Abstracts
Age-related macular degeneration (AMD) is the leading cause of central vision impairment among the elderly. Geographic atrophy is a defining characteristic of AMD, but the detailed mechanism for massive loss of retinal pigment epithelium (RPE) cells is not fully understood. In this study, we found that Z-DNA binding protein 1 (ZBP1), a sensor for dsDNA, is able to induce RPE cell PANoptosis. Silencing ZBP1 efficiently alleviates RPE degeneration and AMD symptoms. Mechanistically, mitochondrial permeability transition pore (mPTP) opening stimulated by Ca2+ overload can trigger the releasing of mtDNA, which leads to ZBP1 activation and PANoptosis. Importantly, our findings reveal a significant role of aberrant formation of mitochondria-associated ER membranes (MAMs) in AMD. MAMs act as conduits for transferring Ca2+ from the ER to mitochondria through the VDAC1/GRP75/IP3R1 complex. Furthermore, our results indicate that GRP75 O-GlcNAcylation is involved in MAM formation. Genetic suppression of GRP75 attenuates PANoptosis and AMD progression. In summary, our study sheds light on the intricate organelle interplay underlying AMD and presents insights into potential avenues for AMD intervention.
Similar content being viewed by others
Introduction
Age-related macular degeneration (AMD) is a disease affecting the macular region of the retina, leading to progressive loss of central vision in older individuals1. By 2020, the global number of people with AMD was expected to reach around 200 million, a figure projected to rise to nearly 300 million by 20402, posing a significant public health challenge with notable socio-economic implications. The disease is categorized into two stages based on underlying biological processes: neovascular AMD and atrophic AMD. Neovascular (“wet”) AMD is distinguished by choroidal neovascularization, fluid leakage, retinal hemorrhage, and eventual fibrous scarring3. Atrophic (“dry”) AMD is characterized by dysfunction and progressive atrophy of the retinal pigment epithelium (RPE)4. While anti-VEGF therapies are well-established clinical treatments for wet AMD5, there is currently no practical therapeutic approach or target for dry AMD. Hence, comprehensive studies on the pathogenesis and identification of therapeutic targets for dry AMD are urgently needed.
AMD is generally associated with the formation of pathologic deposits (drusen) between the Bruch’s membrane and the RPE layer. RPE cells perform a variety of vital functions in the eye, including blood-retinal barrier regulation, ocular immune privilege formation, and the release of inflammatory cytokines3. Our previous studies have demonstrated the essential role of RPE cell death in AMD progression6. It has been observed that both apoptosis and necroptosis contribute to RPE cellular death in AMD7. While apoptosis and necroptosis have different characteristics, they also share common features. Both are precisely regulated forms of programmed cell death (PCD), leading to an inflammatory response and cell fragmentation. Additionally, other forms of programmed cellular death, such as pyroptosis and ferroptosis, have been implicated in RPE and photoreceptor degeneration8,9. Recent research indicates that photoreceptors and RPE cells are affected by all these mentioned cellular death modalities in overlapping and complementary ways10. Inhibiting any single cellular death pathway may not fully reverse RPE dysfunction. Therefore, simultaneously targeting multiple cellular death pathways may be essential to achieve the desired clinical benefit.
PANoptosis, a complex inflammatory PCD pathway, is characterized by the involvement of pyroptosis, apoptosis, and necroptosis11. The complete spectrum of biological effects observed in PANoptosis cannot be only attributed to any one of these pathways. PANoptosis is driven by a cascade of upstream receptor and signaling molecules that assemble to form a multifaceted protein complex termed the PANoptosome. This complex involves numerous molecules, including apoptotic markers such as caspase-3, caspase-8, and caspase-1, as well as necroptotic markers like mixed lineage kinase domain-like (MLKL) and receptor-interacting serine/threonine-protein kinase 1 (RIP1) and RIP312,13. Recent evidence indicates that individual deletion of these three PCD pathways does not fully protect cells from influenza A virus (IAV)-induced cell death. However, deletion of Z-DNA-Binding protein 1 (ZBP1) and Zα2 domains reduces PANoptosis14. Mechanistically, ZBP1 serves as a sensor in the ZBP1-PANoptosome complex, initiating PANoptosis. Moreover, ZBP1 has been reported to be upregulated by cytosolic mitochondrial DNA (mtDNA) and is involved in sensing torsional mtDNA stress and stabilizing Z-form mtDNA15,16. PANoptosis and the PANoptosome play crucial roles in various inflammatory and neurodegenerative diseases. For instance, recent studies have shown that Doxorubicin induces mitochondrial malfunction and mtDNA damage, leading to PANoptosis in cardiomyocytes17. In glaucoma, a prevalent neurodegenerative disease, increased mitochondrial division mediated by Drp1 has been associated. Inhibiting Drp1 was found to downregulate retinal ganglion cell (RGC) PANoptosis by regulating caspase-3, NLRP3, and RIP signaling under pathologically high intraocular pressure (ph-IOP), suggesting that ph-IOP induces mitochondrial dysfunction and RGC PANoptosis in glaucoma18. Mitochondrial dysfunction also plays significant roles during RPE aging and AMD progression19. However, the relationships between mitochondria, PANoptosis, and AMD remain to be fully elucidated. Additionally, while PANoptosome complexes such as ZBP1-PANoptosome and their downstream effectors have been characterized, much remains to be discovered regarding the upstream regulatory elements that recruit and/or activate the PANoptosome.
In this study, our findings suggest that ZBP1-mediated PANoptosis may contribute to RPE dysfunction in AMD. Furthermore, mitochondria-associated endoplasmic reticulum membranes (MAMs) formation could be a potential driver of ZBP1 upregulation. Taken together, these findings provide insights into strategies for preventing RPE damage and suggest potential therapeutic avenues for AMD.
Results
ZBP1 is highly expressed in NaIO3-treated mice and AMD patients
NaIO3-induced RPE cell death is frequently employed in establishing AMD models20. To validate the involvement of PANoptosis in AMD, we initially assessed cell viability using CCK8 and observed partial suppression of NaIO3-induced cell death by several cell death inhibitors (Fig. 1A). Following this, we investigated the key players of PANoptosis in the pathogenesis of dry AMD patients. We used the expression profile from GSE29801, comprising 41 macular RPE-choroid samples from AMD patients (including dry AMD, wet AMD, and geographic atrophy (GA)) and 50 normal macular RPE-choroid samples from donors. We identified 522 upregulated and 563 downregulated genes (Fig. 1B, C). Then, 3 hub genes were obtained by overlapping differential genes and PANoptosis-related genes (Fig. 1D). Subsequently, to further explore the key genes, ROC curves were constructed for the 3 hub genes to demonstrate the predictive ability of individual genes to distinguish AMD from normal samples. The results showed that ZBP1, a PANoptosis initiator, exhibited optimal diagnostic efficacy (Fig. 1E).
A ARPE-19 cells were pretreated with the indicated inhibitors for 2 h in DMEM/F12 medium supplemented with 2% FBS before treatment of 10 mM NaIO3 for 24 h, CCK-8 assay showing ARPE-19 cell viability in the indicated groups (n = 6). B Visualization of principle component analysis (PCA) of GSE29801 dataset (purple: AMD samples; green: normal samples). C Heatmap of RNA-seq data displaying the differential expression gene (DEG) between AMD and normal macular RPE-choroid samples in GSE29801 dataset. D Venn diagram illustrating the overlapped targets of upregulated DEG, downregulated DEG and PANoptosis-related genes (obtained from GeneCards database). E The ROC curve of ZBP1, PYCARD, CASP1. ZBP1 exhibits optimal diagnostic efficacy. F ZBP1 protein expression in primary mouse RPE cells by immunoblots analysis. Quantification of the protein levels relative to β-actin expression are shown. G PANoptosis-related protein expressions (p-MLKL, MLKL, cleaved-GSDMD, GSDMD, cleaved-Caspase3, Caspase3) in primary mouse RPE cells by immunoblots analysis. Quantification of the protein levels are shown. H CCK-8 assay showing ARPE-19 cells viability in the indicated groups (n = 6). I RPE65 and ZO-1 expressions in primary mouse RPE cells by immunoblots analysis. Quantification of the protein levels relative to β-actin expression are shown. J ARPE-19 cells stained with ZO-1 (green) and DAPI (blue) showing the ARPE-19 cells dysfunction treated with NaIO3, and these changes can be reversed significantly by ZBP1 siRNA. Scale bars = 25 μm. Data are mean ± SD, the p values were obtained using one-way ANNOVA. *p < 0.05 versus control group. #p < 0.05 versus NC group.
Given the potential role of ZBP1, we established AMD models of primary mouse RPE cells, ARPE-19 cells, and C57 mice using NaIO3 at a concentration of 10 mM and 50 mg/kg, respectively. The results showed increased ZBP1 protein expression in all models (Figs. 1F and S1A). These findings strongly suggest the involvement of ZBP1 in the pathogenesis of dry AMD.
