Abstract
Pathogenic variants in MED13L cause MED13L syndrome, a well-defined neurodevelopmental disorder characterized by motor deficits, intellectual disability, and language delay, yet its underlying developmental and molecular mechanisms remain largely elusive. To address this gap, we generated a gene knockout mouse model of MED13L syndrome. While homozygous Med13l knockout exhibit neonatal lethality accompanied by reduced brain volume and cortical thickness, heterozygous mice are viable and display hallmarks of MED13L syndrome, including impaired learning and memory, reduced motor coordination, and heightened anxiety. Additionally, heterozygous mice show microcephaly with simplified neuronal morphology in the motor cortex. Single-cell transcriptomics and immunofluorescence reveal severe cortical neurogenesis deficits in Med13l knockout embryos, driven by impaired neural progenitor differentiation. Integrative multi-omics analyses reveal that MED13L orchestrates cortical neurogenesis by priming the transcriptional activation of key developmental genes, including Neurod2, Sox5, Auts2, and Nfib. This priming effect is mediated by MED13L binding to the core mediator complex, which facilitates the complex’s association with RNA Pol II and subsequent dissociation from MED13L. These findings uncover a pivotal role for MED13L in transcriptional regulation during brain development and highlight potential targets for restoring normal transcriptional programs in MED13L syndrome.

Similar content being viewed by others
Introduction
The mediator complex plays a central role in the regulation of gene transcription in eukaryotic organisms1. It consists of highly conserved subunits forming four functional modules: the head and middle modules that interact with RNA polymerase II (Pol II), the tail module that interacts with transcriptional activators and repressors, and the CDK8 kinase module (CKM) that dynamically regulates the mediator complex’s activity2. The CKM, comprising cyclin C, CDK8, MED12, and MED13 (or their paralogs CDK19, MED12L, and MED13L), was reported to disrupt the mediator complex’s interaction with Pol II, thereby repressing Pol II-mediated gene transcription3. This complex interplay between mediator’s various modules and their interactions with transcriptional regulators is crucial for finely tuning gene expression in response to diverse cellular signals.
Mutations in mediator genes are associated with various developmental disorders and cancers4,5. Among these, MED13L is notably one of the most frequently mutated in neurodevelopmental disorders6. MED13L haploinsufficiency syndrome, also known as Asadollahi-Rauch syndrome or MED13L syndrome, is characterized by a spectrum of symptoms, including moderate to severe intellectual disability (ID), delayed speech, motor developmental challenges (such as motor delay and ataxia), hypotonia, distinct facial features, and autism. Some patients also present with cardiac anomalies7,8. Despite extensive clinical documentation, the neurodevelopmental and molecular causes of MED13L syndrome remain largely unexplored.
MED13L, a large gene located on chromosome 12q24.21 of the human genome, spans 317 kb with 31 exons. It encodes a protein that includes the MID_MedPIWI domain, which is essential for binding double-stranded nucleic acids9. MED13L is considered to trigger the conformational switch of the CKM, thereby suppressing Pol II transcriptional activity3,10. However, this view contrasts with findings that CKM components can also enhance gene transcription in Arabidopsis11. Additionally, research indicates that the CKM—mediator complex can bind to CpG islands for developmental genes, priming them for transcriptional activation12,13.
Notably expressed in the nervous system14,15, MED13L’s fundamental role in brain development is not yet fully understood. Pioneer studies in zebrafish indicated that Med13b (the zebrafish equivalent of MED13L) knockdown disrupts cranial neural crest cell migration, upregulates the transcription factor SP8, and alters FGF8 expression in the anterior neural ridge14. In human embryonic stem cell-derived neural progenitor cells (NPCs), MED13L knockdown increased cell proliferation, suggesting involvement in Wnt and FGF signaling14. In cancer cells, MED13L inhibited Rb/E2F-induced cell growth16 and disrupted the CDK8-Mediator association by targeting FBW7, a component of the tumor-suppressing SCF ubiquitin ligase17,18. Recent observations have linked MED13L mutations to mitochondrial dysfunction due to abnormal nuclear release of cyclin C (CCNC) in cultured fibroblasts and skeletal muscles19. However, how MED13L deficiency affects brain development and function, particularly in relation to MED13L syndrome’s clinical traits, remains unanswered.
In this study, we investigated the neurodevelopmental and molecular mechanisms of MED13L syndrome using gene knockout (KO) mouse models. We discovered that heterozygous Med13l KO mice exhibit behavioral and anatomical deficits consistent with MED13L syndrome. Med13l KO impairs cortical neurogenesis and dendrite development. Proteomic, transcriptomic, and single-cell RNA sequencing analyses suggested that MED13L serves as a priming factor for the transcriptional activation of key genes involved in neurogenesis in the developing cerebral cortex.
Results
Med13l gene knockout causes microcephaly
We generated a Med13l KO mouse allele by CRISPR/Cas9-mediated ablation of exon 2, resulting in a frameshift and premature stop codon after exon 1 (Fig. 1A and Supplementary Fig. 1A, B). Med13l gene KO in these mice was confirmed through RT-PCR and Western blotting (Supplementary Fig. 1C–E). Newborn homozygous KO mice exhibited several phenotypic abnormalities, including an absence of stomach milk (Fig. 1B), death within 24 hours (h) of birth (Fig. 1C), and reduced body weight compared to wild-type (WT) and heterozygous KO (Het) littermates (Fig. 1D). These mice had noticeably smaller brains, as indicated by measurements of brain width, diameter, and weight at birth (Fig. 1E–G). Coronal brain sections revealed reduced thickness in the somatosensory and motor cortices, and decreased striatum volume in KO mice at postnatal day 0 (P0), with similar but less severe reductions in Het mice (Supplementary Fig. 1F–J). Het mice survived to adulthood with minor reductions in brain weight and width (Supplementary Fig. 1K–M). They exhibited reduced volumes in the cerebellum and striatum (Supplementary Fig. 1N, O) and decreased thickness in the primary motor cortex (M1), whereas no changes were observed in the somatosensory cortex (S1ULP, S1BF) (Supplementary Fig. 1P–S).
A Schematic diagram of the Med13l knockout strategy. Guide RNAs (gRNAs) were designed to target exon 2 to delete a 238 bp fragment, generating an early stop codon (magenta arrow and star). B Representative photographs of wild-type (WT), heterozygous (Het), and Med13l-knockout (KO) mouse littermates at birth (P0). Arrows indicate the presence or absence of milk in the stomach. Scale bar, 10 mm. C Survival rate of mice at P1. N = 33 (WT), 42 (Het), and 28 (KO). D Body weight of mice at P0. N = 11 (WT), 16 (Het), and 8 (KO). E Dorsal view images of brains from mice of different genotypes at P0. LC longitudinal cortex length, WC1, transverse cortex width; WC2, transverse cerebellum width. Scale bar, 2 mm. F Brain weight of P0 mice of different genotypes. N = 11 (WT), 18 (Het), and 12 (KO). G Comparison of the LC, WC1, and WC2 of P0 mice of different genotypes. N = 15 (WT), 19 (Het), and 10 (KO). H Representative images of P0 WT and KO brain sections stained for TBR1 (magenta) and CTIP2 (green), and with DAPI (blue). White dashed trapezoidal and rectangular boxes indicate the dorsolateral M1 region and dorsomedial ACC region, respectively, which are shown at higher magnification in (I). Scale bar, 500 μm. I High magnification images of the dorsolateral M1 region and dorsomedial ACC region of P0 mice stained for TBR1 and CTIP2, and with DAPI. Scale bar, 250 μm. J Quantitative analysis of the thickness of cortical layers of the dorsolateral M1 and dorsomedial ACC regions of the cortex. K Representative images of the VZ/SVZ region of P0 mouse brains stained for TBR2 (magenta) and DAPI (blue). Scale bar, 250 μm. L Quantitative analysis of the thickness of TBR2+ cell layer in the cortical VZ/SVZ at P0. Quantitative data are presented as Mean ± SEM and analyzed using one-way ANOVA followed by Bonferroni test in (D, F, G), and two-tailed Student’s t-test in (J, L). *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant. Error bars represent SEM.
Immunofluorescent staining of P0 cortical sections using specific cell markers (CTIP2 for layer V pyramidal neurons, TBR1 for layer VI pyramidal neurons, and TBR2 for intermediate neural progenitors (IPCs)) revealed significant reductions in the thickness of both upper (layers I-IV) and deep layers (layers V and VI) in KO mice (Fig. 1H–J). The subventricular zone (SVZ), rich in TBR2+ IPCs, was significantly thicker in KO mice compared to WT littermates at P0 (Fig. 1K, L), suggesting impaired differentiation of IPCs into post-mitotic neurons in KO mice.
Med13l knockout impairs differentiation of cortical neural progenitors
Embryonic cortical development in mice spans approximately embryonic day 11.5 to 18.5 (E11.5–E18.5), during which radial glial cells (RGCs) in the ventricular zone (VZ) drive neurogenesis through self-renewal and differentiation into neurons or intermediate progenitors (IPs)20. To address how MED13L regulates cortical development, we performed bulk RNA sequencing on cortical tissues from Med13l KO mice at E13.5, a stage that represents the peak period of deep-layer neuron generation and neural progenitor differentiation21. Altogether, we discovered 237 upregulated genes and 244 downregulated genes in homozygous KO mice compared to WT littermates (Fig. 2A; Supplementary Data 1). Several DEGs were selected for qPCR validation of their altered expression (Supplementary Fig. 2A; Supplementary Data 1). Most DEGs exhibited a similar but less prominent alteration in their expression in Het mice (Fig. 2B; Supplementary Fig. 2A–C), reflecting a dosage effect of MED13L on transcriptional regulation. Gene ontology (GO) enrichment analysis of DEGs highlights neurodevelopment-related pathways such as “cell differentiation”, “cell proliferation”, “cell fate commitment”, and “regulation of transcription by Pol II” (Fig. 2C; Supplementary Data 1), suggesting an important role of MED13L in the control of the proliferation, differentiation, and cell fate commitment of cortical NPCs.
