Abstract
Marine viral communities harbor an astounding diversity of phages infecting diverse marine bacteria. The double-stranded DNA phage family Autographiviridae is among the most abundant phage families in the ocean. However, the current understanding of marine Autographiviridae phages is predominantly derived from isolates infecting cyanobacteria, SAR11, and Roseobacter. To achieve a more comprehensive understanding of the diversity, ecological traits, and functional profiles of this phage family, we recovered 1253 complete marine Autographiviridae uncultivated viral genomes (UViGs). Comparative genomic analysis showed that marine-derived Autographiviridae phages display genome synteny and share a conserved core of seven genes. The core gene-based phylogenomic analysis classified them into 14 groups, 6 of which were previously undescribed. These groups varied in G + C content, genome size, and presence of specific genes. Metagenomic recruitment analysis demonstrated that Autographiviridae phages are globally distributed and enriched in the upper ocean layers of tropical and temperate zones. The differential distribution patterns among these groups mirror the ecological niches of their potential hosts, emphasizing the top-down control these phages exert on their host populations. Collectively, our study substantially expands knowledge regarding the diversity, potential hosts, functional capacity, and ecological distribution of Autographiviridae phages in the ocean, emphasizing their ecological implications in marine environments.
Similar content being viewed by others
Introduction
As the smallest and most abundant biological entities in the ocean, viruses play essential roles in shaping the structure, function, and evolution of the microbial communities, thereby influencing the biogeochemical cycling of carbon and nutrients1,2,3,4. The enormous diversity of marine viruses has been investigated using culture-dependent and culture-independent approaches. Over the last decade, an unexpected diversity of marine viruses has been discovered through extensive culture-independent viromic studies5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. Billions of uncultivated viral populations have been reconstructed, promoting the understanding of the diversity and population composition of marine viruses as well as their potential to regulate the metabolism of their hosts5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21. Moreover, marine viromic datasets are of great value in providing the basis for elucidating the geographical distribution characteristics of phages22,23,24,25,26,27,28,29. However, the majority of marine viral populations have not been well characterized because they belong to novel viral species without cultured representatives, and accurately determining the host of most viral populations is challenging. To characterize the huge amount of viral dark matter, the combined use of culture-dependent and metagenomic mining approaches, which elucidate the voluminous information contained in metagenomic datasets, has been applied to identify and characterize some important marine phage groups26,28,30,31,32,33,34. These highly abundant marine phage groups have been isolated in the laboratory, and culture-independent approaches have further enhanced the understanding of their diversity and ecological distribution.
Within class Caudoviricetes, Autographiviridae is the largest family of double-stranded DNA (dsDNA) viruses, containing at least 9 defined subfamilies and 132 defined genera. This family was initially termed “T7-like phages” after the first isolate Escherichia coli phage T7 was discovered35. T7-like phages are characterized by the possession of an RNA polymerase (RNAP) gene and were initially classified under the subfamily Autographivirinae within the family Podoviridae36. In 2019, Autographivirinae was updated to the family Autographiviridae based on comparative genomic, proteomic, and marker gene phylogenetic analyses37. Elevation in the taxonomy of T7-like phages to the level of a family reflects their great diversity and increasing importance in bacteriophagology. Currently, members of Autographiviridae infect a myriad of bacterial hosts from diverse environments (marine, freshwater, and terrestrial ecosystems). The genomes of more than 70 marine Autographiviridae isolates, most of which were isolated from Cyanobacteria (Synechococcus and Prochlorococcus), SAR11 (Pelagibacterales order), and Roseobacter strains, have been reported22,25,38,39,40,41,42,43. A novel Autographiviridae cyanophage group without DNA polymerase (DNAP) genes was discovered recently, thereby expanding the known diversity of marine Autographiviridae phages44. Currently, studies on the diversity and evolutionary relationships of marine Autographiviridae phages are restricted to the above-mentioned isolates22,41,42,43, and limited understanding has been gained regarding their geographic distributions22,25,27,44. In addition, several other marine Autographiviridae phages infecting Marinomonas, Citrobacter, Alteromonas, Stappia, and Vibrio are also available in NCBI, highlighting the broad host range and important ecological significance of Autographiviridae phages in the ocean. Metagenomic-based analysis has also provided evidence for the dominance and prevalence of Autographiviridae phages in the ocean. For example, in the GOV (Global Ocean Virome) study, 358 metagenomic viral populations were classified into a viral cluster (VC_9) with Autographiviridae isolates7. VC_9 is among the most abundant and ubiquitous marine viral clusters7.