ZBP1 deficiency alleviates the PANoptosis of RPE cells
To better understand the role of ZBP1 in PANoptosis and dry AMD, we knocked down ZBP1 in primary mouse RPE cells and ARPE-19 cells (Fig. S1B), which resulted in reduced expressions of p-MLKL, cleaved-GSDMD, and cleaved-caspase-3 (Figs. 1G and S1C). Through CCK-8 assay and immunofluorescence experiments (Figs. 1H and S1E), we found that ZBP1 siRNA could reverse RPE cell injury caused by NaIO3, suggesting that silencing ZBP1 could enhance RPE cell function. Immunostaining with the ZO-1 and Occludin antibody revealed that ARPE-19 cells displayed swelling and deformation with a loss of tight junctions under NaIO3 stimulation, indicating impaired outer blood-retinal barrier function, a hallmark of AMD (Figs. 1I, J and S1F). Remarkably, silencing ZBP1 reversed these effects, highlighting its potential as a therapeutic target for AMD.
Considering the potential role of ZBP1 in PANoptosis, we investigated its involvement in AMD progression in vivo. Adeno-associated virus-shZBP1 (AAV8-shZBP1) vectors were engineered to downregulate ZBP1 in mouse RPE with AMD. The mice received intravitreal injection of either AAV8-shZBP1 or AAV8-vector on day 2, and the AMD mouse model was established three weeks later by intravenous injection of NaIO3 (Fig. 2A). The efficiency of AAV8-shZBP1 in silencing ZBP1 expression is shown (Fig. S2A). Notably, downregulation of p-MLKL, cleaved-GSDMD, cleaved-caspase-1, and cleaved-caspase-3 were observed in AAV8-shZBP1-treated mice (Fig. 2B). HE staining of NaIO3-treated mice revealed the characteristic features of dry AMD, impaired RPE cells and subsidence of ONL, which could be reversed following the administration of shZBP1 (Fig. 2C). SS-OCTA was used to capture retinal ultra-wide-angle en-face images and demonstrated a significant reduction in punctate hyperreflective signals (impaired RPE cells or misalignment of the ONL) in the RPE layer of NaIO3-treated mice upon shZBP1 treatment (Figs. 2E and S2B). Low expressions of Rhodopsin and Arrestin in the retina are indicative of photoreceptor degeneration. Here, we observed decreased expression of Arrestin and Rhodopsin in NaIO3-treated mice, which was significantly reversed in shZBP1 mice, suggesting that ZBP1 knockdown partially alleviated photoreceptor degeneration (Fig. 2H). Additionally, NaIO3-treated mice exhibited reduced expression of ZO-1 and RPE65, which was reversed in the ZBP1 knockdown group (Fig. 2D, F, G), further indicating that PANoptosis of the RPE cell contributes to RPE dysfunction and retinal pathological alterations akin to dry AMD.
A Flowchart of the in vivo experiments. B PANoptosis-related protein expressions (p-MLKL, MLKL, cleaved-GSDMD, GSDMD, cleaved-Caspase1, Caspase1, cleaved-Caspase3, Caspase3) in mice RPE by immunoblots analysis. Quantification of the protein levels are shown. C The H&E staining of mice retinas. white triangle: impaired RPE cells; black asterisk: subsidence of outer nuclear layer (ONL). Scale bars = 50 μm. D RPE65 and ZO-1 expressions in mice RPE by immunoblots analysis. Quantification of the protein levels relative to β-actin expression are shown. E Retinal OCT and ultra-wide angle en face images obtained from SS-OCTA showing an abundant number of punctate hyperreflective signals (red circle: impaired RPE cells or misalignment of the ONL layer) and thinning of ONL in the NaIO3-treated mice and administration of AAV8-shZBP1 reversed the above abnormalities. F, G Immunofluorescent staining of RPE65 and ZO-1 in the indicated groups of mice retinas. Scale bars = 50 μm (NC: staining without primary antibody). H Immunofluorescent staining of photoreceptor functional indicators (Arrestin, Rhodopsin) in the indicated groups of mice retinas. Scale bars = 50 μm. Data are mean ± SD, the p values were obtained using one-way ANNOVA (B, D). *p < 0.05 versus Ctrl group, #p < 0.05 versus NC group.
RIP1 and RIP3 are recruited to ZBP1-PANoptosome for activation of PANoptosis
Next, we aimed to understand how ZBP1 orchestrates PANoptosome assembly and initiates downstream PANoptosis signaling. A PPI network was constructed and visualized by STRING, revealing “RIPK1” and “RIPK3” as the most prominent nodes (Fig. 3A and Supplementary Table 6). Subsequently, we investigated whether NaIO3 treatment induced interaction between ZBP1 and RIPK1, RIPK3. We found that both RIPK1 and RIPK3 proteins were co-precipitated with ZBP1 after 24 h of NaIO3 treatment, indicating the formation of ZBP1/RIPK1 and/or ZBP1/RIPK3 complexes (Fig. 3B, C). Additionally, previous studies have linked PANoptosis signaling activation with NLRP3 inflammasome activation21. Therefore, we hypothesized that ZBP1 recruits NLRP3 inflammasome as a catalytic effector in PANoptosis activation. Co-immunoprecipitation studies using a ZBP1 antibody confirmed that NLRP3 forms a complex with ZBP1 in ARPE-19, with enhanced interaction upon NaIO3 induction (Fig. 3D). Moreover, ZBP1 is a well-characterized driver of caspase8-dependent apoptosis22. Consistently, we observed interaction between ZBP1 and caspase8 in ARPE-19 cells, with NaIO3 stimulation increasing total levels of ZBP1 and caspase8, thereby augmenting their interaction (Fig. 3E). In addition, we validated the above findings in primary mouse RPE cells (Fig. S3A). Knockdown experiments further confirmed ZBP1’s role as a sensor protein crucial for assembling RIPK1, RIPK3, caspase8, and NLRP3 into the PANoptosome (Fig. 3F, G).
A Visualization of PPI network among ZBP1 and its predicted functional partners, RIP1 (0.998) and RIP3 (0.996), exhibits the top 2 interaction scores. B–E ZBP1 immunoprecipitation (IP) with anti-RIPK1, anti-RIPK3, anti-NLRP3, and anti-Caspase8 showing the formation of ZBP1-panoptosome in ARPE-19 cells. F, G Panoptosome-related protein (p-RIPK1, RIPK1, p-RIPK3, RIPK3, NLRP3, cleaved-Caspase8, Caspase8) expressions in both ARPE-19 cells and NaIO3-induced mice by immunoblots analysis. Quantification of the protein levels are shown. Data are mean ± SD, the p values were obtained using one-way ANNOVA (F, G). *p < 0.05 versus control (CON/Ctrl) group, #p < 0.05 versus NC group.
Calcium dyshomeostasis activates ZBP1 via mPTP-mediated mtDNA release
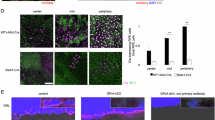
Under physiological conditions, ZBP1 can recognize double-stranded DNA (dsDNA) from either mitochondrial or nuclear origin23. To understand the activation of ZBP1 in AMD, we isolated cytosolic (cyto) and mitochondrial (mito) fractions from ARPE-19 cells and quantified mtDNA expression using quantitative real-time PCR (qRT-PCR). Upon NaIO3 stimulation, we observed a significant increase in cytosolic levels of MTND1, MTND2, ATP6, CO2, and DLOOP (Figs. 4A and S6A), indicating elevated levels of mtDNA in the cytosol. Additionally, we investigated the presence of dsDNA in NaIO3-stimulated ARPE-19 cells. The results showed a decrease in dsDNA and mitochondria colocalization (Fig. 4B), while the colocalization of ZBP1 and mitochondria was enhanced upon NaIO3 stimulation (Fig. 4C), providing additional evidence supporting the activation of the DNA sensor ZBP1 by cytoplasmic mtDNA (Fig. S4A, B). Additionally, we silenced ZBP1 using siRNA (Fig. S1B) and overexpressed it via plasmid transfection in ARPE-19 cells (Fig. S5A). Our results showed that neither ZBP1 knockdown nor overexpression significantly altered cytosolic mtDNA levels (Fig. S5B–D). ATP levels were also measured under ZBP1-modulated conditions. No significant differences in ATP levels were observed between ZBP1-silenced, ZBP1-overexpressed, and NC groups (Fig. S5E, F). This indicates that ZBP1 does not directly influence mitochondrial function under NaIO3 treatment. To further understand the mechanisms underlying mtDNA release, we investigated the effects of NaIO3 exposure on mitochondrial stability in ARPE-19 cells. Increased mitochondrial reactive oxygen species (ROS) levels (Fig. S7B) and decreased membrane potential (△Ψm) indicated mitochondrial destabilization (Fig. S7A), potentially leading to the opening of the mPTP (Fig. 4D) and subsequent release of mtDNA. Next, we investigated this hypothesis by performing pharmacological inhibition of the mPTP. Treatment with Cyclosporin A (CsA) was found to effectively reduce NaIO3-induced mtDNA leakage, leading to a decrease in cytosolic levels of mtDNA, including MTND1, MTND2, ATP6, CO2, and DLOOP (Figs. 4E, G and S6B, C). The enhanced colocalization of dsDNA and mitochondria confirmed these findings (Fig. 4F). Given the close relationship between calcium dyshomeostasis and mitochondrial dysfunction, we examined mitochondrial calcium levels using the mitochondrial calcium probe Rhod-2 in NaIO3-treated cells. The results showed that NaIO3 enhanced Rhod-2 signal and colocalization with the mitochondrial tracker probe in ARPE-19 (Fig. 4H) and that the Ca2+ chelator BAPTA-AM (5 μM) rescued mitochondrial destabilization via JC-1 and mtROS detection (Fig. S7A, B). In addition, BAPTA-AM prevented mPTP opening (Fig. 4J) and subsequent mtDNA release (Figs. 4I, K and S7C), strongly indicating that mitochondrial calcium overload triggers mPTP opening and mtDNA leakage (Fig. S7D, E), thereby upregulating ZBP1 expression.
