. At the same time, TTK antibody or RPA2 antibody alone was used as negative control (Fig. 3E). Additionally, when exogenous pMyc-RPA2 and Flag-TTK plasmids were co-transfected into HEK293T cells for 48 h, Co-IP assays confirmed the binding of TTK and RPA2 exogenously (Fig. 3F, G). Moreover, under CFI treatment, the interaction between TTK and RPA2 was weakened (Fig.3H and Supplementary Fig. 4A). In contrast, an increased interaction occurred with olaparib treatment (Fig.3I and Supplementary Fig. 4B), which was impaired when the cells were treated with CFI (Fig.3J and Supplementary Fig. 4C).
A The MS-based analysis of TTK targeted genes. B, C Co-IP assay was performed to demonstrate the endogenous interaction between TTK and RPA2 in OVCAR8 and OV90 cells. D Immunofluorescence staining showed the intracellular localization of TTK (green) and RPA2 (red). The nucleus was stained with DAPI (blue). Scale bar: 10 µm. E TTK and RPA2 interaction detected by Duo-link PLA in OVCAR8 and OV90 cells. TTK antibody or RPA2 antibody alone was used as negative control. Scale bar: 5 µm. Data are mean ± SD, n = 3, one-way ANOVA, **P < 0.01. F, G HEK293T cells were co-transfected with pMyc-RPA2 and Flag-TTK vectors for 48 h. Co-IP assays verified the exogenous interaction between pMyc-RPA2 and Flag-TTK. F Anti-Myc antibody was used for Co-IP, and anti-Flag antibody detected Flag-TTK expression in the immunoprecipitated proteins. G Anti-Flag antibody was used for Co-IP, and anti-Myc antibody detected pMyc-RPA2 expression in the immunoprecipitated proteins. H OVCAR8 cells were treated with CFI for 24 h, and Co-IP assays were performed to detect the effect of CFI on the interaction between TTK and RPA2. I OVCAR8 cells were treated with olaparib for 24 h, and Co-IP assays were performed to evaluate the effect of olaparib on the interaction between TTK and RPA2. J OVCAR8 and OV90 cells were treated with olaparib alone or in combination with CFI for 24 h. Co-IP assays were conducted to detect the interaction between TTK and RPA2. (Data are mean ± SD, **P < 0.01, n = 3).
In conclusion, these findings provide strong evidence that TTK directly interacts with RPA2 in OC cells.
TTK phosphorylates RPA2 at the S33 residue
The phosphorylation of RPA2, particularly at the Ser33 (S33), Ser4, and Ser8 (S4/8) residues, is known to play a key role in HR. To explore whether TTK can phosphorylate RPA2, we assessed the phosphorylation levels of RPA2 at specific residues. We observed that RPA2 phosphorylation at S33 (pRPA2(S33)) was significantly reduced in TTK knockdown cells (Fig. 4A and Supplementary Fig. 5A) and upon CFI treatment (Fig. 4B and Supplementary Fig. 5B). However, neither TTK knockdown nor inhibitor treatment affected the phosphorylation level of RPA2 at Ser4/8 (pRPA2(S4/8)) (Fig. 4A, B and Supplementary Fig. 5A, B). Exogenous overexpression of TTK led to an increase in the pRPA2(S33) levels in endogenous RPA2 but had no effect on the pRPA2(S4/8) levels in OVCAR8 and OV90 cells (Fig. 4C and Supplementary Fig. 5C). Similarly, the pRPA2(S33) levels of exogenous RPA2 in 293T cells were increased following the transfection of Flag-TTK (Supplementary Fig. 5D). These results suggest that TTK can indeed phosphorylate RPA2 at Ser33. Moreover, an in vitro kinase assay demonstrated that TTK induced the phosphorylation of RPA2 at S33 (Fig. 4D). Furthermore, olaparib treatment increased the level of pRPA2(S33), but this increase was attenuated by either TTK knockdown or CFI treatment (Fig. 4E, F, and Supplementary Fig. 5E, F), indicating that TTK plays a key role in the olaparib-induced phosphorylation of RPA2 at S33. To validate the importance of S33 phosphorylation, we constructed a S33A mutant of RPA2, in which the serine residue was replaced with alanine, rendering it phosphorylation deficient. Our results revealed that serine phosphorylation (p-Ser) increased after olaparib treatment in wild-type (WT) RPA2 but not in the S33A mutant cells, indicating that the mutated RPA2 S33A cannot be phosphorylated (Fig. 4G).