Despite previous studies, a comprehensive understanding of the diversity, evolution, and distribution of the Autographiviridae family in marine environments has been hindered by the scarcity of representative genomes. To address these gaps, we recovered 1253 Autographiviridae uncultivated viral genomes (UViGs) using metagenomic data mining and performed a suite of phylogenomic and comparative genomic analyses. Phylogenetic analyses uncovered substantial diversity and identified several previously unrecognized groups within the Autographiviridae family. Genome comparison revealed both conservation and divergence across distinct Autographiviridae groups. Furthermore, metagenomic read-mapping provided a detailed view of their global distribution, highlighting widespread and clade-specific biogeographic patterns.
Materials and methods
Autographiviridae UViGs retrieval
To retrieve marine-derived Autographiviridae genomes, approximately 7 million UViGs were downloaded from IMG/VR v421, Global Ocean Viromes (GOV and GOV 2.0)7,9, MedDCM fosmid library5, Station ALOHA assembly-free virus genomes11, ALOHA 2.0 virome14, San Pedro Ocean Time-series Viromes13, Red Sea Viromes15, and viromes from the oxic surface (10 m) and oxygen-starved basin (200 m) waters of Saanich Inlet6.
The open reading frames (ORFs) of the UViGs were downloaded from the databases or predicted using Prodigal v2.6.3 (-p meta)45. Three Autographiviridae core genes, including genes encoding RNAP, phage capsid, and terminase large subunit (TerL) were used as baits to retrieve Autographiviridae UViGs. The RNAP, capsid, and TerL genes of the Autographiviridae phages (from RefSeq V215) were aligned using MAFFT v7.50546, and their hidden Markov model (HMM) profiles were constructed using hmmbuild in HMMER v3.3.247. Afterwards, the hmmsearch in HMMER47 was used to identify the three genes from the UViGs (e-value ≤10−3 and score ≥50). A total of 2,839 UViGs containing all three marker genes were retrieved as Autographiviridae UViGs. CheckV v0.9.0 was used to estimate the completeness and quality of the UViGs48. The UViGs with genome end redundancy were self-aligned to identify the redundant regions. Based on the alignment, the redundant regions were manually trimmed to avoid potential biases in subsequent analyses. A total of 1253 UViGs with 100% completeness were utilized for subsequent analyses.
To identify Autographiviridae populations (roughly correspond to species-level taxonomy), the 1253 Autographiviridae UViGs and 98 known marine Autographiviridae isolates were clustered using CD-HIT program (nucleotide identity of ≥95%, ≥80% alignment of the short genome) (-c 0.95 -aS 0.8)49, resulting in a total of 1143 Autographiviridae populations.
Genome annotation and comparative genomic analysis
Prodigal45 was used to predict the ORFs of the Autographiviridae genomes. The translated ORFs were annotated using BLASTP against the NCBI non-redundant and NCBI Refseq v215 databases (BLASTP option: E-value ≤10−3, amino acid identity ≥25%, and alignment length ≥50%). The ORFs were searched against the Pfam database using HMMsearch to identify conserved Pfam domains (-E 1e-3 -T 50)47,50. OrthoFinder v2.5.4 was used to identify the orthogroups based on sequence similarities (BLASTP option: E-value ≤10−3, amino acid identity ≥25%, alignment length ≥50%)51. Representative Autographiviridae genomes were compared and visualized using Easyfig v2.2.252.