A, E, K qRT-PCR assay showing the cytoplasmic mtDNA (MTND1, MTND2, ATP6, CO2, DLOOP) level. A Cytoplasmic mtDNA levels in ARPE-19 cells were elevated by NaIO3 (n = 3). E, K CsA (mPTP inhibitor) and BAPTA-AM (Ca2+ chelator) reversed the elevated cytoplasmic mtDNA induced by NaIO3 (n = 3). B, F, I NaIO3 induces (B) relocation of dsDNA (anti-dsDNA, green) from mitochondria (anti-TOMM20, red) into cytoplasm and was abolished by CsA (F) or BAPTA-AM (I). Nucleus were stained with DAPI. Scale bars = 25 μm. C Immunofluorescence staining shows elevated colocalization between ZBP1 (anti-ZBP1, red) and dsDNA (anti-dsDNA, green) under stimulation of NaIO3. Scale bars = 25 μm. D, J The mPTP was opened in NaIO3 treated ARPE-19 cells (D) and the abnormal phenomena was abolished by administration of BAPTA-AM (J). G CCK-8 assay showing ARPE-19 cells viability in the indicated groups (n = 6). H Representative immunofluorescence staining images showing elevated colocalization (yellow) between Rhod-2 (red) and mitochondria (Mito-Green, green) in ARPE-19 cells under NaIO3 stimulation. Scale bars = 25 μm. Data are mean ± SD, the p values were obtained using one-way ANNOVA (E, G, K) or unpaired t-test (A). *p < 0.05 and #p < 0.05 versus NaIO3 group.
MAMs control mitochondrial Ca2+ homeostasis by regulating IP3R1-GRP75-VDAC1 complex
Here, we investigated the mechanisms underlying mitochondrial calcium overload in AMD. Considering the significance of ER in calcium regulation and its involvement in ER stress24, a common feature in various neurodegenerative diseases, we hypothesized that MAMs might play a crucial role in modulating mitochondrial calcium homeostasis. Therefore, we divided transcriptome data from AMD patients into high and low ZBP1 expression groups based on ZBP1 expression levels and found that ZBP1 highly correlated with ER and mitochondria (Fig. 5A). To investigate this, we assessed the colocalization of ER-Tracker and Mito-Tracker in ARPE-19 cells, revealing an increase in MAM formation upon NaIO3 treatment (Fig. 5B). Previous research from our group highlighted the involvement of the IP3R1-GRP75-VDAC1 axis in calcium transport from the ER to mitochondria in diabetic retinopathy. In this study, we observed elevated GRP75 expression both in vitro and in vivo (Figs. 5C and S8A). Subsequently, we investigated whether NaIO3 treatment enhanced the interaction between GRP75 and IP3R1/VDAC1. The findings demonstrated that GRP75 indeed formed a complex with IP3R1 and VDAC1 in ARPE-19 cells, and this interaction was intensified upon NaIO3 induction (Figs. 5D, E and S8B). To elucidate the role of GRP75 in mitochondrial Ca2+ homeostasis, we conducted GRP75-specific siRNA transfection experiments and observed that decreased GRP75 expression led to reduced binding between the ER and mitochondria (Figs. 5F and S8D), consequently lowering mitochondrial calcium levels, as evidenced by Rhod-2 staining (Figs. 5G and S8E). Overall, our data suggest that GRP75 silencing significantly inhibits the formation of the IP3R1-GRP75-VDAC1 complex, thereby mitigating mitochondrial Ca2+ levels.
A Correlation between ZBP1 and the enrichment scores of top 10 cellular components (based on ssGSEA analysis) shown in bottom left. Correlation between ZBP1 and the enrichment scores of mitochondria-related cellular components shown in top right. B Immunofluorescence staining showing colocalization (yellow) between ER (ER-Tracker, green) and mitochondria (Mito-Tracker, red). Scale bars = 25 μm. C GRP75 protein expression in both primary mouse RPE cells and NaIO3-induced mice by immunoblots analysis. Quantification of the GRP75 levels relative to β-actin expression is shown. D, E VDAC1 and IP3R1 immunoprecipitation (IP) with anti-GRP75 showing the interaction between GRP75 and VDAC1, IP3R1 in ARPE-19 cells. Immunofluorescence staining showing elevated colocalization (yellow) between F ER (ER-Tracker, green) and mitochondria (Mito-Tracker, red) and G Rhod-2 (red) and mitochondria (Mito-Green, green) in ARPE-19 cells under NaIO3 stimulation and GRP75 siRNA significantly reversed the above phenomenon. Scale bars = 25 μm. Data are mean ± SD, the p values were calculated using unpaired t-test between control (CON/Ctrl) and NaIO3-induced ARPE-19 cells or mice. *p < 0.05 versus control group.
GRP75 silencing ameliorates ZBP1 activation and PANoptosis
Our findings suggest that elevated GRP75 levels contribute to mitochondrial calcium dysregulation and instability when exposed to NaIO3. To investigate GRP75’s role in NaIO3-induced PANoptosis, we examined mPTP opening in GRP75-silenced cells using confocal imaging (Figs. 6A and S8C, S9A, B). Additionally, we assessed cytosolic mtDNA levels through qRT-PCR analysis of specific markers (MTND1, MTND2, ATP6, CO2, DLOOP) in ARPE-19 cells (Figs. 6B and S9C). The results showed that GRP75 siRNA markedly decreased cytosolic mtDNA levels, indicating its potential to prevent mtDNA leakage. Notably, GRP75 silencing promoted the colocalization of dsDNA with mitochondria upon NaIO3 stimulation (Figs. 6C and S9D) while reducing its colocalization with ZBP1 (Figs. 6D nd S9E).
ARPE-19 cells were transfected with GRP75 siRNA under NaIO3 (10 mM, 24 h) stimulation. A Immunofluorescence staining reveals that mPTP opening in ARPE-19 cells were decreased significantly by GRP75 siRNA. Scale bars = 25 μm. B qRT-PCR assay showing that elevated cytoplasmic mtDNA (MTND1, MTND2, ATP6, CO2, DLOOP) level stimulated by NaIO3 were reversed after treatment with GRP75 siRNA (n = 3). Immunofluorescence staining showing that NaIO3 induces C relocation of dsDNA (anti-dsDNA, green) from mitochondria (anti-TOMM20, red) into cytoplasm and D sensed by ZBP1 (anti-ZBP1, red). Nuclei were stained with DAPI. GRP75 siRNA decreased C the dsDNA leakage and D colocalization of dsDNA with ZBP1. Scale bars = 25 μm. E Calcein-AM/PI assay reveals dead (red) and live (green) ARPE-19 cells. Scale bars = 50 μm. F CCK-8 assay showing ARPE-19 cells viability (n = 6). G PANoptosis-related protein expressions (p-MLKL, MLKL, cleaved-GSDMD, GSDMD, cleaved-Caspase3, Caspase3) in primary mouse RPE cells by immunoblots analysis. H RPE65 and ZO-1 expressions in primary mouse RPE cells by immunoblots analysis. I ARPE-19 cells stained with ZO-1 (green) and DAPI (blue) showing ARPE-19 cells dysfunction treated with NaIO3 and was reversed significantly by GRP75 siRNA. Scale bars = 25 μm. Data are mean ± SD, the p values were obtained using one-way ANNOVA (F–H) or unpaired t-test (B). *p < 0.05 versus control group, #p < 0.05 versus NC group.