A OVCAR8 and OV90 cells were transfected with PLKO.NC or TTK shRNA (shTTK1 and shTTK2). Western blotting was performed to detect the levels of TTK, phosphorylation on RPA2 S33 (pRPA2 S33), S4 and S8 (pRPA2 S4/8), RPA2, and β-actin. B OVCAR8 and OV90 cells were treated with CFI for 48 h. Western blotting was used to assess the levels of TTK, pRPA2 S33, pRPA2 S4/8, RPA2, and β-actin. C OVCAR8 and OV90 cells were transfected with or without Flag-TTK. Western blotting was performed to evaluate the levels of TTK, pRPA2 S33, pRPA2 S4/8, RPA2, and β-actin. D In vitro translated pMyc-RPA2 was incubated with TTK and ATP, and the pRPA2 S33 level was detected by IP assay. E OVCAR8 and OV90 cells were transfected with NC or shTTK for 24 h, followed by treatment with olaparib for 24 h. Western blotting was performed to detect the levels of TTK, pRPA2 S33, RPA2, and β-actin. F OVCAR8 and OV90 cells were treated with CFI, olaparib alone, or a combination of CFI and olaparib for 48 h. Western blotting was conducted to assess the levels of TTK, pRPA2 S33, RPA2, and β-actin. GWild-type or mutant RPA2 was transfected into OVCAR8 cells, and the cells were treated with olaparib for 24 h before a co-immunoprecipitation (Co-IP) assay was performed. The serine phosphorylation of RPA2 was then detected. (n = 3).
In conclusion, these findings suggest that the S33 residue of RPA2 is a critical phosphorylation site for TTK.
RPA2 silencing enhances olaparib sensitivity in OC cells
To investigate whether RPA2 is associated with olaparib resistance in OC, we silenced RPA2 expression in OVCAR8 and OV90 cells (Fig. 5A and Supplementary Fig. 6A). MTT and colony survival assays revealed that RPA2 knockdown increased the sensitivity of OVCAR8 and OV90 cells to olaparib (Fig. 5B, C and Supplementary Fig. 6B, C). Immunofluorescence analysis revealed that, compared with control treatment, RPA2 silencing resulted in an increase in the number of γH2AX foci in the nucleus following olaparib treatment (Fig. 5D and Supplementary Fig. 6D). To investigate the impact of RPA2 silencing on homologous recombination (HR) activity, we co-stained RAD51 and PCNA, a S-phase marker. The results revealed that RPA2 silencing led to a reduction in RAD51 foci in PCNA-positive S-phase cells, particularly after olaparib treatment (Fig. 5E and Supplementary Fig. 6E). Furthermore, the alkaline comet assay demonstrated that the combination of olaparib and RPA2 knockdown significantly increased the tail moment relative to that of the olaparib-only treatment group (Fig. 5F and Supplementary Fig. 6F). These findings suggest that RPA2 silencing enhances the sensitivity of OC cells to olaparib.
OVCAR8 and OV90 cells were stably transfected with PLKO.NC or RPA2 shRNA (shRPA2 1# and shRPA2 2#). A The protein levels of RPA2 in OVCAR8 and OV90 cells were detected by Western blotting. B MTT assays were performed to assess the effect of RPA2 knockdown on the sensitivity of olaparib in OVCAR8 and OV90 cells. Data are mean ± SD, n = 3, Student’s t-test, **P < 0.01. C The colony survival assay was conducted to evaluate the influence of RPA2 on olaparib sensitivity. D Immunofluorescence was used to evaluate the number of γH2AX foci (Scale bar: 10 µm). E Representative images of costaining for PCNA (red) and RAD51 (green) foci (Scale bar: 10 µm). F The alkaline comet assay was performed to detect the DNA damage levels (Scale bar: 10 µm). (Data are mean ± SD, **P < 0.01, n = 3, Scale bar: 10 µm.).
TTK regulates olaparib resistance through phosphorylating RPA2 at the S33 residue
We further explored the role of RPA2 in olaparib resistance induced by TTK in OC cells. OVCAR8 and OV90 cells were transfected with PCMV, TTK, shRPA2 1# or a combination of TTK and shRPA2 1# for 48 h (Fig. 6A and Supplementary Fig. 7A). MTT and colony survival assays showed that TTK overexpression conferred olaparib resistance, while RPA2 silencing increased the sensitivity of OC cells to olaparib. Furthermore, RPA2 knockdown partially reversed the olaparib resistance induced by TTK overexpression (Fig. 6B–D and Supplementary Fig. 7B). Immunofluorescence analysis demonstrated that TTK overexpression decreased the number of γH2AX foci, while RPA2 silencing increased γH2AX foci in the nucleus, especially following olaparib treatment. Furthermore, RPA2 silencing partially reversed the γH2AX foci formation reduced by TTK overexpression (Fig. 6E and Supplementary Fig. 7C). Co-staining of RAD51 and PCNA revealed that TTK overexpression enhanced RAD51 foci formation in the nuclei of PCNA-positive S-phase cells. Conversely, RPA2 silencing reduced RAD51 foci in these cells, particularly after olaparib treatment. Additionally, RPA2 silencing partially reversed the RAD51 foci formation induced by TTK overexpression (Fig. 6F and Supplementary Fig. 7D). The comet tail assay revealed that TTK overexpression alleviated DNA damage, while RPA2 knockdown induced DNA damage. Additionally, RPA2 knockdown partially reversed the DNA damage reduction caused by TTK overexpression (Fig. 6G and Supplementary Fig. 7E). To assess whether TTK-induced phosphorylation of RPA2 at the S33 contributes to TTK-mediated olaparib resistance in OC cells, we treated cells with CFI for 24 h, followed by transfection with either wild-type or mutant RPA2 plasmids. The response to olaparib was then evaluated. As shown in Supplementary Fig. 8A–D, cells transfected with wild-type RPA2 and the RPA2 S33E mutant presented increased cell viability compared to CFI treatment alone, whereas the RPA2 S33A mutant did not.