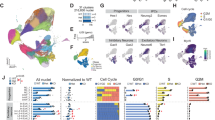
A Volcano plot displaying differentially expressed genes (DEGs) in the cortex of WT and KO mice at E13.5, as revealed by bulk RNA-seq. The vertical axis represents −Log10(p-value), while the horizontal axis shows the log2 fold change (KO/WT). DEGs: |log2 (fold change)| > 1, p < 0.05. B Heatmap showing the clustering of WT, Het, and KO samples based on the expression of DEGs in these samples. C The Sankey diagram showing the representative GO entries of DEGs. Criteria for significant GO terms: FDR < 0.05. D UMAP graph showing clustering of cortical cells of E13.5 mice identified by single-cell sequencing (scRNA-seq). Cortical cells from both WT, Het, and KO mice are included in the UMAP graph. E Bubble chart showing the expression of marker genes in the six cell clusters of the cortical pyramidal lineage at E13.5. Color represents the average expression level, and size represents the percentage of cells expressing each gene. F Pseudo-time trajectory of the six cell clusters of the cortical pyramidal neuronal lineage in WT and KO mice at E13.5. Black arrows indicate the developmental trajectory. G Comparison of the relative fractions of different cell clusters within the pyramidal lineage in WT and KO mice at E13.5. Chi-square test, ****p < 0.0001. H Significant GO terms related to cell proliferation and differentiation for differentially expressed genes in each cell cluster of the pyramidal lineage at E13.5. Gene filtering criteria: |log2 (foldchange)| > 1, p < 0.05. Criteria for significant GO terms: FDR < 0.05. NE neuroepithelia, RGC radial glial cell, IPC intermediate progenitor cell, PyN pyramidal neuron.
We further compared WT and KO cortical tissues at single-cell resolution using single-cell RNA sequencing (scRNA-seq) at E13.5. Altogether, we obtained the single-cell transcriptome of 25,606 cells (8133 cells from 5 WT embryos, 8972 cells from 5 Het embryos, and 8501 cells from 5 KO embryos) and discovered 23 cell clusters in E13.5 mouse cortex (Fig. 2D; Supplementary Data 2). We annotated the cell fate of each cluster based on the expression of marker genes (Fig. 2E; Supplementary Data 2). Pseudotime trajectory analysis (Monocle3) was performed on the six cell clusters of the cortical pyramidal lineage, including neuroepithelial cells (NEs), cycling and non-cycling radial glial cells (RGCs), cycling and non-cycling intermediate progenitor cells (IPCs), and post-mitotic pyramidal neurons (PyNs). Consistent with the well-established developmental process of cortical neurogenesis, the pseudotime trajectory analysis indicated a differentiation pathway of NEs → RGCs → IPCs → PyNs in both WT and KO embryos (Fig. 2F). Chi-square test indicates a significant difference in the relative proportion of different cell clusters between WT and KO mice (Chi-square = 49.33, df = 5, p < 0.0001), with a marked decrease in the proportion of post-mitotic pyramidal neurons in the cortex of KO mice compared to WT counterparts (Fig. 2G). This result suggests that MED13L deficiency impaired cortical neurogenesis.
We analyzed DEGs in each pyramidal lineage cell type at single-cell resolution. GO enrichment analysis of DEGs in each cell cluster revealed significant GO terms related to cell proliferation and differentiation, specifically in cycling and non-cycling RGCs and pyramidal neurons (Fig. 2H; Supplementary Data 2). These findings highlight the crucial role of MED13L in regulating cell proliferation and differentiation within the cortical pyramidal lineage. Additionally, other significant GO terms, such as “regulation of the Wnt signaling pathway,” “axon development,” and “neuron projection development,” indicate a developmental role of MED13L across different pyramidal lineage populations (Fig. 2H; Supplementary Data 2).
At E13.5, the surface area of the cortex was significantly reduced in KO embryos compared to WT counterparts (Fig. 3A, B). Immunofluorescent staining of cortical sections was carried out to further examine whether cortical neurogenesis was impaired by Med13l deficiency at E13.5, a developmental period crucial for the generation of deep-layer neurons of the cortex. Cortex from KO mice displayed a significant reduction in the density of TBR1+ layer VI neurons (cell number within a 250 μm-wide column, Fig. 3C, D) and the thickness of the layer of DCX+ newborn neurons (Fig. 3E, F), consistent with reduced neurogenesis revealed by the scRNA-seq. There was also a significant reduction in the density of TBR2+ IPCs in the VZ/SVZ (Fig. 3E, G), along with an increase in the thickness of PAX6+ NPC layer in the VZ of KO mice (Fig. 3H, I). These results suggest impairments in the differentiation of NPCs into either IPCs or deep-layer neurons at E13.5 caused by Med13l deficiency.
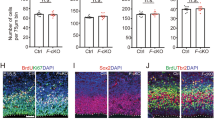
A Representative images of the dorsal view of the whole brains from WT and KO mice at E13.5. Scale bars, 1 mm. B Quantitative analysis of surface area of the cerebral cortex (outlined by the dashed lines in (A) of WT and KO mice at E13.5. N = 8 (WT), 4 (KO). C Representative images of WT and KO mouse brains at E13.5, stained for TBR1 (magenta) and DAPI (blue). Scale bars, 500 μm for the panoramic view and 100 μm for the local view. D Comparison of the number of TBR1+ cells in the cortex of WT and KO mice at E13.5. N = 9 (WT), 9 (KO). E Representative images of WT and KO mouse brains at E13.5, stained for DCX (magenta), TBR2 (green), and DAPI (blue). Scale bars, 500 μm for the panoramic view and 100 μm for the local view. F, G Quantitative analysis of the thickness of DCX+ cell layer and the density of intermediate neural progenitor cells (TBR2+) in the cortex of WT and KO mice at E13.5. N = 6 (WT), 6 (KO) in (F); N = 7 (WT), 10 (KO) in (G). H Representative images of PAX6 (magenta) and DAPI (blue) staining in the cortex of WT and KO mice at E13.5. The selected regions (white rectangular dashed box) are shown at high magnification on the right. Scale bars, 500 μm and 100 μm for the low and high magnification images, respectively. I Quantitative analysis of the thickness of the neural progenitor cell layer (PAX6+) in the cortex at E13.5. N = 9 (WT), 8 (KO). J Representative images of Ki67 (green), BrdU (magenta), and DAPI (blue) staining in the cortex of mouse brains at E13.5, 24 h after the BrdU injection. Scale bar, 100 μm. K Percentage of cell-cycle exit (Q-fraction) in the VZ/SVZ of WT and KO mice at E13.5. N = 4 (WT), 4 (KO). Data are presented as mean ± SEM and analyzed by two-tailed Student’s t-test in (B, D, F, G), and one-tailed Student’s t-test in (I) and (K). *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant. Error bars represent SEM.
We further examined whether Med13l deficiency affects the cell-cycle exit of neural progenitors. Pregnant mice were injected with 5-bromo-2’-deoxyuridine (BrdU, 50 mg/kg) at E12.5, and embryos were harvested at E13.5 for immunostaining of both BrdU and the cell cycle marker Ki67 in the cortex. KO embryos exhibited a significant decrease in the percentage of BrdU+ cells that had exited the cell cycle (Ki67-BrdU+/BrdU+ × 100%, Q-fraction) in the VZ/SVZ compared to WT embryos (Fig. 3J, K), suggesting impaired cell-cycle exit of neural progenitors.
At embryonic day 16.5 (E16.5), a crucial stage for upper-layer neuron generation, KO embryos exhibited decreased cortical surface area (Supplementary Fig. 3A, B) and reduced thickness in both dorsomedial and dorsolateral cortical regions compared to WT embryos (Supplementary Fig. 3C, D, G). Further analysis showed a significant reduction in the thickness of the TBR1+ layer VI and CTIP2+ layer V (Supplementary Fig. 3D, H, I) in the cortex of KO embryos. Additionally, there was a significant reduction in the thickness of DCX+ cortical plate (CP) (Supplementary Fig. 3E, J), as well as a significant reduction in the density of TBR2+ IPCs in the intermediate zone (IZ), but not in the VZ/SVZ (Supplementary Fig. 3E, K), indicating compromised differentiation of IPCs into upper-layer neurons. A significant reduction in the cell-cycle exit was also observed in KO embryos at E16.5 with BrdU administration at E15.5 (Supplementary Fig. 3F, L), confirming the detrimental effect of Med13l deficiency on the differentiation of neural progenitors at this developmental stage.
Pyramidal lineage-specific knockout of Med13l impairs cortical neurogenesis
Given Med13l’s high expression in the pyramidal lineage of developing cortex22, we further examined the effects of Med13l deficiency in this lineage using a conditional knockout mouse line, in which LoxP sites were inserted around exon 2 of the Med13l gene. We crossbred these Med13l-floxed mice with Emx1-Cre mice, achieving pyramidal lineage-specific knockout in the forebrain (Emx1-Cre::Med13lflox/flox, cKO) (Supplementary Fig. 4A, B). The efficiency of the conditional knockout was confirmed through qPCR and Western blotting (Supplementary Fig. 4C–F). Adult cKO mice showed a similar reduction in the thickness of M1 cortex as KO mice, while there was no difference in the somatosensory cortex (S1ULP) (Supplementary Fig. 4G, H). At E13.5, cKO mice showed a reduction in the density of Tbr1+ deep-layer neurons (Supplementary Fig. 4I, J) and the thickness of DCX+ CP (Supplementary Fig. 4K, N) compared to control mice, indicating an impairment in cortical neurogenesis. The density of TBR2+ IPCs was also significantly reduced in the cKO cortex (Supplementary Fig. 4K, O). In contrast, there was a significant increase in the number of PAX6+ NPCs and an increase in the percentage of proliferating NPCs (BrdU+, Supplementary Fig. 4L, P, Q), with a concomitant reduction in the differentiation of NPCs at the VZ/SVZ of cKO mice than control mice (Q-fraction, Supplementary Fig. 4M, R). These results suggest a cell-autonomous role of MED13L in the differentiation of NPCs of the pyramidal lineage.