Since the cytoplasmic mtDNA required for ZBP1 activation can be reduced by GRP75 siRNA, this raises the possibility that GRP75 silencing could suppress PANoptosis. To validate this hypothesis, we then utilized NaIO3-treated RPE cells and observed GRP75 siRNA led to significant attenuation of PANoptosis-related proteins (p-MLKL, cleaved-GSDMD, cleaved-caspase-3) via western blot (Figs. 6G and S9F). Based on these results, we investigated the impact of GRP75 silencing on cell viability and death. We found that GRP75 siRNA enhanced RPE cell viability (Fig. 6F), mitigated NaIO3-induced cell death (Fig. 6E) and reversed the downregulation of ZO-1 and RPE65 expression induced by NaIO3 (Figs. 6H, I and S9G). These findings collectively indicate that GRP75-mediated mtDNA leakage activates ZBP1, subsequently initiating PANoptosis.
O-GlcNAcylation of GRP75 regulates its ubiquitination and degradation
Next, we performed qRT-PCR and western blot analyses to assess GRP75 mRNA and protein levels both in vitro and in vivo. Under NaIO3 stimulation, GRP75 protein levels increased compared to the control group (Fig. 5C). However, qRT-PCR results showed no significant increase in GRP75 mRNA (gene name: HSPA9) (Fig. 7A). We hypothesized that NaIO3 might affect GRP75 levels by altering the stability of the GRP75 protein. To test this, we treated cells with cycloheximide (CHX) to inhibit protein synthesis before extracting proteins at specified intervals in both control and NaIO3-treated groups. Remarkably, NaIO3 substantially prolonged the half-life of the GRP75 protein (Fig. 7B), indicating that NaIO3 enhances GRP75 protein stability by inhibiting protein degradation. To further understand the predominant degradation pathway involved in this process, we treated NaIO3-exposed ARPE-19 cells with either a proteasome inhibitor (MG132) or a lysosome inhibitor (chloroquine). As depicted in (Fig. 7C), MG132 significantly increased GRP75 expression, indicating that NaIO3 mainly enhances GRP75 protein stability by impeding its degradation through the ubiquitin/proteasome pathway. O-GlcNAcylation, a widespread post-translational modification essential for cellular localization and protein stability, may also play a role25,26. The results showed increased expressions of O-GlcNAc transferase (OGT) and O-GlcNAc induced by NaIO3 both in vivo and in vitro (Figs. 7D, E and S10A, B), suggesting the involvement of O-GlcNAcylation in stabilizing the GRP75 protein. Then, we utilized the OGT inhibitor OSMI-1 to reduce O-GlcNAc levels in ARPE-19 cells. Western blot experiments confirmed that OSMI-1 inhibited GRP75 expression (Fig. 7H, I). Furthermore, IP assays using a GRP75 antibody demonstrated the formation of a complex between O-GlcNAc, OGT, and GRP75 in ARPE-19 cells, with increased binding observed under NaIO3 challenge (Fig. 7F, G), reinforcing the idea that O-GlcNAcylation contributes to stabilizing GRP75 expression. Recent research has unveiled that O-GlcNAc inhibits protein degradation by competitively antagonizing protein ubiquitination. Consistently, pharmacological inhibition of the ubiquitin-proteasome system using MG132 prevented OSMI-1-mediated GRP75 downregulation (Fig. 7J). Additional Co-IP experiments confirmed the decreased ubiquitin interacted with GRP75 in NaIO3-treated ARPE-19 cells, whereas pharmacological blockade of O-GlcNAc by OSMI-1 enhanced this interaction (Fig. 7K). Furthermore, we observed that OSMI-1 reduced the colocalization of ER-tracker and mito-tracker (Fig. 7L), consequently lowering mitochondrial Rhod-2 signaling under NaIO3 stimulation (Fig. 7M). Overall, O-GlcNAc upregulates GRP75 expression levels by competitively antagonizing GRP75 ubiquitination degradation, thereby promoting MAMs formation.
A qRT-PCR assay showing mRNA levels of HSPA9 (n = 3). B, I Half-life and quantitative analysis of GRP75 in APRE-19 cells treated with the indicated agents. Cells were treated with 10 μM cycloheximide (CHX) for the indicated time and immunoblotted with anti-GRP75 and anti-β-actin antibodies. Quantification of the GRP75 levels relative to β-actin expression are shown. C, H, J GRP75 protein expression in ARPE-19 cells treated with indicated agents by immunoblots analysis. D, EO-GlcNAcylation level in both ARPE-19 cells and NaIO3-induced mice by immunoblots analysis. F, G GRP75 immunoprecipitation (IP) with anti-O-GlcNAc and anti-OGT showing the O-GlcNAcylation of GRP75 in ARPE-19 cells. K Co-IP of GRP75 and Ubiquitin in ARPE-19 cells. L Representative images of ARPE-19 cells treated with vehicle or OSMI-1 and stained with Mito-Tracker (red) and ER-Tracker (green) showing the binding between the ER and mitochondria in ARPE-19. Scale bars = 25 μm. M ARPE-19 cells stained with Rhod-2 (red) and Mito-Green (green) showing the mitochondrial Ca2+ level in ARPE-19 cells treated with vehicle or OSMI-1. Scale bars = 25 μm. Data are mean ± SD, the p values were obtained using one-way ANNOVA (C, H, J) or unpaired t-test (A, B, I).
GRP75 deficiency alleviates PANoptosis and AMD progression in vivo
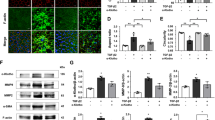
Next, adeno-associated virus-shGRP75 (AAV8-shGRP75) was generated to downregulate GRP75 in AMD mouse RPE (Fig. 8A). The efficiency of AAV8-shGRP75 in silencing GRP75 expression is shown (Fig. S11A). We observed downregulation of p-MLKL, cleaved-GSDMD, and cleaved-caspase-3 in AAV8-shGRP75-treated mice (Fig. S11B). Remarkably, impaired RPE cells and subsidence of ONL in NaIO3-treated mice were improved in the shGRP75 group (Fig. 8B). Additionally, our en-face images displayed an abundance of punctate hyperreflective signals (impaired RPE cells or misalignment of the ONL) and thinning of ONL in the NaIO3-treated mice, whereas shGRP75 reversed these changes (Fig. 8C). The ERG recordings showed that quenched waveforms in NaIO3-treated mice were reversed in shGRP75 mice (Fig. 8D). Furthermore, we observed decreased expression of Arrestin and Rhodopsin in NaIO3-treated mice, whereas a remarkable reversal was detected in shGRP75 mice (Fig. 8H). Moreover, NaIO3-treated mice exhibited reduced expression of ZO-1 and RPE65, which was reversed in the GRP75 knockdown group (Fig. 8E–G). Collectively, these results reveal that GRP75 knockdown mitigates PANoptosis and halts the progression of AMD in vivo.
A Flowchart of the in vivo experiments. B The H&E staining of mice retinas. White triangle: impaired RPE cells; black asterisk: subsidence of outer nuclear layer (ONL). Scale bars = 50 μm. C Retinal OCT and ultra-wide angle en face images obtained from SS-OCTA showing decreased number of punctate hyperreflective signals (red circle: impaired RPE cells or misalignment of the ONL layer) and thicken ONL by administration of AAV8-shGRP75 in the NaIO3-treated mice. D ERG recordings of mice retinas under scotopic condition. E RPE65 and ZO-1 expressions in mice RPE by immunoblot analysis. Quantification of the protein levels relative to β-actin expression is shown. F, G Immunofluorescent staining of RPE65 and ZO-1 in the indicated groups of mice retinas. Scale bars = 50 μm. (NC: staining without primary antibody). H Immunofluorescent staining of photoreceptor functional indicators (Arrestin, Rhodopsin) in the indicated groups of mice retinas. Scale bars = 50 μm. Data are mean ± SD, the p values were obtained using unpaired t-test (C–E). *p < 0.05 versus AAV8-vector group.
Discussion
Our research uncovered the essential role of ZBP1-mediated RPE PANoptosis in AMD pathogenesis. Mechanistically, O-GlcNAcylation of GRP75 promoted the formation of MAMs, leading to mitochondrial calcium overload, mPTP opening, and increased cytosolic mtDNA, which then activates ZBP1, initiating PANoptosis. The study findings are summarized in Fig. 9.