Autographiviridae prophage and host prediction
Marine bacterial genome databases, including manually curated metagenome-assembled genomes (MAGs) from Tara Oceans, MarDB (MAR databases)53, GORG (Global Ocean Reference Genomes)54, GEM (Genomes from Earth’s Microbiomes)55, the Genome Taxonomy Database (GTDB)56, and approximately 25,000 newly reconstructed draft genomes57 were downloaded to detect Autographiviridae prophage. The integration sites of Autographiviridae prophages in the phage genomes are commonly located between genes encoding integrase and RNAP43,58. When phage integration occurs, these two genes are adjacent to the genes of their hosts. To identify the putative integration sites, the nucleotide sequences of integrase and RNAP genes were extracted from marine Autographiviridae genomes. These sequences were used as queries for BLASTN against the marine bacterial genome sequences and MAGs (e-value ≤10−5, nucleotide identity ≥95%, and coverage ≥50%). The recovered sequences were subjected to BLASTN against the NCBI-nt database (e-value ≤10−5, nucleotide identity ≥80%, and query match length ≥500 bp). The sequences that also contain bacterial genes were considered bacterial genomes containing Autographiviridae prophages.
The potential hosts of Autographiviridae UViGs were predicted using the RaFAH tool59 with default settings. The training and validating random forest model for RaFAH was built with 4269 host-known phages downloaded from the NCBI RefSeq (v215).
Phylogenetic analyses
A genome-wide proteomic tree was constructed using ViPTree60 based on whole-genome sequence similarities calculated by tBLASTx. Seven core genes from marine Autographiviridae UViGs and isolates were used for the phylogenomic analysis. The amino acid sequences of the seven core genes were aligned using MAFFT v7.505 (--maxiterate 1 000 --localpair) and trimmed using trimAl v1.4.rev1561. Thereafter, alignment was used to construct the phylogenomic tree using IQ-TREE v2.2.0.362 under the LG + F + I + I + R10 substitution model with 1000 bootstrap replicates. The phylogenetic trees were visualized and annotated using the Interactive Tree Of Life (iTOL) v663.
Viromic read recruitment analysis and statistical analysis
The relative abundance of the Autographiviridae phages and UViGs was estimated using the viromic read-mapping analysis. A total of 220 marine viromic datasets, including Global Ocean Viromes9, Pearl River Estuary Virome19, Mariana Trench Virome18, Eastern Tropical North Pacific Virome20, Delaware Bay and Chesapeake Bay Viromes17, Black Sea Virome16, Red Sea Virome15, and South China Sea DNA Virome10, were used for viromic read-mapping analysis.
Viromic reads were mapped to the non-redundant set of analyzed Autographiviridae genomes using CoverM (-p bwa-mem --min-read-percent-identity 95 --min-read-aligned-length 50 --min-read-aligned-percent 80, https://github.com/wwood/CoverM). The relative abundance was normalized by mapped reads per kilobase pair of genomes per million reads (RPKM). A heatmap of the RPKM values was generated using the pheatmap package in R. Linear-regression analysis was performed using R to test the relationship between environmental parameters and the relative abundance of these phages. Statistical significance was set at p < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results and discussion
Identification and phylogenomic analyses of marine-derived Autographiviridae UViGs
Metagenomic mining analysis identified 1253 Autographiviridae UViGs with 100% genome completeness. These UViGs originated from diverse marine environments, including coastal waters, open oceans, deep oceans, estuaries, and marine sediments (Supplementary Data 1). The genome size of these UViGs ranged from 35.6 to 86.8 kb, and their G + C content ranged from 22.3% to 70.8% (Supplementary Data 1). Genomic comparison revealed that Autographiviridae UViGs are largely syntenic with Autographiviridae phages, and the taxonomic classification based on the ViPTree showed these UViGs are located among known Autographiviridae phages (Supplementary Fig. S1). In combination with known marine Autographiviridae phages, a total of 1143 species-level Autographiviridae populations were identified by using the ≥95% nucleotide identity threshold.