Interactome of MED13L in developing cortex
To understand how MED13L regulates cortical neurogenesis, we performed co-immunoprecipitation (Co-IP) followed by mass spectrometry on cortical protein lysates from WT mice, using lysates from KO mice as a negative control. This analysis identified 39 proteins that interact with MED13L in E13.5 cortex (Fig. 4A; Supplementary Data 3). Selected MED13L-interacting proteins were validated through Western blotting after Co-IP (Fig. 4B). We found that most MED13L-interacting proteins are components of the mediator complex, or proteins known to interact with the mediator complex to regulate gene transcription initiation or elongation, supported by GO analysis of the interacting proteins (Supplementary Data 3). Notably, MED13, MED11, and MED26 were not detected in the MED13L interactome (Supplementary Data 3), consistent with a mutual exclusive relationship between MED13 and MED13L within the CKM3 and between MED26 and the CKM in interacting with the core mediator complex23.
A Volcano plot showing differentially presented proteins that co-immunoprecipitated (Co-IP) with MED13L in the cortex of WT and KO mice at E13.5. The vertical axis represents −Log10 (p-value), and the horizontal axis represents the ratio of KO/WT. Red dots indicate differentially presented proteins meeting the filtering criteria: KO/WT < 0.5, p < 0.05. B Western blotting of MED13L, MED12, TASP1, Pol II, and MED1 before (Input) and after immunoprecipitation (IP) in cortical protein lysates from WT and KO mice. C, G Co-IP using an anti-MED1 antibody showing reduced MED1-MED12 (C) and MED1-Pol II (G) associations in Med13l KO versus WT mice. Blots were probed with anti-MED12, anti-MED1, and anti-Pol II antibodies. Uncropped blots are shown in Supplementary Fig. 8. D, J, K Quantitative analysis of MED12, MED1 and Pol II in lysates of the cortex (input) from WT and KO mice at E13.5. N = 7 (WT), 7 (KO) in (D) and (K); N = 5 (WT), 5 (KO) in (J). E, F, H, I Quantitative analysis of MED12, MED1 and Pol II levels in the MED1-IP products. N = 5 (WT), 5 (KO) in E and F; N = 3 (WT), 3 (KO) in (H); N = 7 (WT), 7 (KO) in (I). L Co-IP using an anti-Pol II antibody showing no interaction with MED12 in either WT or Med13l KO mice. IP products were probed with anti-Pol II and anti-MED12 antibodies. Uncropped blots are shown in Supplementary Fig. 8. M Schematic diagram illustrating the MED13L-dependent priming of developmental genes by the mediator complex, facilitating the complex’s subsequent association with Pol II and the initiation of gene transcription. (Created by the authors). N Top-ranked transcription factors revealed by the ChEA for downregulated (left) and upregulated (right) genes in E13.5 cortex after Med13l KO. O The heatmap illustrates the spatio-temporal correlation between the mRNA level of MED13L and ChEA-predicted transcription factors in the developing human cortex, with color coding representing the strength of their correlation. P The number of potential target genes of Med13l in different cell clusters of the pyramidal lineage. Q Clustering of ChEA-predicted transcriptional regulators of MED13L target genes based on their relative enrichment level across cell clusters. Data are presented as Mean ± SEM and analyzed by two-tailed Student’s t-test in (D–F, H–K). *p < 0.05; **p < 0.01; ns not significant. Error bars represent SEM.
In addition to mediator components, several unexpected proteins were discovered among MED13L-interacting proteins, such as FBXL19 (F-Box and Leucine-Rich Repeat Protein 19), TASP1 (Threonine Aspartase 1), ATF7IP (Activating Transcription Factor 7 Interacting Protein), ERCC1 (Excision Repair Cross-Complementation Group 1), and SENP2 (SUMO Specific Peptidase 2), as shown in Fig. 4A and detailed in Supplementary Data 3. Of particular interest is FBXL19, a class II CXXC domain protein that specifically binds unmethylated CpG-containing DNA24 and recruits the CKM-mediator complex to promoters of silent developmental genes, leading to their induction during the differentiation of embryonic stem cells12,13. Another MED13L-interacting protein TASP1 plays a role in proteolytic activation of the histone methyltransferase MLL1, which promotes the formation of the active gene transcription marker H3K4Me325. Notably, MLL1 is also a class II CXXC domain protein that binds to unmethylated CpG islands24. These findings suggest that MED13L may coordinate the epigenetic regulation of gene transcription.
MED13L promotes the association of core mediator complex with Pol II
Western blot analysis revealed a significant reduction in the protein level of the CKM component MED12 in embryonic cortical tissues from Med13l KO mice (Input; Fig. 4C, D), suggesting that MED13L may play a role in stabilizing MED12.
To investigate how MED13L regulates the interaction between the core mediator complex and Pol II in the embryonic cortex, we performed immunoprecipitation targeting MED1, a subunit of the core mediator. We found that Med13l knockout significantly decreased the co-immunoprecipitation of Pol II by MED1 (Fig. 4G–K), indicating that MED13L facilitates rather than suppresses the association of Pol II with the core mediator complex. Notably, neither MED13L nor MED12 interacts with Pol II (Fig. 4B, L), suggesting that the CKM is released from the core mediator complex once it is associated with Pol II. Moreover, Co-IP using the anti-MED1 antibody demonstrated decreased association of MED12 with MED1 in Med13l KO cortices (IP; Fig. 4C, E). The MED12 IP/Input ratio showed a trend of reduction (Fig. 4F; p = 0.15), suggesting a potential weakening of the MED1-MED12 interaction following Med13l KO. Collectively, these co-immunoprecipitation data and the MED13L interactome suggest that the association of MED13L/CKM with the core mediator complex is necessary for engaging Pol II, after which the CKM is subsequently released (Fig. 4M).
MED13L enhances the transcription of neurogenic genes
This finding, along with the essential role of Pol II–mediator complex in transcriptional initiation, suggests that MED13L is more likely to enhance transcriptional activation rather than inhibit it. To further test this hypothesis, we investigated potential downstream targets of MED13L. We first identified genes that show significant spatiotemporal co-expression with MED13L in the developing human brain, using the BrainSpan human brain transcriptome dataset26 (correlation coefficient r > 0.3, p < 0.05). We next assessed how the expression of these co-expressed genes changes in the mouse cortex following Med13l KO, noting those that were significantly downregulated (for positively co-expressed genes) or upregulated (for negatively co-expressed genes) (Supplementary Data 1). These genes were categorized as either MED13L-activated (positively co-expressed and downregulated upon KO) or MED13L-suppressed (negatively co-expressed and upregulated upon KO). We then performed ChIP-X Enrichment Analysis (ChEA) to predict transcription factors (TFs) regulating these genes (Supplementary Data 1). For MED13L-activated genes, significantly enriched TFs included FEZF2, NEUROD1/2/6, SCRT1, and MYT1L, known for roles in neuronal fate specification, differentiation, or maturation. Conversely, for MED13L-suppressed genes, key TFs were E2F1/2/8, MYBL2, and CENPA, which are associated with cell cycle regulation (Fig. 4N, Supplementary Data 1). Further analysis of the BrainSpan dataset revealed that neurogenic TFs like FEZF2, MYT1L, NEUROD2/6, and SCRT1 positively co-express with MED13L, while cell-cycle regulators such as E2F1/2/7/8, MYBL2, and CENPA negatively co-express with MED13L in the developing human cortex (Fig. 4O; Supplementary Data 1). Therefore, MED13L synergizes with a suite of neurogenic TFs to activate transcription of their target genes during cortical neurogenesis.
Notably, MED13L-activated target genes include several key genes implicated in neurodevelopmental disorders, such as Neurod2 (an epilepsy-associated regulator of neuronal differentiation), Cntn2 (an axon guidance gene linked to epilepsy), Tbr1 (a cortical neuron identity specifier associated with ASD and intellectual disability), Sox5 (a layer specification factor implicated in ASD and ID), Nfib (a gliogenesis regulator linked to ASD and ID), and Auts2 (an ASD-associated gene involved in neuronal migration) (Supplementary Table 1). The convergent dysregulation of these genes in Med13l KO cortices provides mechanistic insights into MED13L-related pathologies.
Single-cell analysis of MED13L target genes
We next analyzed potential target genes of MED13L in each cell cluster of the pyramidal lineage based on the scRNA-seq. Potential downstream target genes of MED13L were identified as those positively correlated with Med13l expression in WT cells and whose expression reduced after Med13l KO, or those negatively correlated with Med13l and whose expression increased after Med13l KO (Fig. 4P). We found that most downstream target genes in each cell cluster were positively correlated with Med13l and showed reduced expression after Med13l knockout (Fig. 4P; Supplementary Data 2), supporting the notion that MED13L primarily enhances the expression of its target genes rather than suppressing them, during cortical neurogenesis, potentially through a priming effect of MED13L on the transcriptional activation.
For each pyramidal cell cluster, ChEA analysis was performed on potential MED13L target genes that were downregulated in KO mice to predict the transcriptional regulators governing their expression (Supplementary Data 2). Distinct transcription regulators were identified for each cell cluster, with well-established neurogenic TFs enriched in NEs, NPCs, and IPCs (Fig. 4Q). This is consistent with the idea that MED13L promotes the expression of genes regulated by these neurogenic transcription factors during the neurogenic differentiation of these progenitor populations.
Taken together, the integrated multi-omics and biochemistry studies suggest that MED13L orchestrates with a cohort of neurogenic TFs to prime their target genes for subsequent transcriptional activation during the differentiation and neuronal fate specifical of cortical neural progenitors.