OVCAR8 and OV90 cells were transfected with PCMV, TTK, shRPA2 1# or TTK combined with shRPA2 1# for 48 h. A Western blot analysis was performed to detect the levels of TTK, RPA2, and β-actin. B MTT assays were performed to detect the relative cell viability after olaparib treatment. Data are mean ± SD, n = 3, one-way ANOVA. C, D The colony survival assay was conducted to assess the colony formation ability under olaparib treatment. Data are mean ± SD, n = 3, Student’s t-test. E Immunofluorescence was used to evaluate the number of γH2AX, Scale bar: 10 µm. F Representative images of costaining for PCNA (red) and RAD51 (green) foci, Scale bar: 10 µm. G The alkaline comet assay was performed to detect the DNA damage induced by olaparib treatment, Scale bar: 10 µm. (Data are mean ± SD, *P < 0.05, **P < 0.01, Scale bar: 10 µm, n = 3).
Together, these results suggest that TTK inhibition can overcome olaparib resistance in HR-proficient OC cells, in part by inhibiting RPA2-S33 phosphorylation.
TTK activates the ATR signaling pathway through RPA2
The phosphorylation of RPA2 at S33 has been established as a marker for ATR activation. After we verified that TTK phosphorylates RPA2 at S33, we aimed to determine whether TTK activates the ATR signaling pathway through RPA2. First, OVCAR8 and OV90 cells were transfected with either PLKO.NC (control) or RPA2 shRNA. After RPA2 knockdown, the level of phosphorylated ATR (pATR) was reduced (Fig. 7A and Supplementary Fig. 9A). As shown in Fig. 7B and Supplementary Fig. 9B, ATR was activated to a greater extent in cells transfected with wild-type RPA2 than in those transfected with the RPA2 S33A mutant. In addition, TTK knockdown and CFI treatment reduced the pATR level in OC cells (Fig. 7C, D and Supplementary Fig. 9C, D), whereas TTK overexpression increased the phosphorylation of ATR (Fig. 7E and Supplementary Fig. 9E). Olaparib treatment further enhanced ATR activation (Fig. 7E and Supplementary Fig. 9E). Transfecting RPA2 into cells treated with CFI reversed the decrease in ATR activity caused by TTK inhibition (Fig. 7F and Supplementary Fig. 9F). To explore whether TTK promotes olaparib resistance through the RPA2/ATR signaling pathway, OVCAR8 and OV90 cells transfected with shTTK2 and RPA2 were treated with VE-821, an ATR inhibitor. MTT and colony survival assays revealed that TTK knockdown increased sensitivity to olaparib and that RPA2 overexpression reversed the effects of TTK knockdown. Moreover, blocking ATR with VE-821 counteracted the protective effect of RPA2 overexpression (Fig. 7G, H, and Supplementary Figs. 9G and 10A).
A OVCAR8 and OV90 cells were transfected with PLKO.NC or RPA2 shRNA (shRPA2 1# and shRPA2 2#). Western blot assays were conducted to detect the levels of RPA2, pATR, ATR, and β-actin. B OVCAR8 and OV90 cells were transfected with pcDNA3.1, pcDNA3.1-RPA2, and pcDNA3.1-RPA2 S33A plasmids for 48 h. Western blot analysis was used to measure the protein levels of RPA2, pATR, ATR, and β-actin. C OVCAR8 and OV90 cells were transfected with PLKO.NC or TTK shRNA (shTTK1 and shTTK2). Western blot assays were performed to assess the levels of TTK, pATR, ATR, and β-actin. D OVCAR8 and OV90 cells were treated with CFI for 48 h, then the levels of TTK, pATR, ATR, and β-actin were evaluated. E OVCAR8 and OV90 cells transfected with PCMV-TTK were treated with or without olaparib. Western blot assays were used to detect the protein levels of TTK, pRPA2 S33, RPA2, pATR, ATR, and β-actin. F OVCAR8 and OV90 cells were treated with CFI for 24 h, followed by transfection with pcDNA3.1 or pcDNA3.1-RPA2. The protein levels of TTK, pRPA2 S33, RPA2, pATR, ATR, and β-actin were assessed. G, H OVCAR8, and OV90 cells were transfected with shTTK2 and RPA2 for 48 h, followed by treatment with or without VE-821 (an ATR inhibitor) for 24 h. Afterward, olaparib was added for 72 h. MTT and colony survival assays were performed to demonstrate that TTK inhibition overcame olaparib resistance in HR-proficient OC cells, partly through the RPA2/ATR axis. Data are mean ± SD, n = 3, one-way ANOVA. (Data are mean ± SD, *P < 0.05, **P < 0.01, n = 3).
In conclusion, these results suggest that TTK inhibition can overcome olaparib resistance in HR-proficient OC cells, in part by suppressing RPA2-S33 phosphorylation and attenuating ATR signaling.