Marine Autographiviridae are phylogenetically highly diverse
OrthoFinder protein clustering analysis identified 4560 orthologous protein groups (≥ 2 members) in marine Autographiviridae genomes; however, only 435 proteins have been assigned putative biological functions. The accumulation curves of the pan protein groups reached saturation, indicating that our study captured the majority of the genetic diversity within marine Autographiviridae phages (Supplementary Fig. S2A). Core genome analysis revealed that Autographiviridae phages share a common set of seven core genes. The accumulation curve of core genes leveled off (Supplementary Fig. S2B). These core genes include those involved in phage replication and development, such as genes encoding RNAP, portal protein, scaffolding protein, capsid protein, tail tubular protein A and B, and TerL. Highly conserved core genes and flexible pan genes have also been observed in other phage groups, such as HMO-2011-type and HTVC010P-type phages26,28. In the Autographiviridae family, the core genes are involved in DNA metabolism and replication, morphogenesis, and DNA packaging, suggesting a conserved framework underpinning infection and propagation mechanisms. By contrast, distinct subgroups tend to encode specific accessory genes, highlighting pronounced pan-genomic divergence across subgroups. The conservation of core genes likely arises from their indispensable roles in the phage life cycle, while the high diversity observed in the accessory genome is potentially driven by long-term phage-host interactions and environmental adaptation, facilitated through mechanisms such as horizontal gene transfer and mutation.
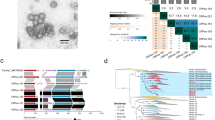
To resolve the genetic diversity and evolutionary relationships among marine Autographiviridae phages, a phylogenomic analysis based on the concatenated alignment of seven core genes was performed. Phylogenomic analysis revealed that marine Autographiviridae phages are remarkably diverse (Fig. 1A). Based on the tree topology and genomic features, Autographiviridae phages formed at least 14 well-supported groups, referred to as Autographiviridae groups AG-1 to AG-14 (Fig. 1A). The remaining 59 UViGs did not form well-separated groups and thus remained unclassified. Most marine Autographiviridae groups were distinct from previously defined non-marine Autographiviridae subfamilies, except for AG-9, which clustered with Studiervirinae, and AG-14, which grouped with multiple non-marine subfamilies (Supplementary Fig. S3). We further compared the genomic features of these groups, and found significant variations in their G + C content and genome sizes (Fig. 1B and Supplementary Data 1).
A A maximum-likelihood phylogenetic tree was constructed using concatenated sequences. Phylogenetic inference was performed using the maximum-likelihood method implemented in IQ-TREE. Marine Autographiviridae phages were clustered into 14 groups based on the phylogeny and genome content. Shading indicates the distinct groups. Reference isolates are shown with colored dashed lines. Gray circles on the nodes indicate bootstrap values of >80%. B Boxplots showing the genome size and G + C content of each Autographiviridae group along the x-axis.
The number of members in these groups varies from 8 to 350 (Supplementary Data 1). Eight (AG-1, 2, 4, 6, 8, 9, 13, and 14) of the 14 groups comprised cultivated representatives, and the remaining 6 groups comprised only UViGs. AG-1 is the largest Autographiviridae group with 350 members. Their G + C content ranges from 32.4% to 63.2% (Fig. 1B). All previously isolated Autographiviridae cyanophages (Synechococcus and Prochlorococcus phages) were classified into this group and most AG-1 UViGs were predicted to infect Synechococcus or Prochlorococcus (discussed below). Most AG-1 members originated from the upper ocean ( < 200 m) (Supplementary Data 1), where marine cyanobacteria are highly abundant. AG-1 can be further divided into four closely related subgroups (AG-1.1 to 1.4). AG-1.1 corresponds to the previously defined cyanophage clades MPP-A and MPP-B41, whereas AG-1.3 and 1.4 contained the newly discovered MPP-C cyanophages without the DNAP gene (Fig. 1A)44. These results suggest that Autographiviridae cyanophages are evolutionarily closely related and form a monophyletic group. AG-2 consists of 24 UViGs and 5 cultivated representatives that infect SAR11 bacteria. The G + C content of AG-2 members ranges from 31.3% to 35.0%, similar to the G + C content of SAR11 and previously reported pelagiphages22,26,27,28,43,64,65. AG-4 included nine UViGs and six cultivated representatives that infect Roseobacter strains. AG-6 contained 27 UViGs and 3 cultivated representatives that infect Roseobacter strains. AG-8 consisted of 69 UViGs and 25 cultivated Autographiviridae pelagiphages. The G + C content of AG-8 members ranged from 31.9% to 47.5%; more than 65% of the AG-8 members had a G + C content below 36%, similar to that of pelagiphages22,25,26,27,28,30,43,66. Most AG-2 and AG-8 members were predicted to infect SAR11 (discussed below). AG-9 included 63 UViGs and Citrobacter phage phiCFP-1. AG-13 contained 20 UViGs and roseophage CRP-143. AG-14 contained 54 UViGs and 14 cultivated representatives that infect Alteromonadaceae, Vibrio, Marinomonas, and Stappia.