The primary pathogenic mechanism in dry AMD involves RPE death and loss, with various forms of PCD contributing to disease development. In a mouse model of AMD, activation of RIPK1/RIPK3 has been linked to maintaining necroptosis in the RPE7. Additionally, the NLRP3 inflammasome has been associated with pyroptosis in the RPE27, and targeting the pyroptotic pore-forming protein GSDMD has shown promise in alleviating retinal degeneration by suppressing the release of inflammatory factors such as IL-1β encapsulated in extracellular vesicles (EVs)28,29. In human eyes with GA, RPE cells accumulate toxic Alu RNA, leading to caspase-8-mediated apoptosis through a Fas ligand-dependent mechanism30. However, these different forms of PCD do not fully represent the spectrum of RPE cell death modalities. Our research also demonstrates that several PCD inhibitors only partially reverse NaIO3-induced RPE mortality. Given the extensive interplay between different programmed death pathways, focusing on PANoptosis could provide important insights on neurodegenerative diseases such as AMD. PANoptosis, mediated by ZBP1, is implicated in various neurodegenerative diseases, including Parkinson’s disease and Alzheimer’s disease31, and can be activated by the recognition of cytoplasmic dsDNA32, triggering downstream PANoptosis-related proteins. ZBP1, identified as a crucial innate immune receptor and the innate immune response to inducing agents causing inflammatory death of RPE, is a recognized pathogenic mechanism of AMD3,33,34. The present study revealed a significant increase in ZBP1 expression in AMD patients, the NaIO3-induced mouse model, and in vitro cell culture models. Elevated ZBP1 levels in RPE cells promote the expression of p-MLKL, cleaved-caspase-3, cleaved-caspase-1, and cleaved-GSDMD by recruiting RIPK1, RIPK3, Caspase8, and NLRP3 to assemble the PANoptosome. Inhibiting ZBP1 reduced the expression of PCD-related molecules and the incidence of related diseases. Studies have shown that ZBP1 knockdown in mice increased resistance to doxorubicin-induced cardiotoxicity15 and protected against mortality during IAV infection by reducing inflammatory responses and epithelial damage14. Our findings demonstrated that ZBP1 siRNA effectively suppressed PANoptosis-related protein expression in RPE cells, thereby reducing cell death and improving RPE layer function. Furthermore, through the injection of AAV8-shZBP1, we confirmed the essential role of ZBP1 in an AMD mouse model. Despite these advancements, the redundancy observed in different PANoptosis pathways raises the intriguing question of whether all forms of cell death can be more effectively inhibited upstream.
In line with previous findings, ZBP1 activation occurs in response to cytoplasmic dsDNA. Elevated cytoplasmic dsDNA levels have been implicated in various ocular diseases, including AMD19. For instance, in a dry eye model, mtDNA-mediated cGAS-STING activation exacerbates ocular surface inflammation35. However, the specific role of dsDNA in AMD pathogenesis remains unexplored. Cytoplasmic dsDNA comprises genomic DNA from the nucleus and mtDNA. Our investigation revealed that NaIO3 exposure increases cytoplasmic mtDNA levels in RPE cells, a process facilitated by mPTP opening. Nonetheless, the precise mechanisms underlying NaIO3-induced mPTP opening and mtDNA release require further elucidation. Mitochondrial calcium homeostasis plays a crucial role in regulating mitochondrial aerobic metabolism and membrane channel opening36. Calcium interacts with cyclophilin D and adenine nucleotide translocase (ANT) to form mPTP, with excessive mitochondrial Ca2+ uptake triggering mPTP opening37. Notably, our experiments demonstrated a marked rise in mitochondrial calcium levels following NaIO3 exposure, suggesting that NaIO3-induced mitochondrial calcium overload leads to mtDNA release, thereby initiating PANoptosis. It is important to note that, apart from mPTP, VDAC1 may also contribute to mtDNA release38, potentially explaining why treatment with CsA only partially restored cytoplasmic DNA levels in RPE cells.
Interestingly, bioinformatics analyses suggest that ER is essential in AMD pathogenesis. For instance, Elovl2 deficiency, a long fatty acid elongase in the ER, disrupts lipid synthesis, leading to increased ER stress and mitochondrial dysfunction, ultimately contributing to AMD39. Moreover, in the RPE, PERK/EIF2AK3 mediates ER stress-induced apoptosis40. However, emerging research suggests that the ER and mitochondria do not function independently but rather synergistically in disease processes41. MAMs, which are specialized cellular compartments, serve as contact sites between the ER and mitochondria and mediate various events, including calcium signal transduction, lipid metabolism, and mitochondrial dynamic changes42. In Parkinson’s disease, the PARKIN protein regulates mitochondrial-ER interactions in an ATF4-dependent manner to promote cell survival43. The IP3R1-GRP75-VDAC1 complex mediates ER stress-mitochondrial oxidative stress, exerting a pivotal influence on the remodeling of the atrium of diabetic cases44. Nonetheless, the role of MAMs in AMD has not been reported. Calcium ions are essential for communication between the ER and mitochondria, and their abundance is strictly controlled by the ER. IP3R1, an important calcium ion efflux channel located on the ER surface, and VDAC1, an ion channel embedded in the outer mitochondrial membrane (OMM), participate in calcium ion transport. GRP75 facilitates the structural integrity of the MAM by orchestrating the interaction between IP3R1 and VDAC1. Moreover, GRP75, a member of the HSP protein family, is mainly located in mitochondria but also distributed in the ER. Studies have identified GRP75 as one of the 5 important proteins (Grp75, nucleolin, Grp94, calnexin, and calreticulin) involved in the pathogenesis of Parkinson’s disease45. Our findings demonstrate that NalO₃ triggers aberrant upregulation of GRP75, which in turn promotes IP3R1–GRP75–VDAC1 coupling. The enhanced coupling leads to mitochondrial calcium overload, promotes the cytosolic release of mitochondrial DNA, and ultimately activates PANoptosis. GRP75 gene ablation in cardiomyocytes reduces intracellular ROS levels and alleviates mitochondrial calcium overload and apoptosis44. Consistently, we found that knocking down GRP75 with siRNA significantly reversed the effects of NaIO3, including increased MAMs, mitochondrial calcium overload, mtDNA leakage, and PANoptosis. Additionally, silencing GRP75 in vivo and in vitro ameliorates AMD phenotypes, further confirming the critical role of calcium crosstalk between the ER and mitochondria in AMD.
We also found that NaIO3 stimulation failed to increase the level of GRP75 mRNA as expected, suggesting that the upregulation of GRP75 in RPE may be associated with a reduction in its degradation. In eukaryotic cells, the main intracellular pathways for protein degradation are the ubiquitin-proteasome pathway and the autophagy-lysosome pathway. Our findings confirm the inhibition of the ubiquitin-proteasome degradation pathway of GRP75 in AMD models. There is accumulating evidence suggesting that protein stability is intricately linked to its post-translational modifications (PTMs). O-GlcNAcylation is a common type of PTM known to primarily influence PPIs and subcellular localization. Importantly, O-GlcNAcylation is closely associated with disorders in glycolipid metabolism and oxidative stress, which are also implicated as pathogenic mechanisms in AMD46,47. Moreover, O-GlcNAcylation has been shown to enhance the activity of phosphoglycerate kinase 1 (PGK1), leading to increased lactate production and the translocation of PGK1 to the mitochondria, which has been linked to oncogenesis48. Herein, our findings indicate a significant increase in O-GlcNAcylation levels in RPE under AMD conditions. Importantly, we found that O-GlcNAcylation stabilizes GRP75 and thereby promotes the binding of mitochondria and ER through the IP3R1-GRP75-VDAC1 axis. Emerging evidence suggests that O-GlcNAcylation is closely linked to ubiquitin degradation pathways. For instance, O-GlcNAcylation of FAM134B stabilizes the protein and reduces its affinity for ubiquitin49. Similarly, glycosylation modification of YTHDF2 at Ser263 competitively inhibits its ubiquitination, extending the protein’s half-life50. Moreover, GFPT1 enhances the O-GlcNAcylation of YAP, which mitigates SCFβ-TRCP E3 ubiquitin ligase-mediated degradation, thereby prolonging YAP’s half-life51. Our study provides insights by demonstrating that increased O-GlcNAcylation significantly promotes the formation of MAMs through the regulation of GRP75.
In conclusion, this study highlights the role of ZBP1-mediated PANoptosis in driving AMD development and sheds light on the intricate interplay between mitochondria and the ER in PANoptosis induction. However, these findings should be viewed with consideration of the following limitations. Firstly, while our study is based on clinical sample databases, further validation using human clinical samples is necessary. Secondly, identifying the specific O-GlcNAcylation sites on GRP75 and elucidating its specific E3 ubiquitin ligase demand further investigation. Thirdly, the current experimental design did not incorporate direct observation of the c-wave in ERG and permeability studies, which serve as functional indicators of RPE integrity. In addition, the utilization of RPE-specific Cre models in subsequent studies would enable targeted observation of cellular changes with minimum off-target effects. Addressing these limitations through additional research could further enhance our understanding of organelle interactions in AMD pathogenesis and facilitate the development of therapeutic strategies.