TTK facilitates olaparib resistance in vivo
We used a mouse model to investigate whether TTK contributes to olaparib resistance in vivo. OVCAR8 cells were subcutaneously injected into the left armpit of 5-week-old female mice. Once the tumor volume reached ~100 mm³, the mice were randomly divided into four groups: Control (Ctr), CFI, Olaparib (OLA), and CFI combined with Olaparib (CFI + OLA). The indicated drugs were administered for 2 weeks. As shown in Fig. 8A–C, both CFI and olaparib treatment alone inhibited tumor growth, but the combination of CFI and olaparib resulted in more pronounced inhibition. Western blot analysis revealed that CFI treatment decreased the levels of pRPA2(S33) and pATR. However, olaparib treatment increased the levels of pRPA2(S33) and pATR, with the increase being attenuated by CFI in the mouse model (Fig. 8D, E). Furthermore, CFI and olaparib treatment alone decreased the Ki67 level, but increased the cleaved caspase 3 level. The combination of CFI and olaparib resulted in more significant changes in Ki67 and cleaved caspase 3 levels (Fig. 8D, E). These results suggest that TTK inhibition can overcome olaparib resistance in vivo, likely by suppressing RPA2-S33 phosphorylation and attenuating ATR signaling.
OVCAR8 cells were subcutaneously injected into the left armpit of each 5-week-old female mouse. The mice were randomly divided into four subgroups (Control, CFI, OLA, and CFI + OLA) when the tumor volumes reached ~100 mm3, and the indicated drugs were administered for 2 weeks. A Photographs of tumors from each group. B Tumor weights in each group. Data are mean ± SD, n = 6, one-way ANOVA. C Body weight of the mice in each group. Data are mean ± SD, n = 6, one-way ANOVA. D, E The protein levels of TTK, pRPA2 S33, RPA2, pATR, ATR, Ki67, cleaved caspase 3, pro-caspase 3, and β-actin were measured in tumor tissues from each group. Data are mean ± SD, n = 6, one-way ANOVA. (Data are mean ± SD, #P>0.05, *P < 0.05, **P < 0.01, ***P < 0.001, n = 6).
Discussion
TTK has been identified as an oncogene, and its inhibition has been explored as a potential targeted therapy for various cancers23,24,25,26,27. In basal-like breast cancer, TTK inhibition enhances sensitivity to radiotherapy by impairing HR capability23. Similarly, both genetic and pharmacological suppression of TTK induce G2/M cell cycle arrest, increase DNA damage, and promote apoptosis and necrosis through p21-mediated pathways in vitro, thereby improving radiotherapy sensitivity in liver cancer cells24. In glioma, TTK inhibition enhances radiotherapy sensitivity by reducing the HR and non-homologous end joining (NHEJ) repair capacity26. Moreover, our previous study demonstrated that TTK-specific siRNAs and inhibitors increase the toxicity of cisplatin in OC cells27. In the present study, we show that TTK inhibition overcomes olaparib resistance in HR-proficient OC cells, whereas TTK promotes olaparib resistance in HR-deficient OC cells.
TTK is a dual-specificity protein kinase that plays a crucial role as a mitotic checkpoint protein, regulating cell proliferation, the cell cycle, and the DNA damage response8,9,10. TTK is activated and phosphorylates CHK2 at Thr68, thereby activating CHK246. In turn, CHK2 can phosphorylate and stabilize TTK following DNA damage47. These interactions suggest a potential cross-talk between the spindle assembly checkpoint and the DNA damage checkpoint, with TTK being proposed as a DNA damage sensor in mitotic cells46. We observed that olaparib treatment increased TTK levels, likely due to replication stress and DNA damage induced by olaparib. Moreover, the induction of TTK during olaparib treatment may represent an adaptive resistance mechanism, although the precise mechanism requires further investigation. Additionally, TTK phosphorylates MDM2, promoting H2B ubiquitination and chromatin decompaction, which enhances oxidative DNA damage repair through the ATR-CHK1 signaling pathway48. In this study, we identified RPA2 as a novel substrate of TTK, with TTK directly interacting with RPA2 to promote its phosphorylation at S33.
Phosphorylated RPA2 plays a significant role in cell cycle regulation and the DNA damage response (DDR). RPA binds to single-strand DNA (ssDNA) and forms an essential structure (RPA-ssDNA) for activating ATR kinase49,50. Once activated, ATR phosphorylates multiple substrates to prevent further DNA replication origin activation, stabilize stalled replication forks, and activate cell-cycle checkpoints51. Additionally, RPA2 and ATR are key players in HR repair. During HR-mediated repair, RPA2 activates ATR and recruits RAD51 or directly interacts with RAD5133. For example, following hydroxyurea (HU) treatment, cells with an RPA2-(7xA) phospho-mutant formed fewer RAD51 foci and exhibited reduced HR capacity28. Furthermore, cells expressing RPA2 S8D/S33D presented significantly lower HR efficiency than did cells expressing wt-RPA252,53. In addition, ATR enhances HR and RAD51 functions54. In our study, we demonstrated that RPA2 is involved in the regulation of olaparib resistance in OC, with TTK playing a role in promoting this resistance, partly through RPA2. Specifically, silencing RPA2 resulted in increased γH2AX expression and decreased RAD51 accumulation. Conversely, TTK overexpression significantly reduced γH2AX expression and increased the number of RAD51 foci following olaparib treatment. These changes were reversed by RPA2 knockdown, indicating that the DNA repair function of TTK and RPA2 is partly dependent on HR. Additionally, we showed that TTK activates the ATR signaling pathway in OC through RPA2. However, it remains unclear whether TTK inhibition primarily acts by potentiating replication stress, suppressing HR, or both. To fully understand these mechanisms, future studies should examine replication fork dynamics and HR efficiency. Furthermore, it should be confirmed whether TTK inhibition and the subsequent abrogation of ATR activity during Olaparib-mediated replication stress led to the uncontrolled firing of origins, excessive accumulation of ssDNA, DSBs, and collapsed replication forks.