Med13l deficiency leads to abnormal dendrite development
In addition to its role in embryonic cortical neurogenesis, we investigated the impact of Med13l heterozygous KO on adult cortical tissues. We conducted proteomic analysis on the adult M1 cortex of WT and Het mice, revealing that the differentially expressed proteins are enriched in axons, dendrites, and synapses (Fig. 5A-D; Supplementary Data 4). Golgi staining revealed that layer III pyramidal neurons in the M1 cortex of Het mice exhibited reduced dendritic length, branching, and complexity compared to WT mice, while the neuronal soma size remained unchanged (Fig. 5E–I). Moreover, there was a significant reduction in the density of dendritic spines on basal dendrites but not apical dendrites of layer III pyramidal neurons in Het mice compared to WT mice (Fig. 5J–L; Supplementary Fig. 5A–C). Conditional knockout of Med13l specifically in the forebrain pyramidal lineage (cKO) resulted in similar dendritic simplification in layer III pyramidal neurons of the M1 cortex (Supplementary Fig. 5D–G).
A Three-dimensional PCA plot of differentially expressed proteins from the motor cortex of adult WT and Het mice. B Volcano plot illustrating differentially expressed proteins from motor cortex of WT and Het mice. Criteria for differentially expressed protein: p-value < 0.05, |log2Foldchange| ≥ 0.26. C Heatmap depicting the clustering of differentially expressed proteins in the motor cortex from WT and Het mice. D GO enrichment analysis of differentially expressed proteins in the motor cortex of WT and Het mice. Criteria for significant GO terms: FDR < 0.05. E Representative traces of pyramidal neuron dendrites revealed by Golgi staining in adult WT and Het mice. Scale bar, 100 μm. F–H Statistical analysis of total dendritic length (F), dendritic branch number (G), and soma area (H) of pyramidal neurons in adult WT and Het mice. I Sholl analysis showing the dendritic complexity of cortical pyramidal neurons from WT and Het male mice. J Representative microscopic images (left) and schematic diagrams (right) of basal dendrites of layer III pyramidal neurons in adult WT and Het mice based on Golgi staining. Scale bars, 10 μm. Different types of dendritic spines are coded by different colors in the schematic diagrams. K, L Statistical analysis of the density of total dendritic spines (K) and the densities of different subtypes of dendritic spines (L) based on Golgi staining. For F–I and K, L, data are presented as mean ± SEM and analyzed by two-tailed Student’s t-test (F–H and K, L) or two-way ANOVA (I). N = 53 (WT), 48 (Het). Each point represents a neuron from 3 WT and 3 Het mice in (F–I). N = 39 (WT), 40 (Het). Each point represents a dendrite segment from 4 WT and 4 Het in (K, L).**p < 0.01; ***p < 0.001; ****p < 0.0001; ns not significant. Error bars represent SEM.
Med13l deficiency and behavioral deficits associated with MED13L syndrome
To determine if Het mice exhibit behavioral characteristics indicative of MED13L syndrome, we conducted behavioral assays on both male and female mice. In the open field test (OFT), adult male Het mice exhibited a significant reduction in center exploration and a markedly lower frequency of vertical exploration (up rearing) compared to their WT counterparts (Fig. 6A), suggesting heightened anxiety levels. In contrast, adult female Het mice did not display any significant behavioral changes in the OFT (Fig. 6A). In the balance beam test, male Het mice required significantly more time to traverse an ascending balance beam, whereas female Het mice performed similarly to WT controls (Fig. 6B). Both male and female Het mice showed significantly reduced performance in the rotarod test, further indicating impairments in motor coordination (Fig. 6C). In the Morris water maze (MWM) test, male Het mice demonstrated slower learning of the spatial location of the hidden platform and poorer memory retention compared to WT mice. Female Het mice, however, performed comparably to WT littermates in the MWM (Fig. 6D).
A Representative traces (left) and quantitative analyses (right) of the locomotion distance, time in the central zone, and the up-rearing events of male and female mice of different genotypes in the open field arena. B Schematic diagram of the ascending balance beam (left, created by the authors) and the comparison of the total latency for the mice to cross the ascending balance beam between control and Het mice of different sexes (right). C Schematic diagram of the rotarod test (left, created by the authors) and the maximal time of maintaining balance on the accelerating rotarod over seven consecutive days for male and female mice of different genotypes (right). D Schematic diagram of the Morris Water Maze (left, created by the authors) and the quantitative analyses of the latency to reach the hidden platform during the four consecutive training days (middle) as well as the number of target quadrant crossings during the test day for male and female mice of different genotypes. Each dot in histograms represents data from an individual mouse. Data are presented as mean ± SEM and analyzed by two-way ANOVA for (A, C), and (D) or two-tailed Student’s t-test for (B) and (D). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns not significant. Error bars represent SEM.
In other behavioral assays, including the three-chamber sociability test, self-grooming test, grip strength test and hindlimb clasping test, both male and female Het mice performed normally (Supplementary Fig. 6). Overall, Het mice replicate the core behavioral deficits associated with MED13L syndrome, such as intellectual disability and motor impairment.
Pyramidal lineage-specific knockout of Med13l impairs motor function
We further analyzed the impact of pyramidal lineage-specific knockout of Med13l (cKO) on mouse behavior. We found that male cKO mice displayed significantly prolonged center exploration during the OFT compared to their littermates without gene ablation (Ctrl) (Supplementary Fig. 7A). In contrast, female cKO mice demonstrated decreased locomotion and a diminished frequency of rearing behavior relative to Ctrl mice (Supplementary Fig. 7A). Both male and female cKO mice exhibited significantly impaired performance in the balance beam and rotarod tests, indicative of substantial deficits in motor coordination and motor learning (Supplementary Fig. 7B, D). Additionally, a notable proportion of cKO mice displayed rear paw clasping when suspended by the tail, further evidence of motor dysfunction (Supplementary Fig. 7C). Grip strength and spatial learning and memory were unaffected in cKO mice (Supplementary Fig. 7E, F). Collectively, these findings suggest that cKO mice manifest more severe motor deficits compared to Het mice but do not demonstrate the learning and memory impairments characteristic of Het mice.
Discussion
Behavioral and functional implications
The MED13L syndrome is mostly caused by haploinsufficiency of the gene. The present study showed that heterozygous Med13l KO mice (Het) exhibited heightened anxiety, impaired motor coordination, and deficits in learning and memory, mirroring core behavioral traits of MED13L syndrome (Supplementary Table 2). Med13l gene deficiency, particularly the homozygous KO, caused microcephaly, another anatomical abnormality associated with a subgroup of the MED13L syndrome. Thus, Het mice could serve as a valuable animal model for studying the neuropathological mechanisms underlying MED13L syndrome and for exploring therapeutic strategies for this developmental disorder.
Several brain structures engage in motor control, including the motor cortex, the striatum, and the cerebellum. We found that conditional knockout of Med13l in the forebrain pyramidal lineage (cKO) caused more pronounced motor deficits compared to Het mice, indicating that aberrant cortical circuitry likely plays a significant role in the motor impairments associated with MED13L syndrome. Therefore, targeting the motor cortex could be a promising intervention strategy to mitigate motor deficits in MED13L syndrome, a hypothesis that warrants further investigation. However, since Med13l is also expressed in both the striatum and cerebellum, and dysplasia of these two brain regions occurred in homozygous KO mice (Supplementary Fig. 1N, O), the potential involvement of developmental deficits in the cerebellum and striatum in these motor deficits in Het mice and in MED13L syndrome cannot be excluded.
Interestingly, cKO mice did not exhibit the same learning and memory impairments as Het mice, suggesting that the neuronal basis of intellectual disability in MED13L syndrome is more complex than the motor deficits and may involve different neuronal populations from brain regions beyond the cortical pyramidal lineage. This complexity highlights the need for further studies to identify the specific brain regions and neuronal circuits contributing to the cognitive deficits observed in MED13L syndrome.
Although MED13L is considered a high-risk gene for ASD, only 23% of individuals with MED13L gene deficiency exhibit an autistic phenotype27. We did not see a statistically significant defect in the social interaction of Het mice, although a few such mice indeed displayed abnormal sociability. Considering the multigenic nature of ASD, it is likely that MED13L gene deficiency per se is not sufficient for causing ASD but could significantly increase the risk.
Role of MED13L in cortical neurogenesis
Our findings underscore the essential role of MED13L in regulating neural progenitor proliferation and differentiation, which are pivotal processes for normal brain development. We found that KO mice are neonatally lethal with severe deficits in cortical development, including microcephaly and reduced cortex surface area and cortical thickness. Heterozygous KO mice survive to adulthood but display significant reductions in the sizes of multiple brain regions and cortical thinning, particularly in the primary motor cortex. These anatomical abnormalities parallel the functional impairments (motor deficits, intellectual disability) that characterize MED13L syndrome, suggesting that defects in neural progenitor proliferation and differentiation may underlie the syndrome’s brain malformations.
Several lines of evidence support a critical role of MED13L in the differentiation of cortical progenitors. The decreased presence of neurons in both deep and upper cortical layers at different developmental stages suggests that MED13L is necessary for the proper development of these layers. At E16.5, TBR2+ IPCs in IZ reflect a population of differentiating IPCs migrating toward the CP. This IPC population is reduced in KO mice (Supplementary Fig. 3F, K), indicating impaired differentiation of IPCs into post-mitotic neurons. Additionally, the thicker SVZ rich in Tbr2+ cells in neonatal KO mice (Fig. 1K, L) and the increased PAX6+ NPC layer in the E13.5 VZ of KO and cKO mice (Fig. 3H, I; Supplementary Fig. 4L, P, Q) suggest an accumulation of progenitor cells that failed to differentiate into neurons or IPCs. This further reinforces the role of MED13L in promoting the differentiation of neural progenitors.
Our scRNA-seq and immunofluorescent staining of E13.5 cortex showed a marked reduction in the proportion of pyramidal neurons (Fig. 2G; Fig. 3C–F), supporting impaired differentiation of neural progenitors into neurons. Cell cycle exit assays provided additional evidence of aberrant differentiation of progenitors in the embryonic cortex of KO and cKO mice (Fig. 3J, K; Supplementary Fig. 3F, L; Supplementary Fig. 4M, R). Altogether, these findings suggest that MED13L promotes the fate switch from progenitor cells to differentiated cell types during embryonic development of the cortex, including the transitions from NPCs to IPCs and from IPCs to post-mitotic neurons. These results are consistent with previous studies indicating the involvement of MED13L in cell proliferation and differentiation pathways in cell culture14.