Methods
Materials and reagents
Sodium iodate (NaIO3) was bought from Sigma-Aldrich (St. Louis, USA). Various compounds and reagents were bought from different suppliers: BAPTA-AM, Chloroquine (CQ), OSMI-1, Dideoxycytidine (ddC), Cyclosporin A (CsA), Necrostatin-1, Z-VAD-FMK, MCC950, Ferrostatin-1, 3-Methyladenine, and Protein A/G magnetic beads were obtained from MedChemExpress (NJ, USA), and 2-D08, MG132 and cycloheximide were acquired from Selleck (USA); Rhod-2 AM was obtained from YEASEN (Shanghai, China). Kits such as the Cell Counting Kit-8, JC1 assay kit, mPTP assay kit, Calcein-AM/PI assay kit, Cell Mitochondria Isolation kit, Mito-Tracker Red, Mito-Tracker Green, and ER-Tracker Green were bought from Beyotime (Nantong, China). MitoSOX™ Red was purchased from Thermo Fisher (USA), while the HE staining kit, IHC staining kit, and DAB were bought from BOSTER (Wuhan, China). The riboFECT CP Transfection Kit was obtained from Guangzhou, China, and the HiScript III RT SuperMix kit was purchased from Vazyme (Nanjing, China). Primary antibodies were acquired from various suppliers: GRP75, cleaved-caspase-3, caspase-3, cleaved-Gasdermin D, Gasdermin D, cleaved-caspase-1, caspase-1, Caspase 8, NLRP3, p-RIPK1, RIPK1, p-RIPK3, RIPK3, p-MLKL, and MLKL were sourced from CST (USA); TOMM20 from Abcam (USA), and GRP75, ZBP1, Ubiquitin, Caspase 8, Rhodopsin, Arrestin, dsDNA, RPE65, IP3R1, VDAC1, O-GlcNAc, and IgG from Santa (USA); ZBP1, ZO-1, OGT, and β-actin from Proteintech (China). The details of reagents and antibodies are provided in Supplementary Tables 1 and 2.
Animals and NaIO3-induced mice
Male C57BL/6J mice (weight 24–26 g, 6–8 weeks, n = 12, each group, bought from Changzhou Cavans Experimental Animal Co., Ltd). All the animals were housed in the Experimental Animal Center of Wuxi People’s Hospital under controlled environmental conditions at a temperature of 24 ± 2 °C, a 12-h light/dark cycle, and humidity maintained at 45 ± 5%. The mice were provided ad libitum access to tap water and standard chow. To establish the AMD model, the mice were intravenously administered with 50 mg/kg of NaIO3. All animal experiments were conducted following the Guide of the Animal Care and Use Committee of Nanjing Medical University (approval No: SYXK(SU)2020-0010). This study was approved by the Ethics Committee of the Affiliated Wuxi People’s Hospital of Nanjing Medical University (approval No: DL2022229).
Isolation of primary mouse RPE cells
C57BL/6J mice (age: 2–3 weeks) were euthanized, and 12 eyes of six mice were enucleated and rinsed in cold PBS containing 1% penicillin/streptomycin (Gibco). Surrounding tissues, including the conjunctiva, extraocular muscles, and optic nerve, were carefully removed. A circumferential incision was made along the limbus to remove the cornea and lens, followed by gentle detachment and discarding of the retina. The remaining eyecup, containing the RPE-choroid-sclera complex, was incubated in Dispase II at 37 °C for 15 min. After digestion, the tissue was transferred to DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS; Gibco) to neutralize enzymatic activity. The RPE layer was gently brushed off the choroid using an iris repositioner under a stereomicroscope. The dissociated RPE cells were collected by centrifugation (2200 rpm, 3 min) and resuspended in culture medium (DMEM/F12, 10% FBS, 1% penicillin/streptomycin). The cells were seeded into T25 flasks and cultured at 37 °C. Subculturing was performed after 5 days of culture.
Mitochondrial ROS and Ca2+ Detection
The ARPE-19 cells were treated with MitoSOX™ Red (5 μM, M36008, Thermo Fisher) or Rhod-2 (2 μM, 40776ES72, YEASEN) for 30 min, followed by washing with PBS, then incubated with MitoTracker-Green (200 nM, C1048, Beyotime) for an additional 30 min. Fluorescence imaging was conducted using a confocal microscope (40× magnification, Leica, Germany).
Measurement of mitochondria–endoplasmic reticulum (ER) interactions
To investigate mitochondria-ER interactions, ARPE-19 cells were initially treated with ER-Tracker Green (1 μM, C1042, Beyotime) for 20 min, subsequently incubated with Mito-Tracker Red (200 nM, C1035, Beyotime) for an additional 30 min.
Measurement of mitochondrial membrane potential (MMP) and ATP
The JC-1 staining kit (C2006, Beyotime) was used to assess MMP. A decrease in ratio of red (aggregate) fluorescence to green (monomer) fluorescence indicated mitochondrial depolarization. The cells were incubated with JC-1 working solution for 20 min. The cellular ATP levels were measured using a firefly luciferase-based ATP assay kit (S0026, Beyotime) according to the manufacturer’s protocol.
Determination of mitochondrial permeability transition pore (mPTP)
To perform the assay, the cells were incubated with a mixture of calcein AM (2 μM) and CoCl2 (250 μM) for 30 min. Then, the cells were cultured in DMEM/F12 medium (10% FBS) for 40 min. Then treated with MitoTracker Red (200 nM, C1035, Beyotime) for an additional 30 min.
Calcein-AM/Propidium iodide (PI) staining
Dead cells were identified using a Calcein-AM/PI double staining kit (C2015M, Beyotime). ARPE-19 cells were incubated with Calcein-AM (2 μM) and PI (5 μM) for 30 min at 37 °C. The observation of live (green) and dead (red) cells was conducted using a laser scanning confocal microscope (20× magnification, Leica, Germany).
Intravitreal injection
The mice were anesthetized with sodium pentobarbital (50 mg/kg, i.p.). To dilate the pupils, 1% tropicamide eye drops were administered, followed by disinfection of the eyes with 0.5% povidone-iodine and rinsing of the ocular surface with artificial tears. Intravitreal injection was performed under a microscope. A disposable 32G needle was used to create a puncture hole through the sclera at the temporal part of the corneoscleral limbus. Then, a Hamilton microsyringe with a 33G blunt needle was slowly inserted into the vitreous cavity through the puncture hole (care was taken to avoid injury to the lens and retina), and 1 μL of the drug solution was gently injected.
AAV8-mediated ZBP1/GRP75 knockdown in mice
Adeno-associated virus serotype 8 (AAV8) vectors containing either AAV8-shZBP1 or AAV8-shGRP75, capable of RNAi targeting ZBP1 or GRP75, respectively, were applied in this experiment. Briefly, the mice were categorized into groups dependent on body weight randomly, and intravitreal injections of 1 μL of AAV8-vector, AAV8-shZBP1, or AAV8-shGRP75 were administered using a 33-G needle. The concentrations of shZBP1 (3.6 × 1012 viral genomes/mL) and shGRP75 (7.6 × 1012 viral genomes/mL), and details regarding shZBP1 or shGRP75 are provided in Supplementary Table 3.
Cell culture
Human retinal epithelial cells (ARPE-19, Zhong Qiao Xin Zhou Biotechnology, Shanghai, China) were cultured in DMEM/F12 medium supplemented with 10% FBS (Gibco, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C with 5% CO2. ARPE-19 cells were treated with NaIO3 (10 mM) for 24 h to establish AMD model. The cells were pretreated with various inhibitors for 2 h prior to NaIO3 stimulation, including BAPTA-AM (5 μM), Chloroquine (2 μM), OSMI-1 (5 μM), Dideoxycytidine (100 μM), Cyclosporin A (5 μM), Z-VAD-FMK (5 μM), 3-Methyladenine (2.5 mM), MCC950 (1 μM), Ferrostatin-1 (2 μM), 2-D08 (5 μM), MG132 (1 μM), Cycloheximide (10 μM) and Necrostatin-1 (5 μM).
Cell viability
The Cell Counting Kit-8 (C0043, Beyotime) was used to assess cell viability. 100 μL of CCK-8 solution was added to each well (96-well plate), and then the cells were incubated at 37 °C in a CO2 incubator for 1.5 h. A microplate reader was used to determine the absorbance of each well at 450 nm.
Western blotting
ARPE-19 cells and RPE tissues (all mouse eye cups used for protein extraction had the peripheral fascial tissue, optic nerve, cornea, lens, and neuron retina removed) were lysed on ice in RIPA buffer containing protease inhibitors, phosphatase inhibitors, and PMSF (1 mM, ST506, Beyotime). Equal amounts of proteins were separated on 10% or 12% SDS polyacrylamide gels and transferred to 0.45 mm PVDF membranes. Then, after 2 h of blocking with 5% skimmed milk, the membranes were incubated with the following antibodies: ZBP1 (1:200, sc-271483, Santa), GRP75 (1:200, sc-133137, Santa), Ubiquitin (1:100, sc-8017, Santa), O-GlcNAc (1:100, sc-59623, Santa), RPE65 (1:200, sc-390787, Santa), IP3R1 (1:200, sc-271197, Santa), VDAC1 (1:200, sc-390996, Santa), cleaved-caspase-3 (1:1000, 9664S, CST), caspase-3 (1:1000, 14220S, CST), cleaved-Gasdermin D (1:1000, 36425S, CST), Gasdermin D (1:1000, 39754S, CST), cleaved-caspase-1 (1:1000, 4199S, CST), caspase-1 (1:1000, 83383S, CST), Caspase 8 (1:1000, 4790S, CST), Caspase 8 (1:1000, 9746S, CST), NLRP3 (1:1000, 15101S, CST), p-RIPK1 (1:1000, 65746S, CST), RIPK1 (1:1000, 3493S, CST), p-RIPK3 (1:1000, 93654S, CST), RIPK3 (1:1000, 13526S, CST), p-MLKL (1:1000, 91689S, CST), MLKL (1:1000, 14993S, CST), OGT (1:2000, 11576-2-AP, Proteintech), ZO-1 (1:5000, 21773-1-AP, Proteintech), β-actin (1:5000, 66009-1-Ig, Proteintech). After washing with TBST for 10 min, the membranes were incubated with secondary antibodies. Lastly, the signals were detected using a commercial imaging system (34580, Thermo Fisher).