Our study demonstrates that TTK inhibition effectively overcomes olaparib resistance in HR-proficient OC cells. Mechanistically, TTK interacts directly with RPA2 to promote its phosphorylation at S33, which in turn activates the ATR signaling pathway. On the basis of these findings, combining TTK inhibitors with olaparib as treatment for OC can lead to increased efficacy. This approach could particularly benefit OC patients with high TTK expression or resistance to olaparib alone.
Materials and methods
Cell lines and cell culture
OVCAR8 cells were obtained from the M.D. Anderson Cancer Center’s Characterized Cell Line Core. OV90 and UWB1.289 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), while olaparib-resistant SKOV3 (SKOV3/OLA) cells were established by gradually exposing parental SKOV3 cells to increasing concentrations of olaparib in our laboratory. HEK293T cells were purchased from the Chinese Academy of Sciences (Shanghai, China).
OV90 and HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 15% fetal bovine serum (FBS). OVCAR8 cells were maintained in RPMI-1640 medium with 10% FBS. SKOV3/OLA cells were cultured in DMEM with 10% FBS. UWB1.289 cells were grown in a 50:50 mixture of RPMI-1640 and Mammary Epithelial Growth Medium (MEGM), supplemented with 5% FBS. All cultures were incubated at 37 °C in a humidified atmosphere with 5% CO2. All cells were absence of mycoplasma contamination and routinely tested using Short Tandem Repeat (STR) profiling.
Cell culture reagents
Olaparib (S1060), CFI402257(S7552), and BAY1161909 (S8214) were obtained from Selleck Chemicals (Houston, TX, USA).
Stable cell lines establishment
The TTK overexpression plasmid and corresponding control vectors were obtained from GeneChem (Shanghai, China). Knockdown plasmids were constructed by inserting the indicated sequences into PLKO.NC plasmid, which served as the control. Lentivirus was generated co-transfecting HEK293T cells with the psPAX2, pMD2.G, and PCMV/TTK (PLKO.NC/shTTK or shRPA2) plasmids. OVCAR8, OV90, UWB1.289, and SK/OLA cells, at appropriate confluence, were treated with the corresponding lentivirus for 24 h and subsequently selected with 2 μg/ml puromycin (Merck Millipore, USA) for 1 week.
The sequences of the short hairpin RNAs (shRNA) were as follows:
shTTK1: 5′-AATGAACAAAGTGAGAGACAT-3′
shTTK2: 5′-GCACAATTTGAACTGTCACAA-3′.
shRPA2 1# (TRCN0000231924) and shRPA2 2# (TRCN0000005990) were sourced from Sigma -Aldrich.
Cell viability assay
To assess the relative toxicity of olaparib in different groups, stable cell lines were constructed as described above. Gradient concentrations of olaparib were then added to the corresponding wells of a 96-well plate. Each well was seeded with 3000 cells from each group 24 h prior to treatment with olaparib. After 48 h, 20 µL MTT solution was added to each well and incubated for an additional 4 h. Following incubation, 100 µL of DMSO was added to each well to dissolve the formazan crystals. The relative absorbance at 490 nm was measured using a Varioskan Flash microplate reader (Thermo Scientific).
Colony survival assay
Ovarian cancer cells stably overexpressing or knocking down TTK or RPA2, along with their corresponding control cells, were seeded at 1000 cells per well in 6-well plates. The medium was replaced every 3 days until the colonies grew to ~50 cells. Then, gradient concentrations of olaparib were added, and the cells were cultured for an additional 48 h. After treatment, the plates were fixed with methanol and stained with 1% crystal violet. Images of the stained colonies were captured for further analysis.
Protein extraction and Western blotting
Cells were lysed by RIPA (Beyotime, P0013B, China), following with centrifuging at 4 °C for 10 min, and the supernatants were collected. The protein concentration was quantified with BCA Protein Assay kit (Merck Millipore, USA). Equivalent amounts of proteins were loaded onto gels and separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), followed by transfer to PVDF membranes (Merck Millipore, Burlington, MA, USA). PVDF membranes were incubated with specific primary antibodies at 4 °C overnight, followed by the addition of the appropriate secondary antibodies. After a 1-h incubation with the secondary antibodies, an enhanced chemiluminescence detection kit (ECL ORT2655, PerkinElmer, Waltham, MA, USA) was used to visualize the signal, which was captured with a GE Amersham Imager 600 (GE). The primary antibodies used in the analysis were as follows: TTK (1:1000; abcam; ab11108), RPA2(1:1000; proteintech; 10412-1-AP), pRPA2 S4/8 (1:1000; abcam; ab243866), pRPA2 S33 (1:1000; abcam; ab211877), β-actin (1:5000; Sigma–Aldrich; A5441), pATR S428 (1:1000; CST; 2853 T), ATR (1:1000; Selleck; F0773), Flag-tag (1:1000; CST; 8146 s), Myc-tag (1:1000; proteintech; 60003-2-Ig).