The six Autographiviridae groups—AG-3, AG-5, AG-7, AG-10, AG-11, and AG-12—comprise 8, 240, 102, 54, 23, and 32 UViGs, respectively, all of which lack cultured representatives. AG-3 is the smallest group with only 8 UViGs. AG-5 is the sister group of AG-6 and is the second-largest group. AG-7 is the third-largest group, containing 102 UViGs. AG-10 contained 54 UViGs, and was further separated into two subgroups (AG-10.1 and AG-10.2), which varied in genome size and G + C content (Fig. 1B).
Genome variation among Autographiviridae groups
The examination of the distribution of the orthologous groups among the Autographiviridae subgroups revealed clear core-genome conservation and pan-genome differences among various groups. In the DNA metabolism and replication modules, approximately half of the Autographiviridae phages contain a set of T7-like DNA replication related genes located downstream of the RNAP gene, including genes encoding single-stranded DNA-binding proteins (SSB), endonuclease, DNA primase/helicase, DNAP, and exonuclease. The remaining Autographiviridae phages lack at least one of these genes (Fig. 2). Phage-encoded DNAP, DNA primase, and SSB are essential components of the DNA replication machinery. These phages may rely on host genes to compensate for the absence of these genes (Fig. 2). These results suggest that Autographiviridae phages have differences in DNA replication machinery. In the structure and packaging module, all Autographiviridae phages possess a set of conserved genes, including genes encoding head-tail connector, scaffolding protein, capsid protein, tail tubular protein A and B, and TerL, suggesting that they all have a conserved T7-like neck-tail module (Fig. 2). Other structural genes were more varied between the groups.
Each row represents an Autographiviridae group (with parenthetical numbers indicating total group members), while each column corresponds to a specific gene. Color intensity reflects the proportion of gene carriers within each group, with numerical values showing detection counts for each gene in each group. PAPS Reductase: 3’-phosphoadenosine-5’-phosphosulfate reductase.
Genome content and comparative genomic analyses revealed that several groups possess specific genomic features, such as the presence of group-specific genes, absence of DNAP genes, and large genome size, that differentiate them from other groups. These unique features are discussed below.
Autographiviridae subgroups lack DNAP
DNAP was considered the core gene of Autographiviridae until the DNAP-lacking Autographiviridae cyanophages were discovered44. All members of the three AG-1 subgroups (AG-1.2, AG-1.3, and AG-1.4) lack DNAP (Figs. 2 and 3A), with AG-1.2 being a newly identified subgroup without cultured representatives. Except for members of AG-1, DNAP is also absent in AG-11 (Figs. 2 and 3A). As Autographiviridae phages evolved, the phage-encoded DNAP gene may have been lost on several occasions. It is possible that these DNAP-lacking Autographiviridae phages exclusively employ the DNAP of their hosts for DNA replication. AG-11 members also lack homologs of Autographiviridae SSB, exonuclease, and endonuclease (Figs. 2 and 3A). In addition, their DNA primase and helicase are located upstream of the RNAP and share limited identity (< 30% amino acid identity) with those in other Autographiviridae genomes. These results suggest that the DNA replication modules in AG-11 members are evolutionarily distant.