Hematoxylin and eosin (H&E) staining
Retinal paraffin sections (4 μm) were deparaffinized with xylene, followed by dehydration using a series of graded ethanol concentrations (50, 75, 80, 90, 95, and 100%). The sections were then stained with an erythromycin and eosin kit (AR1180, BOSTER).
Co-immunoprecipitation
The cells were lysed using 500 μL of Pierce™ IP lysis buffer (Thermo Fisher, USA) containing protease inhibitors (Roche, Switzerland) for 30 min on ice. The supernatant was then centrifuged at 4 °C and incubated overnight at 4 °C with 2 μg of primary antibodies, including GRP75 (sc-133137, Santa), ZBP1 (sc-271483, Santa) or mouse IgG (sc-2025, Santa). Subsequently, the lysate was incubated with 50 μL of protein A/G beads (HY-K0202, MCE) for 4 h at 4 °C, followed by four washes with IP buffer on a magnetic rack. Lastly, after boiling with 1× SDS buffer, the protein A/G beads were finally eluted.
Immunohistochemistry (IHC)
Retinal paraffin sections (4 μm) were used for IHC staining. Briefly, the sections were deparaffinized in xylene, hydrated in ethanol, and then treated with 3% H2O2 for 30 min to remove endogenous peroxidase activity. They were then placed in a microwave with 1× sodium citrate antigen retrieval solution for 15 min, followed by blocking with cell sealing solution for 2 h and incubated overnight at 4 °C with the following primary antibodies: GRP75 (1:100, Santa, sc-133137), ZBP1 (1:100, Santa, sc-271483), cleaved-caspase-3 (1:500, 9664S, CST), cleaved-GSDMD (1:200, 10137S, CST), p-MLKL (1:200, 37333S, CST). Next, the sections were incubated with a secondary antibody (SV0004, BOSTER). Color development was achieved using DAB (AR1027, BOSTER), followed by counterstaining with Mayer hematoxylin.
Immunofluorescence (IF)
ARPE-19 cells and frozen eye sections (8 μm) were initially fixed with 4% formaldehyde, followed by blocking with 5% bovine serum albumin (BSA) at 37 °C for 2 h. The samples were then incubated with the mentioned primary antibodies: ZO-1 (1:500, 21773-1-AP, Proteintech), RPE65 (1:100, sc-390787, Santa), Arrestin (1:100, sc-271159, Santa), Rhodopsin (1:100, sc-57432, Santa), dsDNA (1:100, sc-58749, Santa), TOMM20 (1:250, ab186735, Abcam), and ZBP1 (1:100, 13285-1-AP, Proteintech). The sections were rinsed with PBS for 15 min before incubation with a secondary antibody (1:400, Thermo Fisher). Finally, the tissue samples were stained with DAPI for 15 min.
RPE layer ZO-1 staining
For mouse RPE layer ZO-1 staining, the cornea, lens, neural retina, and surrounding myofascial tissues of the mice's eyes were carefully removed in 4% paraformaldehyde. The eye globes were then washed with PBS and incubated with ZO-1 primary antibody (1:200, 82870-7-RR, Proteintech) at 4 °C overnight. The eyecups were trimmed into a four-leaf clover shape on a slide and then incubated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:400, Thermo Fisher, A11008) prepared in 1% BSA for 2 h at 37 °C. Lastly, the tissue samples were stained with DAPI for 15 min.
MtDNA extraction and quantitative PCR
After separating the mitochondria using the cell mitochondria isolation kit (C3601, Beyotime), cytoplasmic DNA was extracted using a DNA Extraction Kit (DP304, TIANGEN). Briefly, the ARPE-19 cells were first digested with trypsin and then homogenized with a mitochondrial isolation reagent containing PMSF. The homogenate was centrifuged three times to isolate the mitochondria, following the manufacturer’s instructions. The sediment obtained was further centrifuged at 9000 g for 5 min after a single wash with homogenization buffer, and mtDNA was extracted after resuspending the mitochondrial sediment in PBS.
Relative gene expression was measured by qRT–PCR using HiScript III RT SuperMix kit (Vazyme, R323-01) and Fast SYBR Green Master Mix (Roche, 04913914001). The primer sequences are provided in Supplementary Table 4.
Swept-source OCT(SS-OCT) and SS-OCTA imaging
SS-OCT and SS-OCTA imaging (TowardPi BMizar, TowardPi Medical Technology, Beijing, China) were used to examine the structure of the mouse retina. The SS-OCT/SS-OCTA system offers advanced features, including a significant axial scanning depth (6 mm), high axial optical resolution (3.8 μm), and OCTA imaging capacity over an area of 24 mm × 20 mm. This coverage is equivalent to an imaging range of 120° of the central field of view, facilitating the acquisition of high-resolution fundus images. Herein, the mice were anesthetized as previously described, and the mice's B-scan OCT images were obtained using SS-OCT, while RPE layer images were captured in SS-OCTA en face mode. All data were collected using the integrated software of the instrument.
Mouse electroretinography (ERG)
ERG recordings and data analysis in mice were performed using the Celebris system (Diagnosys, Lowell, MA). Prior to experimentation, each mouse underwent dark adaptation overnight and was subsequently anesthetized. 1% sodium hyaluronate was applied topically to maintain the corneas' moisture. Under scotopic conditions, the overall retinal responsiveness was determined using a fixed intensity stimulus (3.0 cd.s/m2). The b-wave amplitude is measured from the trough of the a-wave to the peak of the b-wave.
Bioinformatic analysis
In this study, gene expression data for AMD and normal samples were retrieved from the GEO database (GSE29801)52. Redundant data were removed, and cases with macular RPE-choroid records were selected because both the RPE and the choroid are the primary pathological sites of AMD. Subsequently, 41 macular RPE-choroid samples from AMD patients and 50 normal macular RPE-choroid samples were included in our analysis. Moreover, PANoptosis-related genes (n = 33) used in this study were obtained from GeneCards database, and the detailed gene information are provided in Supplementary Table 5. Differential expression analysis was conducted using the limma package in R software (version 4.2.1; Lucent Technologies, NJ, USA). The cutoff value was set as log2 |fold change (FC)| > 0.263 and P < 0.05. Additionally, we calculated receiver operating characteristic (ROC) curves and evaluated the diagnostic ability of the gene biomarkers by measuring the area under the curve (AUC) using the glm R package. To determine the main cellular component contributing to differences between the ZBP1 high-expression and low-expression groups, we utilized the R “GSVA” version 1.50.0 package to perform the analysis.
Statistics and reproducibility
The data were analyzed using GraphPad Prism (GraphPad Software version 9.0, San Diego, CA) and are expressed as mean ± standard deviation (SD). Unpaired t-tests and one-way ANOVA were utilized for group comparisons, with differences considered statistically significant at P < 0.05. The sample size was based on previous studies. All experiments were performed with biological replicates at least three times.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
References
Fleckenstein, M., Schmitz-Valckenberg, S. & Chakravarthy, U. Age-related macular degeneration: a review. JAMA 331, 147–157 (2024).
Guymer, R. H. & Campbell, T. G. Age-related macular degeneration. Lancet 401, 1459–1472 (2023).
Datta, S., Cano, M., Ebrahimi, K., Wang, L. & Handa, J. T. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 60, 201–218 (2017).
Fleckenstein, M. et al. The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology 125, 369–390 (2018).
Yashkin, A. P., Hahn, P. & Sloan, F. A. Introducing anti-vascular endothelial growth factor therapies for AMD did not raise risk of myocardial infarction, stroke, and death. Ophthalmology 123, 2225–2231 (2016).
Wei, T. T. et al. Interferon-gamma induces retinal pigment epithelial cell ferroptosis by a JAK1-2/STAT1/SLC7A11 signaling pathway in age-related macular degeneration. FEBS J. 289, 1968–1983 (2022).
Tong, Y. et al. Comparative mechanistic study of RPE cell death induced by different oxidative stresses. Redox Biol. 65, 102840 (2023).
Gupta, U. et al. Increased LCN2 (lipocalin 2) in the RPE decreases autophagy and activates inflammasome-ferroptosis processes in a mouse model of dry AMD. Autophagy 19, 92–111 (2023).