Co-Immunoprecipitation (CO-IP)
Total protein was extracted using Western and IP Lysis Buffer (Beyotime; P0013) and then centrifuged at 4 °C for 15 min. The supernatant (800 µL) was transferred to a new 1.5 mL tube and incubated overnight at 4 °C with the indicated primary antibodies or control (IgG).
On the following day, 40 μL of protein A/G Magnetic Beads for IP (B23202, Selleck) was added to the mixture and incubated for another 1 h at 4 °C with gentle rotation. The beads were then washed three times with cold PBS containing PMSF (1:100). After washing, the beads were resuspended in 25 µL of 3 × SDS loading buffer and boiled for 5 min in a metal bath. Finally, Western blot analysis was performed as described earlier.
Mass spectrometry (MS) assay
A total of 1 × 107 OVCAR8 cells were lysed with Western and IP Lysis Buffer (Beyotime; P0013) and centrifuged at 4 °C for 15 min. The cell supernatant was then incubated with anti-IgG or anti-TTK antibody (ab11108, Abcam) at 4 °C overnight.
On the following day, the proteins were collected as described in the CO-IP part and separated on SDS-PAGE gels. The gels were then stained with Coomassie Brilliant Blue. Afterward, the gel bands were sent for Liquid chromatography-tandem MS (LC-MS/MS) analysis. The LC-MS/MS analysis was performed by Applied Protein Technology (Shanghai, China).
Immunofluorescence assay and Duolink proximity-ligation assays (PLA)
Cells were seeded onto glass coverslips within 24-well plates for 24 h. Afterward, they were fixed with 4% paraformaldehyde for a quarter of an hour and blocked with normal goat serum for half an hour.
For immunofluorescence assay, the cells were incubated with γH2AX (1:100, Abcam; ab26350) alone or RAD51 (1:100, Abcam; ab133534) along with PCNA (1:100, proteintech;10205-2-AP) overnight at 4 °C. On the following day, the cells were gently rinsed with PBS for three times and incubated with the corresponding secondary antibodies for 1 h at 37 °C in the dark. Finally, the cells were stained with DAPI (1:1000) for 5 min at room temperature to label the nuclei. Images were captured using a Zeiss LSM 780 (Carl Zeiss, Jena, Germany).
For PLA assay, TTK (1:100, abcam; ab11108) and RPA2(1:100; proteintech; 10412-1-AP) antibodies were added overnight at 4 °C. While, TTK antibody or RPA2 antibody alone was used as negative control. Then, Duolink® experiments were performed according to the manufacturer’s protocol (navinci, NC.MR.100.RED).
In vitro phosphorylation assay
Flag-TTK, pMyc-RPA2, and the corresponding control proteins were synthesized by the TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, Wisconsin, USA) according to the manufacturer’s instructions. The synthesized proteins were incubated in 10× kinase buffer at 4 °C overnight. The following day, pMyc-RPA2 was mixed with 100 μM ATP and incubated separately with Flag-TTK or the control in 10× kinase buffer at 30 °C for 1 h. After the incubation, an anti-Myc tag antibody was added to the sample, mixed with 500 μL of 1× kinase buffer, and incubated at 4 °C for 1 h. Protein A/G Magnetic Beads for IP (B23202, Selleck) were then added, and the mixture was rotated at room temperature for 1 h. The proteins were collected as described in the CO-IP section and detected by western blot.
Alkaline comet assay
Cells were harvested and resuspended in cold 1×PBS at a concentration of 1 × 105 cells/mL. The cell suspension was mixed with molten CometAssay LMAgarose (at 37 °C) at a 1:10 (v/v) ratio, and 50 μL of the mixture was immediately pipetted onto CometSlide™. The slide was incubated at 4 °C in the dark for 10 min to allow solidification. Following this, the slide was immersed in Lysis Solution at 4 °C for 30 min, then treated with Alkaline Unwinding Solution for 20 min at room temperature. The slide was placed in an electrophoresis tray, immersed in Alkaline Electrophoresis Solution, and electrophoresis was carried out at 21 V for 30 min. After electrophoresis, the slide was washed with deionized water and 70% ethanol, then stained with SYBR® Gold. The slide was visualized using a fluorescence microscope. Finally, the images were analyzed with OpenComet on ImageJ.
Tumor formation assay in nude mice
All animal experiments were conducted with approval from the Shandong University Animal Care and Use Committee. We have complied with all relevant ethical regulations for animal use.
OVCAR8 cells (about 5 × 106) were subcutaneously injected into the left armpit of each female athymic BALB/c nude mouse. Tumor volumes were observed and measured every other day using the formula: volume = length × width2 × 0.5. Once the tumor volume reached ~100 mm³, the mice were randomly assigned to four subgroups (Control, CFI, OLA, and CFI + OLA). CFI-402257 was given at the dose of 6 mg/kg via oral gavage, while olaparib was administered at a dose of 50 mg/kg via intraperitoneal injection. The drugs were given either alone or in combination, once daily, for 14 days. Following the treatment period, the tumors were harvested, photographed, and analyzed.