MED13L and neuronal morphogenesis
In addition to its role in neurogenesis, we found that MED13L is crucial for proper dendrite development. Proteomic and neuronal morphometry analyses of cortical tissues from homozygous KO mice showed significant impairments in dendritic development of pyramidal neurons. These dendritic abnormalities in cortical pyramidal neurons of Het mice may underlie the motor coordination deficits observed in behavioral assays, linking structural brain defects to functional outcomes. Similar dendritic impairments in cortical pyramidal neurons were observed in mice with conditional knockout of Med13l in cKO mice (Supplementary Fig. 5D–G), underscoring the cell-autonomous role of MED13L in neuronal morphogenesis. This cell-autonomous action of MED13L in dendrite development is consistent with a recent report showing impaired dendrite arborization after in-utero electroporation of different mutant forms of MED13L into cortical pyramidal neurons28. Further studies are needed to investigate whether axon projection and synapse formation are also affected by MED13L deficiency, as suggested by our transcriptomic analysis. Understanding these aspects could provide deeper insights into the full spectrum of MED13L’s role in neuronal development and its impact on neurological function.
Molecular mechanisms and pathways
Our findings suggest that MED13L is co-expressed with a group of key neurogenic TFs and is required for gene expression under the control of these neurogenic TFs. The proper developmental expression of MED13L ensures that NPCs differentiate into distinct types of neurons at the appropriate times.
The canonical Wnt/β-catenin pathway plays a well-established role in promoting cortical neurogenesis29,30,31. Crosstalk between mediator complex and the Wnt pathway has been frequently reported. A previous study in cultured human NPCs showed that knockdown of MED13L caused a slight increase in the proliferation of NPCs, a process correlated with significant alteration in the expression of canonical Wnt pathway genes14. Genetic studies in Drosophila and C. elegans have revealed that mutations in other Mediator subunits Med1, Med6, Med12, Med13 and Med23 variously affect developmental processes regulated by Wnt signaling32,33,34,35,36. MED12 has been shown to physically interact with β-catenin and was required for β-catenin-stimulated gene expression37. Notably, we found that MED13L is essential for maintaining MED12 protein levels in the developing cortex, providing a plausible mechanism by which MED13L regulates Wnt signaling via MED12. Although we did not discover β-catenin in the MED13L interactome in E13.5 cortical tissues, it cannot be ruled out that MED13L may regulate its target genes via crosstalk between CKM components and the canonical Wnt pathway. Consistent with this notion, our scRNA-seq at E13.5 cortex highlights aberrant expression of canonical Wnt pathway genes in radial glial cells (Fig. 2H). The detailed molecular machinery underlying MED13L’s crosstalk with the Wnt pathway remains to be investigated.
How does MED13L regulate its target genes during cortical development? We found that MED13L-interacting proteins are predominantly components of the mediator complex. The absence of MED13 and MED26 in MED13L interactome is consistent with the mutual exclusivity between MED13 and MED13L in the CKM3 and between MED26-Mediator and CKM-Mediator23, supporting the reliability of our proteomic findings. MED13L is traditionally considered to suppress Pol II-mediated gene transcription by mediating the binding of CKM to the core mediator complex, thus interfering with its interaction with Pol II. However, components of the CKM, such as MED12 and MED13, have also been discovered to positively regulate gene expression. We discovered that FBXL19 and TASP1, both epigenetic regulators of transcriptional activation, are part of the MED13L interactome. Previous studies have demonstrated that FBXL19 recruits the CDK-Mediator complex to CpG island-associated promoters, priming genes for activation12,13. These findings suggest that MED13L enhances the expression of developmental genes through a priming effect of the CKM-Mediator complex on transcriptional activation during the cell fate specification of cortical NPCs. Consistent with this notion, genes whose expression was significantly reduced in Med13l deficient cortex are predicted to be activated by a group of neurogenetic transcription factors that are known to regulate the neuronal fate specification in developing cerebral cortex. This notion is further supported by our scRNA-seq analysis, which shows that most target genes of MED13L are activated rather than repressed by MED13L.
Conclusion
In conclusion, Med13l deficiency results in profound structural and functional brain abnormalities, underscoring the pivotal role of MED13L in neuronal genesis and maturation within the cerebral cortex. Our findings highlight a priming effect of MED13L for the transcription of key developmental genes in coordination with a group of neurogenic transcription factors and epigenetic factors during the cell fate specification of cortical neural progenitor cells, advancing the understanding of MED13L’s critical function in cortical development and the molecular underpinnings of the MED13L syndrome.
Methods
Animals
Housing, handling, and all experimental protocols for mice were performed in accordance with the Regulations for the Administration of Laboratory Animals of East China Normal University (ECNU) and were approved by the Animal Care and Use Committees of ECNU (Approval ID: 10269). We have complied with all relevant ethical regulations for animal use. Mice were housed in a certified specific-pathogen-free (SPF) facility under standardized conditions: housed in standard filter-top cages (3-5 animals per cage) with free access to water and food. They were maintained on a 12:12-h light/dark cycle (07:00 to 19:00 lighting) at 22°C with relative humidity of 50–60%. The noon of the day when the vaginal plug was found was counted as embryonic day (E) 0.5.
Wild-type C57BL/6 J mice (MGI: 3028467) were purchased from Shanghai Jihui Laboratory Animal Care Co., Ltd. and ICR mice (CD-1, MGI:5649524) were obtained from Hunan SJA Laboratory Animal. Med13l knockout mice were generated by CRISPR/Cas9 in C57BL/6 N zygotes at Cyagen Biosciences (C57BL/6N-Med13lem1cya/Cya, Cyagen Ltd., Nanjing, China; Stock No. S-KO-14807; MGI: 7835672) and backcrossed > 5 generations to C57BL/6 J, then crossed with ICR mice to establish experimental breeding colonies. All KO experiments used littermate controls from these crosses. Med13l-floxed mice (C57BL/6JGpt-Med13lem1Gpt/Gpt; Gempharmatech Ltd., Nanjing, China; Stock No. T010181; MGI: 6426082), and Emx1-Cre mice (B6.129S2-Emx1tm1(cre)Krj/J; Jackson Laboratories, JAX: 005628, MGI: 3617405). Genotyping and sequencing primer sequences for all the aforementioned mouse lines are provided in Supplementary Data 5.
Med13l knockout mice were generated by CRISPR/Cas9-mediated gene editing in C57BL/6 N mouse zygotes38. Guide RNAs (gRNAs) were designed to target Exon 2 (gRNA1: 5′-GCACGTGGGCTGTTCGATACAGG-3′; gRNA2: 5′-GCTTCACACCACCTACGGTTTGG-3′) to delete a fragment of 238 bp. Specificity of the gRNA was evaluated using the online tool available at crispr.mit.edu. A mix of Cas9 mRNA and gRNAs was microinjected into zygotes. An F0 founder carrying a 3187-bp deletion and an 8-bp insertion was identified by Sanger sequencing (Supplementary Fig. 1A; primers listed in Supplementary Data 5), resulting in complete deletion of Exon 2 and a frameshift that introduced a premature stop codon. For the floxed allele, gRNAs targeting Exon 2 (gRNA1: 5′-TAACCATCTGCAACCCAACC-3′; gRNA2: 5′-AGCACTCCCCACGGAGACAT-3′) were used with Cas9 mRNA and Flox Donor DNA; Med13l-floxed mice (Gempharmatech Stock No. T010181) were later crossed with Emx1-Cre (JAX: 005628) mice for forebrain-specific knockout.
Sanger sequencing
Genomic DNA was amplified by polymerase chain reaction (PCR), and the resulting products were purified and directly sequenced using an ABI PRISM 3730 automated sequencer. Sequencing chromatograms were aligned to the mouse reference genome using Chromas software. Due to the 3187-bp deletion, only the junction regions of the deleted allele are presented in Supplementary Fig. 1A.
Histological analysis
For newborn mice (P0), the heads were removed after ice anesthesia, and the brains were dissected and photographed under a stereoscopic anatomical microscope (Leica M165 FC). For adult mice, animals were anesthetized by 0.7% (w/v) pentobarbital sodium solution at 5 mL/kg (35 mg/kg body weight), followed by perfused with precooled phosphate buffer (PBS, pH 7.4) and 4% paraformaldehyde (PFA). The brains were dissected and photographed. Morphological parameters measured included the width of cortex (WC1), longitudinal length of cortex (LC1), oblique length of cortex (LC2), and width of cerebellum (WC2).
For brain slice preparation, brains were post-fixed in 4% PFA (pH 7.4) for 24 h. (P0) or overnight (adult), followed by dehydration in 20% and 30% sucrose solutions in PBS. Brains were embedded in OCT compound (SAKURA, USA). Coronal sections of 40-μm thickness were cut using a cryostat (Leica CM1950). Glass slides with brain sections were treated with Nissl staining solution for 15 min at 55 °C. Microscopic images of Nissl-stained brain slices were collected using the TissueFAXS Plus ST system (TissueGnostics GmbH, Vienna, Austria). The thickness of the motor cortex (M1) and primary somatosensory cortex (S1ULP/S1BF) were measured in microscopic images of Nissl-stained brain slices using Image J software. The volumes of the unilateral cerebellum or striatum were obtained by cumulative integration of areas of all brain slices encompassing the entire brain region.