Zhu, M. et al. Crosstalk between RPE cells and choroidal endothelial cells via the ANXA1/FPR2/SHP2/NLRP3 inflammasome/pyroptosis axis promotes choroidal neovascularization. Inflammation 45, 414–427 (2022).
Upadhyay, M. & Bonilha, V. L. Regulated cell death pathways in the sodium iodate model: insights and implications for AMD. Exp. Eye Res. 238, 109728 (2024).
Pandian, N. & Kanneganti, T. D. PANoptosis: a unique innate immune inflammatory cell death modality. J. Immunol. 209, 1625–1633 (2022).
Sun, X. et al. PANoptosis: mechanisms, biology, and role in disease. Immunol. Rev. 321, 246–262 (2024).
Samson, A. L. & Murphy, J. M. Mapping where and when necroptotic cell death occurs in disease. Cell Death Differ. https://doi.org/10.1038/s41418-024-01318-1 (2024).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, https://doi.org/10.1126/sciimmunol.aag2045 (2016).
Lei, Y. et al. Cooperative sensing of mitochondrial DNA by ZBP1 and cGAS promotes cardiotoxicity. Cell 186, 3013–3032 e3022 (2023).
Szczesny, B. et al. Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci. Rep. 8, 914 (2018).
Bi, Y. et al. FUNDC1 protects against doxorubicin-induced cardiomyocyte PANoptosis through stabilizing mtDNA via interaction with TUFM. Cell Death Dis. 13, 1020 (2022).
Zeng, Z. et al. Pathologically high intraocular pressure induces mitochondrial dysfunction through Drp1 and leads to retinal ganglion cell PANoptosis in glaucoma. Redox Biol. 62, 102687 (2023).
Kaarniranta, K. et al. Mechanisms of mitochondrial dysfunction and their impact on age-related macular degeneration. Prog. Retin. Eye Res. 79, 100858 (2020).
Henning, Y., Blind, U. S., Larafa, S., Matschke, J. & Fandrey, J. Hypoxia aggravates ferroptosis in RPE cells by promoting the fenton reaction. Cell Death Dis. 13, 662 (2022).
Zheng, M. & Kanneganti, T. D. The regulation of the ZBP1-NLRP3 inflammasome and its implications in pyroptosis, apoptosis, and necroptosis (PANoptosis). Immunol. Rev. 297, 26–38 (2020).
Yuan, F. et al. Z-DNA binding protein 1 promotes heatstroke-induced cell death. Science 376, 609–615 (2022).
Jiao, H. et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580, 391–395 (2020).
Xu, Y., Chen, J., Chen, J. & Teng, J. EI24 promotes cell adaption to ER stress by coordinating IRE1 signaling and calcium homeostasis. EMBO Rep. 23, e51679 (2022).
Chatham, J. C., Zhang, J. & Wende, A. R. Role of O-Linked n-acetylglucosamine protein modification in cellular (patho)physiology. Physiol. Rev. 101, 427–493 (2021).
He, X. et al. O-GlcNAcylation and stablization of SIRT7 promote pancreatic cancer progression by blocking the SIRT7-REGgamma interaction. Cell Death Differ. 29, 1970–1981 (2022).
Maran, J. J., Adesina, M. M., Green, C. R., Kwakowsky, A. & Mugisho, O. O. The central role of the NLRP3 inflammasome pathway in the pathogenesis of age-related diseases in the eye and the brain. Ageing Res. Rev. 88, 101954 (2023).
Sekar, R. et al. Impairing gasdermin D-mediated pyroptosis is protective against retinal degeneration. J. Neuroinflamm.20, 239 (2023).
Gao, J., Cui, J. Z., To, E., Cao, S. & Matsubara, J. A. Evidence for the activation of pyroptotic and apoptotic pathways in RPE cells associated with NLRP3 inflammasome in the rodent eye. J. Neuroinflamm. 15, 15 (2018).
Lewis Lujan, L. M. et al. Nutraceuticals/drugs promoting mitophagy and mitochondrial biogenesis may combat the mitochondrial dysfunction driving progression of dry age-related macular degeneration. Nutrients 14, https://doi.org/10.3390/nu14091985 (2022).
Rajesh, Y. & Kanneganti, T. D. Innate immune cell death in neuroinflammation and alzheimer’s disease. Cells 11, https://doi.org/10.3390/cells11121885 (2022).
Cho, M. G. et al. MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature 625, 585–592 (2024).
Zou, M. et al. Inhibition of cGAS-STING by JQ1 alleviates oxidative stress-induced retina inflammation and degeneration. Cell Death Differ. 29, 1816–1833 (2022).
Saada, J. et al. Oxidative stress induces Z-DNA-binding protein 1-dependent activation of microglia via mtDNA released from retinal pigment epithelial cells. J. Biol. Chem. 298, 101523 (2022).
Ouyang, W. et al. The cGAS-STING pathway-dependent sensing of mitochondrial DNA mediates ocular surface inflammation. Signal Transduct. Target Ther. 8, 371 (2023).
Garbincius, J. F. & Elrod, J. W. Mitochondrial calcium exchange in physiology and disease. Physiol. Rev. 102, 893–992 (2022).
Bernardi, P. et al. Identity, structure, and function of the mitochondrial permeability transition pore: controversies, consensus, recent advances, and future directions. Cell Death Differ. 30, 1869–1885 (2023).
Kim, J. et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366, 1531–1536 (2019).
Li, X. et al. Lipid metabolism dysfunction induced by age-dependent DNA methylation accelerates aging. Signal Transduct. Target Ther. 7, 162 (2022).
Saptarshi, N., Porter, L. F. & Paraoan, L. PERK/EIF2AK3 integrates endoplasmic reticulum stress-induced apoptosis, oxidative stress and autophagy responses in immortalised retinal pigment epithelial cells. Sci. Rep. 12, 13324 (2022).
Missiroli, S. et al. Mitochondria-associated membranes (MAMs) and inflammation. Cell Death Dis. 9, 329 (2018).
Hu, Y. et al. The AMPK-MFN2 axis regulates MAM dynamics and autophagy induced by energy stresses. Autophagy 17, 1142–1156 (2021).
Bouman, L. et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ. 18, 769–782 (2011).
Yuan, M. et al. IP3R1/GRP75/VDAC1 complex mediates endoplasmic reticulum stress-mitochondrial oxidative stress in diabetic atrial remodeling. Redox Biol. 52, 102289 (2022).
Liu, Y. et al. DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc. Natl. Acad. Sci. USA 116, 25322–25328 (2019).
Tan, W. et al. Posttranscriptional regulation of de novo lipogenesis by glucose-induced O-GlcNAcylation. Mol. Cell 81, 1890–1904 e1897 (2021).
Gorg, B. et al. O-GlcNAcylation-dependent upregulation of HO1 triggers ammonia-induced oxidative stress and senescence in hepatic encephalopathy. J. Hepatol. 71, 930–941 (2019).
Nie, H. et al. O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat. Commun. 11, 36 (2020).
Luo, R. et al. O-GlcNAc transferase regulates intervertebral disc degeneration by targeting FAM134B-mediated ER-phagy. Exp. Mol. Med. 54, 1472–1485 (2022).
Yang, Y. et al. O-GlcNAcylation of YTHDF2 promotes HBV-related hepatocellular carcinoma progression in an N(6)-methyladenosine-dependent manner. Signal Transduct. Target Ther. 8, 63 (2023).
Zhang, X. et al. Endogenous glutamate determines ferroptosis sensitivity via ADCY10-dependent YAP suppression in lung adenocarcinoma. Theranostics 11, 5650–5674 (2021).
Newman, A. M. et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 4, 16 (2012).
Acknowledgements
This study was supported by National Natural Science Foundation of China (82201220); Natural Science Foundation of Jiangsu Province (BK20241760); Wuxi Taihu Lake Talent Plan (2020-THRC-DJ-1, 2020-THRCTD-1); Top Talent Support Program for young and middle-aged (Double Hundred Medical Youth Professionals Program) from Wuxi Health Committee (BJ2023002); General Program of Wuxi Medical Center (WMCG202324, WMCG202306).
Author information
Authors and Affiliations
Contributions
Y.Y., L.Z., and T.-T. W. conceived and designed the experiment. A. Z. and H. Y. performed the experiments. A.Z., H.Y., C.-.Y.T., and C.H. analyzed the data. A.Z. wrote the manuscript. Y.Y., T.-T.W. and L.Z. provided the overall guidance and helped to revise the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Yao Tong and the other anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Toshiro Moroishi and Joao de Sousa Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, A., Wei, TT., Yin, H. et al. Endoplasmic reticulum-mitochondria coupling prompts ZBP1-mediated RPE cell PANoptosis in age-related macular degeneration. Commun Biol 8, 1118 (2025). https://doi.org/10.1038/s42003-025-08565-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08565-z