Statistics and reproducibility
All experiments were repeated independently at least three times. The sample size (n) is indicated in the figure legends. The data are presented as means ± SD. The significance between two groups was analyzed using Student’s t-test, while one-way ANOVA was applied for comparisons among more than two groups, using SPSS v22.0 (SPSS, Inc., Chicago, IL, USA). A p value of <0.05 was considered statistically significant, with the following designations: #p > 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001. Data and graphics were processed using GraphPad Prism 8.00 (GraphPad Software, La Jolla, CA, USA) and Adobe Photoshop CC 2019 (Adobe, San Jose, CA, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the article and its Supplementary Information files. Uncropped WB images can be found in the Supplementary Fig. 11 within the Supplementary Information. The MS-based analysis of TTK binds proteins is provided as Supplementary Data 1. The source data behind the graphs in the paper are provided in Supplementary Data 2. Additional data and materials are available from the corresponding author upon reasonable request.
References
Bookman, M. Optimal primary therapy of ovarian cancer. Ann. Oncol. 27, i58-i62 (2016).
Vaughan, S. et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer 11, 719–725 (2011).
Armstrong D, A. R. et al. Ovarian cancer, version 1. 2024, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw 22, 43–69 (2024).
Noordermeer, S. & van Attikum, H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol. 29, 820–834 (2019).
Li, H. et al. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol. Cancer 19, 107 (2020).
Dias, M. P., Moser, S. C., Ganesan, S. & Jonkers, J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 18, 773–791 (2021).
Fugger, K., Hewitt, G., West, S. C. & Boulton, S. J. Tackling PARP inhibitor resistance. Trends Cancer 7, 1102–1118 (2021).
Liu, X. & Winey, M. The MPS1 family of protein kinases. Annu. Rev. Biochem. 81, 561–585 (2012).
Jones, M. et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. CB 15, 160–165 (2005).
Lara-Gonzalez, P., Westhorpe, F. & Taylor, S. The spindle assembly checkpoint. Curr. Biol. CB 22, R966–R980 (2012).
Stratford, J. et al. Genetic and pharmacological inhibition of TTK impairs pancreatic cancer cell line growth by inducing lethal chromosomal instability. PLoS ONE 12, e0174863 (2017).
Liang, X. et al. Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS ONE 9, e97739 (2014).
Liu, X., Liao, W., Yuan, Q., Ou, Y. & Huang, J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget 6, 34309–34320 (2015).
Chen, S., Wang, Y., Ni, C., Meng, G. & Sheng, X. HLF/miR-132/TTK axis regulates cell proliferation, metastasis and radiosensitivity of glioma cells. Biomed. Pharmacother. 83, 898–904 (2016).
Chen, S. et al. Silencing TTK expression inhibits the proliferation and progression of prostate cancer. Exp. Cell Res. 385, 111669 (2019).
Huang, H., Yang, Y., Zhang, W., Liu, X. & Yang, G. TTK regulates proliferation and apoptosis of gastric cancer cells through the Akt-mTOR pathway. FEBS Open Bio 10, 1542–1549 (2020).
Zhang, L. et al. Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt pathway. Med. Oncol. 37, 5 (2019).
Chen, F. et al. Protein kinase TTK promotes proliferation and migration and mediates epithelial-mesenchymal transition in human bladder cancer cells. Int. J. Clin. Exp. Pathol. 11, 4854–4861 (2018).
Canadian Cancer Trials, G., Stand Up To, C., Canadian Breast Cancer, F. & Ontario Institute for Cancer, R. CFI-402257 in combination with paclitaxel in patients with advanced/metastatic HER2-negative breast cancer (2021).
Bayer. Phase I dose escalation of Oral BAY1161909 in combination with intravenous paclitaxel (2017).
Bayer. Phase I study of oral BAY 1217389 in combination with intravenous paclitaxel (2018).
Boston, P. Study of paclitaxel in combination with BOS172722 in patients with advanced nonhaematologic malignancies (2021).
Chandler, B. et al. TTK inhibition radiosensitizes basal-like breast cancer through impaired homologous recombination. J. Clin. Investig. 130, 958–973 (2020).
Zhang, H. et al. TTK inhibitor promotes radiosensitivity of liver cancer cells through p21. Biochem. Biophys. Res. Commun. 550, 84–91 (2021).
Maia, A. et al. Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann. Oncol. 26, 2180–2192 (2015).
Maachani, U. B. et al. Targeting MPS1 enhances radiosensitization of human glioblastoma by modulating DNA repair proteins. Mol. Cancer Res. 13, 852–862 (2015).
Qi, G. et al. TTK inhibition increases cisplatin sensitivity in high-grade serous ovarian carcinoma through the mTOR/autophagy pathway. Cell Death Dis. 12, 1135 (2021).
Shi, W. et al. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis 31, 994–1002 (2010).
Byrne, B. & Oakley, G. Replication protein A, the laxative that keeps DNA regular: the importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 86, 112–120 (2019).
Niu, H. et al. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 272, 12634–12641 (1997).
Zernik-Kobak, M., Vasunia, K., Connelly, M., Anderson, C. & Dixon, K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 272, 23896–23904 (1997).
Nuss, J. et al. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry 44, 8428–8437 (2005).