A Comparison of genetic maps of representative Autographiviridae phages that lack the DNA polymerase (DNAP) gene from AG-1.2, AG-1.3, AG-1.4, and AG-11. Predicted open reading frames are represented by arrows and colored based on their putative functions. The scale color bar indicates amino acid identities between homologous genes. B Comparison of genetic maps of representative Autographiviridae phages that possess two RNA polymerase (RNAP) genes from AG-14 and AG-10.1. All RNAP genes are indicated in red and the additional RNAP genes are indicated with red asterisks. C Unrooted maximum-likelihood phylogenetic tree of RNAP in Autographiviridae phages. The typical Autographiviridae RNAPs (located upstream of DNA replication genes) are colored according to groups, and the additional RNAP genes in AG-10.1 and AG-14 are indicated with arrows.
Autographiviridae phages encoding two RNAP genes
Apart from the typical Autographiviridae RNAP gene (approximately 800 amino acids) located upstream of the DNA replication and metabolism module, 23 members in AG-10.1 and 17 members in AG-14 were found to encode an additional RNAP of approximately 650 amino acids in length (Fig. 3B). The additional RNAP genes in AG-10.1 genomes are all located upstream of the typical Autographiviridae RNAP and are surrounded by small, functionally unknown genes. Meanwhile, the additional RNAP genes in AG-14 genomes are all located between the DNA metabolism and the structure modules. Sequence analysis revealed that the additional RNAPs share limited sequence identity with other Autographiviridae RNAPs and are classified into two distinct groups in the phylogenetic tree (Fig. 3C). These results suggest that they may have originated from horizontal gene transfer from other phages. The presence of two RNAPs implies that these phages may require different RNAPs for the transcription of different phage genes. Notably, unlike the concatenated core gene phylogeny, the typical RNAP genes of AG-14 are divided into two distinct clusters (Fig. 3C), with the smaller cluster showing closer evolutionary relationships with the AG-1 members. This incongruence suggests that the RNAPs in AG-14 exhibit greater evolutionary diversity and highlights their unique evolutionary dynamics.
A unique Autographiviridae group with large genome sizes
Notably, AG-10.1 members have a significantly larger genome than the other subgroups (Fig. 1B). A closer analysis revealed that AG-10.1 members harbor several interesting features. AG-10.1 members possess a set of genes upstream of RNAP genes. This region is more varied than other regions. Interestingly, all members in AG-10.1 possess a gene that encodes a large protein (3274 to 5566 amino acids) in this region (Fig. 3B). Most of these large proteins exhibit little or no homology, with very limited similarity in some regions. Sequence analysis revealed the presence of conserved domains within some of these large protein sequences. For example, eight genes contain an RNase_H domain (PF13482), seven genes contain the N terminal domain of SMC (structural maintenance of chromosomes) (PF02463), and four genes contain the RNA_pol domain (PF00940). The RNase_H domain was found in many proteins with unknown functions, such as the hypothetical protein YqgF of Escherichia coli67. The SMC N-terminal and C-terminal domains constitute SMC proteins, but the SMC C-terminal has not been identified in these genes. Therefore, although some domains have been identified in these sequences, the specific functions of these large protein genes remain unclear. AG-10.1 members originate from various marine environments (Supplementary Data 1). It is possible that as the AG-10.1 members evolved, they gained this large gene via gene transfer to adapt to specific hosts. Another notable feature of the AG-10.1 members is the absence of the SSB gene. In addition, several AG-10.1 members possess an additional RNAP gene (discussed above), located downstream of the large protein gene (Fig. 3B).
Variation in metabolic potential
Through the functional annotation of the protein groups, we identified various host-derived auxiliary metabolic genes (AMGs) and revealed that different groups varied in their metabolic potentials.