Immunofluorescence
Glass slides with brain sections were washed with PBS (pH 7.4) three times and incubated for 15 min in 10 mM sodium citrate buffer (pH 6.0) at 99°C for antigen retrieval. Sections were then incubated in blocking buffer (5% BSA, 5% goat serum, and 0.25% Triton X-100 in PBS) for 2 h at room temperature. Subsequently, sections were incubated with specific primary antibodies for 48 h at 4 °C. For primary antibodies, we used mouse anti-BrdU (1:500, Cell Signaling, 5292S), rabbit anti-Pax6 (1:300, MBL, PD022), rabbit anti-Tbr1 (1:1000, Abcam, ab31940), rabbit anti-EOMES (1:1000, Abcam, ab183991), rat anti-EOMES (1:100, Invitrogen, 14-4875-82), rat anti-Ctip2 (1:1000, Abcam, ab18465), rabbit anti-DCX (1:800, Cell Signaling, 4604), rabbit anti-Ki67 (1:200, Cell Signaling, 12202). After rinsing in PBS three times, sections were incubated with secondary antibodies in the dark for 2 hr. at room temperature and then thoroughly rinsed. Secondary antibodies were: goat anti-rabbit, Alexa Fluor 546 (1:1000, Invitrogen, A11035), donkey anti-mouse, Alexa Fluor 647 (1:1000, Invitrogen, A32787), goat anti-rat, Alexa Fluor 647 (1:1000, Cell Signaling, 4418), goat anti-rabbit, Alexa Fluor 488 (1:1000, Invitrogen, A11008), goat anti-rabbit, Alexa Fluor 647 (1:1000, Cell Signaling, 4414). Nuclei were labeled by incubating sections in PBS containing 1 μg/ml 4’,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, 62248), and samples were mounted using ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, P36970).
For BrdU labeling, time-pregnant mice were intraperitoneally injected with 50 mg/kg body weight BrdU (Abcam, ab251467). After 1.5 h. (proliferation assay) or 24 h. (differentiation analysis), mice were anesthetized, and the brains of embryos were dissected on ice and fixed with 4% PFA for 4-6 h. Cryostat sections of 14-μm (1.5-h pulse) or 16-μm (24-h pulse) thickness were prepared. Sections were treated with 2 N HCl for 15 min at 37 °C before immunofluorescence staining.
Fluorescence images of brain sections were collected using the TissueFAXS Plus ST system (TissueGnostics GmbH, Vienna, Austria) and further processed with Adobe Illustrator. For cell counting, a 250 μm wide rectangular column was placed perpendicular to the ventricular surface in the cortical area, and the cell number in the column was counted using the cell counter program of Image J software.
Behavior assessments
Behavioral tests and data analyses were conducted according to previously described protocols39. Briefly, 17- to 20-week-old age-matched male and female mice were used. Mice were handled for 5 min each day for 3 days prior to the behavioral tests. All behavioral tests were conducted during the light cycle by an experimenter blind to the mouse genotypes. The surfaces of the behavioral apparatus were cleaned using 75% ethanol before each experiment and between trials, with at least a 5-min wait before the next test to allow ethanol evaporation and odor dissipation.
Open-field test
The Panlab Infrared (IR) Actimeter system was used for the open-field test. The test mouse was gently placed near the wall-side of a 20 cm × 20 cm × 25 cm open-field arena and allowed to explore freely for 30 min. The movement of the mouse was recorded by a video camera and analyzed with the IR Actimeter. The total locomotion distance, the time exploring the central zone (10 cm × 10 cm), and the number of hindlimb rearing events in the central zone were measured.
Balance beam test
The experimental device consisted of a flat surface rod, 100 cm long and 1 cm wide, placed 50 cm above the ground with an upward angle of 30°. A target cage was placed at the upper end of the rod. The experiment was divided into a 3-day training phase and a testing phase. During the training phase, the tested mouse was guided to walk through the balance beam from the lower end toward the upper end once daily. On the fourth day, the mouse was tested, and the time required for each mouse to traverse the balance beam without guidance was recorded. Mouse movement was video recorded from the front of the balance beam for detailed analysis.
Rotarod test
The test was conducted using a 5-channel rotarod device (DigBehv-RRTM, Shanghai Jiliang Software Technology Co., Ltd., Shanghai, China). The total experiment spanned 7 days, with 4 trials per day and a 20-min interval between trials. The speed of the rotating rod was set to smoothly accelerate from 4 rpm to 40 rpm within 5 min. The maximum time and speed which the testing mice kept balance on the rotating rod were recorded.
Hindlimb clasping test
The hindlimb clasping test followed a 2-level scoring system40. Each mouse was tested three times, with each trial lasting 10 s. The tail of the mouse was gently lifted to suspend the mouse head down to observe whether the hind limbs moved closer to the abdomen. If the hind limbs were always stretched out away from the abdomen, with both legs open, a score of 0 was given. If the hind limbs moved close to the abdomen for more than 50% of the time, it was considered moderate paw clasping, and a score of 1 was given. The average score of each mouse was calculated.
Gripping strength test
The experiment was conducted using a grasping net (100 cm × 50 cm) placed 50 cm above the ground. The mouse was placed on top of the net, which was then quickly and smoothly turned upside down, so the mouse hung under the net. The time from reversing the net to the mouse falling was recorded as the holding latency. Five repeated trials were performed for each mouse, with a 20-min interval between trials. For latencies longer than 120 s, a value of 120 s was recorded. The average latency of the 5 trials was calculated for each mouse.
Morris water maze
The standard Morris water maze was used to assess the spatial learning and memory abilities of the experimental mice. A stainless water-filled circular tank (210 cm in diameter and 50 cm in height) with non-reflective interior surfaces and ample visual cues was used. White edible emulsifier was added to the water to make it non-transparent. The pool was divided into four quadrants, with a circular hidden platform (13 cm in diameter) placed 2 cm below the water surface in the center of the target quadrant. The pool was surrounded by black shading curtains with spatial cues of different shapes and colors. The water temperature was maintained at 22–24 °C with a heating device. A camera above the pool recorded animal behavior, controlled by SuperMaze software. The experiment spanned 5 days, including 4 days for training and 1 day for testing. Each mouse trained with 4 trials per day, being gently released into the tank with its head toward the inner wall from a random quadrant. If the mouse found the platform within 60 s, the time was recorded, and the mouse stayed on the platform for 20 s. If the mouse did not find the platform within 60 s, it was guided to the platform and allowed to stay for 20 s, with a latency of 60 s recorded. On the 5th day, two independent 60-s tests were conducted without the hidden platform, recording the target quadrant entry times and total swimming time inside the target quadrant. After testing, mice were dried and returned to a dry cage to prevent stress from low body temperature.
Three-chamber social test
A modified three-chamber sociability test was conducted in a transparent rectangular chamber (40 cm × 30 cm × 25 cm) divided into three equal-sized chambers (13.3 cm × 30 cm × 25 cm) with two doors (3 cm × 5 cm) connecting the central chamber to the side chambers. The social partner mouse was tied in the corner of a side chamber with a 25 cm polyacrylonitrile string. In stage 1, the test mouse was placed in the central chamber and allowed to explore freely for 10 min to adapt. In stage 2, the test mouse was placed in the central chamber, a stranger mouse (stranger 1) was tied in one side chamber, and a mouse doll was placed in the other side chamber. The mouse’s behavior was recorded for 10 min. In stage 3, the doll was replaced by another stranger mouse (stranger 2), and behavior was recorded again. The total sniffing time of the test mouse on each stranger mouse or the doll was measured.
Self-grooming
Self-grooming was analyzed as described previously39. The test mouse was placed in the center of an open field arena (20 cm × 20 cm × 25 cm) and allowed to explore freely for 20 min. The first 10 min analyzed self-grooming in an unfamiliar environment, and the last 10 min analyzed self-grooming in a familiar environment. The self-grooming time and frequency were determined by manual review of the video recordings.
RNA isolation and qRT-PCR
Mice were sacrificed by cervical dislocation at the indicated time points. Total mRNA from mouse brain tissues was isolated using the RNAiso Plus kit (Takara, #9109) on ice. Phase separation was achieved with 200 µl or 100 µl chloroform. After centrifugation at 12,000 rpm for 20 min at 4 °C, RNA was precipitated by mixing the aqueous phase with equal volumes of isopropyl alcohol and 1 µl of 20 mg/ml glycogen. The RNA precipitates were dissolved in DNase/RNase-free water. cDNA was synthesized using a cDNA reverse transcription kit (KR116-02, Tiangen, China) from 1 µg of total RNA. qPCR was performed using the SYBR Green qPCR kit (AG11701, Accurate Biology, China) on a CFX96 Real-Time System (Bio-Rad). The thermal profile was 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60°C for 20 s, and 70 °C for 30 s. Gapdh gene was used as the internal control. The relative expression level of target genes was normalized to the Ct value of Gapdh using the 2−ΔΔCt relative quantification method. The primers used are listed in Supplementary Data 5.
Bulk RNA-seq
Total RNA was extracted as described above. Bulk RNA-seq was conducted by Shanghai Genergy Bio. Co., Ltd. The concentration and quality of RNA were measured using Qubit fluorometer (Thermo Fisher Scientific, Q33226). RNA-seq libraries were constructed using the VAHTS Universal V10 RNA-seq Library Prep Kit (Premixed Version, NR616). Briefly, mRNA was extracted using poly-A selection with magnetic beads with poly-T and then converted into cDNA through first and second-strand synthesis. The newly synthesized cDNA was purified using AMPure XP beads (1:1) and eluted in 50 μl of nucleotide-free water. RNA-seq libraries were sequenced on NovaSeq XPlus platform with paired-end reads of 150 bp, achieving a sequencing depth of 60 million reads per library.
Quality control of the RNA-seq data was performed using FastQC (version 0.11.9). Paired-end reads were trimmed to remove adapters and low-quality reads using Skewer (version 0.2.2). Clean reads were aligned to UCSC mm10 mouse genome using STAR (version 25.2b) with default parameters. The number of mapped reads was counted using stringtie (version 2.2.1). The resulting read counts were processed with the R package DESeq2 (version 1.18.1) to identify differentially expressed genes (|log2 fold change| > 1 and p-value < 0.05) between groups. Gene expression levels were normalized by fragments per kilobase of transcript per million mapped reads (FPKM). Raw sequencing data were deposited in the Gene Expression Omnibus under accession number GSE277054.