Anantha, R., Vassin, V. & Borowiec, J. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 282, 35910–35923 (2007).
Liu, Y., Kvaratskhelia, M., Hess, S., Qu, Y. & Zou, Y. Modulation of replication protein A function by its hyperphosphorylation-induced conformational change involving DNA binding domain B. J. Biol. Chem. 280, 32775–32783 (2005).
Wu, X., Yang, Z., Liu, Y. & Zou, Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 391, 473–480 (2005).
Bélanger, F. et al. Replication protein A availability during DNA replication stress is a major determinant of cisplatin resistance in ovarian cancer cells. Cancer Res. 78, 5561–5573 (2018).
Manthey, K., Glanzer, J., Dimitrova, D. & Oakley, G. Hyperphosphorylation of replication protein A in cisplatin-resistant and -sensitive head and neck squamous cell carcinoma cell lines. Head. Neck 32, 636–645 (2010).
Zhang, Y. et al. Repression of PRMT activities sensitize homologous recombination-proficient ovarian and breast cancer cells to PARP inhibitor treatment. bioRxiv https://doi.org/10.1101/2024.05.21.595159 (2024).
Lu, X. et al. CBL0137 impairs homologous recombination repair and sensitizes high-grade serous ovarian carcinoma to PARP inhibitors. J. Exp. Clin. Cancer Res. 41, 355 (2022).
Gralewska, P. et al. PARP inhibition increases the reliance on ATR/CHK1 checkpoint signaling leading to synthetic lethality-an alternative treatment strategy for epithelial ovarian cancer cells independent from HR effectiveness. Int. J. Mol. Sci. 21, 9715 (2020).
Wilson, A. J. et al. Panobinostat sensitizes cyclin E high, homologous recombination-proficient ovarian cancer to olaparib. Gynecol. Oncol. 143, 143–151 (2016).
Wilson, A. J., Stubbs, M., Liu, P., Ruggeri, B. & Khabele, D. The BET inhibitor INCB054329 reduces homologous recombination efficiency and augments PARP inhibitor activity in ovarian cancer. Gynecol. Oncol. 149, 575–584 (2018).
Xu, J. et al. Arsenic compound sensitizes homologous recombination proficient ovarian cancer to PARP inhibitors. Cell Death Discov. 7, 259 (2021).
Ma, H. et al. HMGB3 promotes PARP inhibitor resistance through interacting with PARP1 in ovarian cancer. Cell Death Dis. 13, 263 (2022).
Teng, K., Ma, H., Gai, P., Zhao, X. & Qi, G. SPHK1 enhances olaparib resistance in ovarian cancer through the NFkappaB/NRF2/ferroptosis pathway. Cell Death Discov. 11, 29 (2025).
Wei, J. et al. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J. Biol. Chem. 280, 7748–7757 (2005).
Yeh, Y., Huang, Y., Lin, T. & Shieh, S. The cell cycle checkpoint kinase CHK2 mediates DNA damage-induced stabilization of TTK/hMps1. Oncogene 28, 1366–1378 (2009).
Yu, Z., Huang, Y. & Shieh, S. Requirement for human Mps1/TTK in oxidative DNA damage repair and cell survival through MDM2 phosphorylation. Nucleic Acids Res. 44, 1133–1150 (2016).
Meessen, S. et al. A comparative assessment of replication stress markers in the context of telomerase. Cancers 14, 2205 (2022).
Shiotani, B. et al. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 3, 1651–1662 (2013).
Wethington, S. L. et al. Combination ATR (ceralasertib) and PARP (olaparib) inhibitor (CAPRI) trial in acquired PARP inhibitor-resistant homologous recombination-deficient ovarian cancer. Clin. Cancer Res. 29, 2800–2807 (2023).
Lee, D. et al. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 17, 365–372 (2010).
Sleeth, K. et al. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J. Mol. Biol. 373, 38–47 (2007).
Wang, H., Wang, H., Powell, S. N., Iliakis, G. & Wang, Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 64, 7139–7143 (2004).
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (82303896), China Postdoctoral Science Foundation (2022M721961), Natural Science Foundation of Shandong Province (ZR2023QH306, ZR2024MH293).
Author information
Authors and Affiliations
Contributions
Conception and design: K.T. Development and methodology: G.Q., H.M., and P.G.; Acquisition of data: G.Q., H.M., and J.C. Analysis and interpretation of data: G.Q., H.M., P.G., and J.C. Administrative, technical, or material support: K.T., H.M., and P.G., Study supervision: K.T. Writing, review, and/or revision of the manuscript: All authors. Final approval: All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All methods in this study were performed in accordance with the relevant guidelines and regulations. Ethics Committee at Qilu Hospital of Shandong University approved the study. The tumor formation assay was approved by the Shandong University Animal Care and Use Committee.
Informed consent
All authors of the manuscript agree to article's publication.
Peer review
Peer review information
Communications Biology thanks Yi-Hui Wu, Yu-Yi Chu, and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Joanna Timmins and Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qi, G., Ma, H., Chen, J. et al. TTK activates ATR through RPA2 phosphorylation to promote olaparib resistance in ovarian cancer. Commun Biol 8, 1011 (2025). https://doi.org/10.1038/s42003-025-08444-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08444-7