Single-cell RNA sequencing (scRNA-seq)
The scRNA-seq was performed by Shanghai OriginGene-Tech Biotechnology Co., Ltd. Cortices from 5 embryos of the same genotype from 3 litters were pooled into 5 ml tissue storage solution (Miltenyi Biotec, Inc., MD, USA) on ice and carefully cut into small pieces of 1–3 mm. Tissue samples were subjected to enzymatic digestion with 0.25% Trypsin-EDTA (Thermo Fisher Scientific) at 37 °C for up to 30 min. Cell suspensions were centrifuged at 300×g for 5 min at 4 °C. The pellets were further digested in basal media supplemented with 0.2% type II collagenase (Sigma-Aldrich) at 37 °C for up to 4 h. Isolated cells were filtered through 70 μm nylon filters (BD), washed twice with sterile PBS (pH 7.4). Samples with a cell density of 700–1200 cells/μl and a viability higher than 85% were proceeded for scRNA-seq.
Droplet-based scRNA-seq was performed using the 10× Genomics Chromium Single Cell 3′ Reagents Kit (v3.1)41. The single-cell samples were passed through a 40 μm cell strainer and counted using a hemocytometer with trypan blue. Subsequently, the single-cell suspension, Gel Beads and Oil were added to the 10× Genomics single-cell A chip. After droplet generation, samples were transferred into PCR tubes and reverse transcription was performed using a T100 Thermal Cycler (Bio-Rad). After reverse transcription, cDNA was recovered using a recovery agent, provided by 10× Genomics, followed by silane DynaBead clean-up as outlined in the user guide. Before clean-up using SPRIselect beads, cDNA was amplified for 10 cycles. Libraries were submitted to 150 bp paired-end sequencing on an Illumina NovaSeq 6000 platform, yielding 150 Gb of sequencing data per sample. ScRNA-seq data are available in GEO under accession number GSE277054.
Data analysis was conducted according to previous studies42,43. CellRanger 6.6.1 was applied for preliminary data analysis. Raw data from single-cell RNA-seq libraries were first trimmed to remove the template switch oligo (TSO) sequence and poly(A) tail sequence. Reads with adaptor contaminants and low-quality bases were removed. Clean reads were aligned to the reference genome (GRCm39/mm39). Gene expression levels were quantified as transcripts per million (TPM). Valid barcodes and Unique Molecular Identifiers (UMIs) were used to generate the gene-barcode matrix for single cells. Seurat v4.1.1 was used for quality control and data analysis in R. Cells with fewer than 200 genes, more than 6,000 detected genes, or with more than 10% of transcripts from mitochondrial genes, were excluded to maintain data quality.
Principal component analysis with variable genes was used to identify significant principal components (PCs) based on the JackStraw function. Twenty PCs were selected as the input for uniform manifold approximation and projection (UMAP) and t-distributed stochastic neighbor embedding (t-SNE) when statistically significant. Cell clusters were annotated manually based on marker gene expression and developmental information. Defined cell clusters included neuroepithelial cells (NEs), cycling and non-cycling radial glial cells (RGCs), cycling and non-cycling intermediate progenitors (IPCs), pyramidal neurons (PyNs), interneurons (INs), Cajal-Retzius cells, ependymal progenitors (EpPs), oligodendrocyte-astrocyte progenitor cells (OAPCs), and microglia, among others, based on their specific marker gene panels (detailed in Fig. 2 and Supplementary Data 2). These clusters were assigned to broad lineages, such as the interneuron lineage, pyramidal lineage, NG2-oligodendrocyte lineage, or microglial lineage. The Monocle3 algorithm was used to infer the developmental trajectories of cell clusters within the pyramidal lineage, including NEs, cycling RGCs, non-cycling RGCs, cycling IPCs, non-cycling IPCs, and PyNs.
Differential expression analysis was conducted independently for each cell cluster. Only genes with a minimum expression of 0.1 TPM and detected in at least 100 cells were included in the differential expression analysis. Significantly dysregulated genes were identified with p-value < 0.05 and log2 fold change (log2FC) > 1 or log2FC < −1. Gene ontology (GO) analysis was performed using Gene Ontology Resource (https://www.geneontology.org/) with the PANTHER overrepresentation test (Fisher’s exact test with FDR). For redundant/overlapping pathways identified by the GO analysis, only one representative GO entry was shown.
For cell clusters within the pyramidal lineage, the transcriptional regulatory targets of MED13L in each cluster were further analyzed through three steps. Firstly, Med13l-correlated genes were identified as those with significant correlation in expression across all cells from WT mice, based on Spearman correlation analysis (p < 0.05), including both positively and negatively correlated genes. Secondly, for each cell cluster, significantly upregulated or downregulated genes in KO mice compared to WT mice were identified. Finally, the gene lists from Steps 1 and 2 were compared to identify overlapping genes as potential targets of MED13L, including positively correlated & down-regulated genes and negatively correlated & up-regulated genes.
Western blotting and immunoprecipitation
For Western blotting, brain tissues were homogenized in lysis buffer (P0013K, Beyotime Biotechnology, Shanghai, China). Proteins were separated by 6% SDS-PAGE and transferred onto a PVDF membrane. The blots were incubated in blocking buffer (5% non-fat dry milk in TBST) at room temperature for 2 h, followed by overnight incubation with the primary antibody (diluted in 0.05% TBST with 3% BSA). After thorough rinsing with TBST, the blots were incubated with HRP-conjugated secondary antibodies at room temperature for 2 h. Chemiluminescence detection of protein bands was performed using the BeyoECL Plus kit (Beyotime Biotechnology, P0018S). Western blot images were acquired using the UVP ChemStudio System (Analytik Jena GmbH Co. KG, Jena, Germany). The band intensity was analyzed using VisionWorks Software (VisionWorks 8.0 associated with the UVP ChemStudio System.
For co-immunoprecipitation (Co-IP), cortical tissues from E13.5 embryos were homogenized in lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, and 1 mM EDTA) containing protease and phosphatase inhibitors (P10008, Beyotime Biotechnology). Lysates (500 μl, approximately 800 μg total protein) were incubated with 10 μg MED13L antibody at 4 °C overnight to allow antibody-protein binding. Meanwhile, 25 μl protein-A magnetic beads (1008D, ThermoFisher) were incubated with 3% BSA at 4 °C overnight to block non-specific binding sites. The BSA-saturated magnetic beads were then added to the lysate and antibody mixture and incubated for 1 h. to capture the antibody-protein complex. Beads were washed with lysis buffer to remove any unbound proteins and contaminants. Finally, the immunoprecipitated proteins were eluted from the beads for further analysis using mass spectrometry or Western blotting. Primary antibodies used were rabbit anti-MED13L (1:1000, custom-developed via Abclonal, E19406; antigen: aa2000-2090), mouse anti-β-actin (1:5000, Proteintech, 66009-l-Ig), rabbit anti-α-Tubulin (1:5000, Proteintech, 11224-l-Ap), rabbit anti-MED12 (1:2000, Abcam, ab70842), mouse anti-Pol II (1:1000, Biolegend, 664906), rabbit anti-MED1 (1:1000, Cell Signaling, 51613S), rabbit anti-TASP1 (1:50, Santa Cruz, sc514677), rabbit anti-IgG (1:250, Invitrogen, 02-6102). Secondary antibodies were goat anti-mouse IgG(H + L)-HRP conjugate (1:5000, Absin, abs20163), goat ant-rabbit IgG(H + L)-HRP conjugate (1:5000, Absin, abs20147).
4D-DIA proteomics analysis
Purified immunoprecipitated proteins were subjected to 4D-DIA LC-MS/MS analysis performed by PTM Bio. (Hangzhou) Co., Ltd. (Zhejiang, China). Briefly, tryptic peptides were dissolved in solvent A (0.1% formic acid, 2% acetonitrile in water) and directly loaded onto a homemade reversed-phase analytical column (25 cm length, 75/100 μm i.d.). Peptides were separated using a gradient from 6% to 24% solvent B (0.1% formic acid in acetonitrile) over 70 min, 24% to 35% in 14 min, and increasing to 80% in 3 min, holding at 80% for the final 3 min, all at a constant flow rate of 450 nL/min on a nanoElute UHPLC system (Bruker Daltonics, Massachusetts, USA). The peptides were ionized using a capillary source and analyzed with a timsTOF Pro (Bruker Daltonics) mass spectrometer. The electrospray voltage applied was 1.60 kV. Precursors and fragments were analyzed using the TOF detector, with an MS/MS scan range from 100 to 1700 m/z. The timsTOF Pro operated in parallel accumulation serial fragmentation (PASEF) mode, selecting precursors with charge states 0 to 5 for fragmentation, and acquiring 10 PASEF-MS/MS scans per cycle. The dynamic exclusion was set to 30 s.
The DIA proteomics data were aligned to the Mus_musculus_10090_SP_20230103.fasta database using DIA-NN (v1.8) software. Data were filtered to pass quality control for both peptide length (typically 7–20 amino acids) and peptide counts (at least 2 peptides per protein). Pearson’s Correlation Coefficient, Principal Component Analysis, and relative standard deviation were applied to evaluate repeatability. To compare the normalized intensity of each protein between different groups, fold change (FC) was calculated and transformed to log2FC. Significantly different proteins were identified based on two criteria: p < 0.05 and KO/WT Ratio < 0.5. The results were visualized using a volcano plot. Identified proteins underwent Gene Ontology (GO) analysis, with the top 20 MED13L-related pathways presented in a bubble plot.
Proteomics analysis of cortical tissues
Proteomic analysis of mouse motor cortex tissue was carried out by Shanghai Biotree Biotech Co., Ltd. Briefly, tissue samples were collected to perform protein extraction, followed by BCA measurement of protein concentration, acetone precipitation, protein reconstitution, reduction, alkylation, enzymatic hydrolysis, SDC removal, and peptide desalination. Then 200 ng of total peptide were separated using the Evosep One nano UPLC liquid phase system and analyzed using a mass spectrometer (timsTOF Pro2, Bruker Daltonics) with a nanoliter ion source. The mass spectrometer used the DDA PASEF (data dependent acquisition) mode for data acquisition, with a scanning range from 100–1700 m/z. Raw data files were searched using SpectroMine (4.2.230428.5,2329; Biognosys AG) software with the Pulsar search engine, followed by further qualitative and quantitative analyses. Proteomics data have been deposited to ProteomeXchange via PRIDE with identifiers PXD055474 and PXD055844.
Bioinformatics
Gene Ontology (GO) enrichment analysis was performed using Gene Ontology Resource (https://www.geneontology.org/). A Sankey plot of the GO enrichment entries, clustering analysis heatmap, and volcano plots was generated using an online platform for data analysis and visualization (https://www.bioinformatics.com.cn). The bubble plot was created using the online tool Hiplot (https://hiplot.cn/). Transcription factor (TF) enrichment analysis of target genes was performed using the web-based tool ChEA3 (https://maayanlab.cloud/chea3/) as described previously44. The ChEA3 background database contains a collection of gene set libraries generated from multiple sources, including TF-gene co-expression from RNA-seq studies, TF-target associations from ChIP-seq experiments, and TF-gene co-occurrence computed from crowd-submitted gene lists. The analysis results were ranked based on Fisher’s Exact Test p-values (Score), where lower p-values indicate higher statistical significance and greater relevance to the transcriptional regulation of the target genes. For graphic presentation, top 15 TFs were shown in histograms. PCA plots were produced using the online tool BioLadder (https://www.bioladder.cn/web/#/pro/cloud). Gene co-expression analysis was performed as previously described39. Transcriptomes from different cortical regions, ranging from 8 PCW to 13 PCW, were obtained from the BrainSpan human developmental transcriptome dataset. Spearman’s correlation coefficient was computed between gene pairs to assess their co-expression levels. The gene co-expression matrix was generated using Hiplot.
Statistics and reproducibility
Data are presented as the mean ± SEM. All statistical analyses were performed using GraphPad Prism 9. The sample size “n” denotes biologically independent samples, with exact values reported in the figure legends. At least three biological replicates were analyzed in each experimental group to ensure reproducibility. Data normality was determined by the Shapiro-Wilk test. Statistical significance was determined using one-tailed or two-tailed Student’s t-test for single comparisons and one-way ANOVA with two-tailed Bonferroni post-hoc tests for multiple comparisons. For GO enrichment analysis, statistical significance was set at FDR < 0.05 (with Benjamini-Hochberg correction). Statistical significance was set at p < 0.05 for all comparisons. Detailed test types (including tail specification for t-tests), all p-values, and group sizes are reported in the figure legends and Supplementary Data 6.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data of bulk RNA-seq and single-cell RNA-seq have been deposited at Gene Expression Omnibus (GEO) under the accession number GSE277054. Proteomics raw data have been deposited at ProteomeXchange with identifiers PXD055474 and PXD055844. The BrainSpan human brain transcriptome data set used in this study are available in the GEO database under accession number GSE25219. Source data presented in the figures of this paper are available in Supplementary Data 1–6. Uncropped Western blot images are shown in the Supplementary Information. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
Allen, B. L. & Taatjes, D. J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015).
Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 (2018).
Tsai, K. L. et al. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 (2013).
Spaeth, J. M., Kim, N. H. & Boyer, T. G. Mediator and human disease. Semin. Cell Dev. Biol. 22, 776–787 (2011).
Schiano, C. et al. Involvement of Mediator complex in malignancy. Biochim. Biophys. Acta 1845, 66–83 (2014).
Deciphering Developmental Disorders, S. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
Adegbola, A. et al. Redefining the MED13L syndrome. Eur. J. Hum. Genet. 23, 1308–1317 (2015).
Asadollahi, R. et al. Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur. J. Hum. Genet. 21, 1100–1104 (2013).
Burroughs, A. M., Iyer, L. M. & Aravind, L. Two novel PIWI families: roles in inter-genomic conflicts in bacteria and Mediator-dependent modulation of transcription in eukaryotes. Biol. Direct 8, 13 (2013).
Knuesel, M. T., Meyer, K. D., Bernecky, C. & Taatjes, D. J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 (2009).
Liu, Q. et al. The characterization of Mediator 12 and 13 as conditional positive gene regulators in Arabidopsis. Nat. Commun. 11, 2798 (2020).
Dimitrova, E. et al. FBXL19 recruits CDK-Mediator to CpG islands of developmental genes priming them for activation during lineage commitment. Elife 7 (2018).
Feldmann, A., Dimitrova, E., Kenney, A., Lastuvkova, A. & Klose, R. J. CDK-Mediator and FBXL19 prime developmental genes for activation by promoting atypical regulatory interactions. Nucleic Acids Res. 48, 2942–2955 (2020).
Utami, K. H. et al. Impaired development of neural-crest cell-derived organs and intellectual disability caused by MED13L haploinsufficiency. Hum. Mutat. 35, 1311–1320 (2014).
Hamada, N., Iwamoto, I., Nishikawa, M. & Nagata, K. I. Expression analyses of mediator complex subunit 13-like: a responsible gene for neurodevelopmental disorders during mouse brain development. Dev. Neurosci. 43, 43–52 (2021).
Angus, S. P. & Nevins, J. R. A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene 31, 4709–4717 (2012).
Welcker, M. & Clurman, B. E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008).
Davis, M. A. et al. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 27, 151–156 (2013).
Chang, K. T. et al. Aberrant cyclin C nuclear release induces mitochondrial fragmentation and dysfunction in MED13L syndrome fibroblasts. iScience 25, 103823 (2022).
Taverna, E., Gotz, M. & Huttner, W. B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502 (2014).
Greig, L. C., Woodworth, M. B., Galazo, M. J., Padmanabhan, H. & Macklis, J. D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013).
Loo, L. et al. Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 (2019).
Taatjes, D. J. et al. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 (2002).
Xu, C. et al. DNA sequence recognition of human CXXC domains and their structural determinants. Structure 26, 85–95.e83 (2018).
Hsieh, J. J., Cheng, E. H. & Korsmeyer, S. J. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115, 293–303 (2003).
Kang, H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011).
Torring, P. M. et al. Is MED13L-related intellectual disability a recognizable syndrome? Eur. J. Med Genet 62, 129–136 (2019).
Hamada, N., Iwamoto, I. & Nagata, K. I. MED13L and its disease-associated variants influence the dendritic development of cerebral cortical neurons in the mammalian brain. J. Neurochem 165, 334–347 (2023).
Munji, R. N., Choe, Y., Li, G., Siegenthaler, J. A. & Pleasure, S. J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 31, 1676–1687 (2011).
Machon, O., van den Bout, C. J., Backman, M., Kemler, R. & Krauss, S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience 122, 129–143 (2003).
Singh, K. K. et al. Common DISC1 polymorphisms disrupt Wnt/GSK3beta signaling and brain development. Neuron 72, 545–558 (2011).
Janody, F., Martirosyan, Z., Benlali, A. & Treisman, J. E. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130, 3691–3701 (2003).
Yoda, A., Kouike, H., Okano, H. & Sawa, H. Components of the transcriptional Mediator complex are required for asymmetric cell division in C. elegans. Development 132, 1885–1893 (2005).
Treisman, J. Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128, 603–615 (2001).
Zhang, H. & Emmons, S. W. A. C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes Dev. 14, 2161–2172 (2000).
Zhang, H. & Emmons, S. W. The novel C. elegans gene sop-3 modulates Wnt signaling to regulate Hox gene expression. Development 128, 767–777 (2001).
Kim, S., Xu, X., Hecht, A. & Boyer, T. G. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 281, 14066–14075 (2006).
Hall, B. et al. Genome editing in mice using CRISPR/Cas9 technology. Curr. Protoc. Cell Biol. 81, e57 (2018).
Wu, N., Wang, Y., Jia, J. Y., Pan, Y. H. & Yuan, X. B. Association of CDH11 with autism spectrum disorder revealed by matched-gene co-expression analysis and mouse behavioral studies. Neurosci. Bull. 38, 29–46 (2022).
Chou, A. H. et al. Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol. Dis. 31, 89–101 (2008).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
La Manno, G. et al. Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021).
Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e210 (2019).
Keenan, A. B. et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019).
Acknowledgements
We thank the ECNU Multifunctional Platform for Innovation (010 and 011) for their mouse breeding service. This work was supported by the National Key Research and Development Program of China grant 2022YFC2705200, National Natural Science Foundation of China grant 32271022, 31100273, and 81941013, National Natural Science Foundation of China and Israel Science Foundation Cooperative Research Project grant 32061143016, and Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
X.B.Y., Y.L.Z., and X.L.H. conceived the project. X.B.Y. and Y.H.P. acquired funding. X.B.Y. and Y.L.Z. supervised the experiments. J.M. and X.X.C. performed in situ hybridization. J.L. and J.C. conducted behavioral tests. S.Y.X. and N.W. contributed to the generation of transgenic mouse lines. J.L., Y.Z.H., and J.C. performed histology and biochemistry studies. J.L., Y.Z.H., J.M., and Y.H.P. contributed to multi-omics studies. J.L., Y.Z.H., J.C., N.W., and Y.L.Z. analyzed the data. J.L., Y.Z.H., and J.C. prepared figures and tables. J.L. and X.B.Y. designed and created all illustrations, including those in Figs. 4M and 6B, C, graphical abstract, and Supplementary Fig. 6A. X.B.Y., J.L., and Y.L.Z. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Reza Asadollahi and Chad Grueter for their contribution to the peer review of this work. Primary Handling Editors: Rosie Bunton-Stasyshyn & Jasmine Pan. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, J., Hao, Yz., Cui, J. et al. Regulation of cortical neurogenesis by MED13L via transcriptional priming and its implications for MED13L syndrome. Commun Biol 8, 1180 (2025). https://doi.org/10.1038/s42003-025-08532-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08532-8