Photosynthetic-related genes, including genes encoding photosystem II D1 protein (psbA), high light-inducible protein (hli) and phycobilisome proteolysis adapter (nblA) are exclusively present in Autographiviridae cyanophages from AG-1.1 (Fig. 2). psbA and hli are widely distributed in marine cyanophage genomes, and their expression contributes to the maintenance of the host’s photosynthetic machinery during the latent phase of infection68,69. NblA is a key regulator of phycobilisome degradation in cyanobacteria and is highly expressed during nutrient deprivation70,71. Recent studies have shown that marine Autographiviridae phages commonly encode the nblA gene72. NblA may facilitate the degradation of host phycobilisomes, thereby releasing amino acids that serve as building blocks for virion assembly and accelerating the phage replication cycle72. Genes encoding transaldolase are also exclusively present in AG-1.1. The transaldolase in cyanophages is thought to enhance the pentose phosphate pathway of the host, thereby producing more NADPH and ribose 5-phosphate to support phage replication73,74. In addition to the aforementioned AMGs, within the AG-1 group, AG-1.1 contains more AMGs than AG-1.2, 1.3, and 1.4, including those encoding ribonucleotide reductase (RNR), phosphohydrolase, thioredoxin, thymidylate synthase (ThyX), and transglycosylase. This suggests that AG-1.1 may employ a distinct survival strategy by leveraging these AMGs to maximize host metabolic resources utilization, whereas AG-1.2, AG-1.3, and AG-1.4 likely prioritize replication efficiency by minimizing host metabolic burden during virion replication. These divergent evolutionary strategies drive various Autographiviridae subgroups to exploit divergent ecological niches, thereby enabling holistic optimization of resource utilization efficiency.
AG-2 and AG-8 were both predicted to infect SAR11 (as discussed below). Notably, AG-8 contains more AMGs compared to AG-2. AMGs encoding RNR, phosphohydrolase, S-adenosylmethionine decarboxylase (AdoMetDC), Hsp20 heat shock proteins, and DNA methyltransferase (DNMT) were frequently detected in AG-8 but were absent in AG-2. AdoMetDC is crucial for polyamine biosynthesis as it catalyzes the formation of the aminopropyl group donor75,76. Polyamines are crucial for bacterial physiology and are known to be important substrates for the growth of SAR1177,78. We speculate that this enzyme provides advantages for the physiology and survival of the host during phage infection. Previous studies demonstrate that in SAR11 and other bacteria, Hsp20 expression is significantly upregulated under stress conditions79. Hsp20 may help stabilize host protein homeostasis, thereby prolonging the window for efficient viral replication. In addition, studies suggest that Hsp20 might play an important scaffolding role during capsid maturation80.
Lysogenetic life cycle and host taxonomic diversity of marine Autographiviridae phages
The integrase gene (int) is prevalent in marine Autographiviridae phages, with 625 out of 1143 populations encoding a tyrosine integrase gene (Fig. 2 and Supplementary Data 2). Phage-encoded integrase catalyzes the site-specific recombination between phage and host genome81,82. Autographiviridae phages that encode int have been reported previously in cyanophage, pelagiphage, and roseophage genomes39,40,41,43. Autographiviridae pelagiphages have been reported to integrate into their SAR11 host genomes at various tRNA genes43. In addition, it has been reported that Autographiviridae S-TIP37 cyanophage could perform unstable integration with its host58. The int gene was detected in all Autographiviridae groups except for the AG-10 and AG-13 groups, indicating that members of these groups may adopt a strictly lytic lifestyle (Fig. 2). In AG-1, the int gene was present in only 2 of the 50 members in AG-1.3, whereas 35–97% of the members in other subgroups encode this gene. This suggests that AG-1.3 may employ a different lifestyle compared to other AG-1 subgroups. These results suggest that most marine Autographiviridae groups have the capability to perform a lysogenic lifecycle, potentially playing an important role in the evolution of their hosts.
To confirm the lysogenic life cycles of some Autographiviridae phages and identify their potential hosts, we searched for the hybrid phage-host integration sites from marine MAGs and have identified several sequences containing integration sites (Supplementary Data 3). Among these identified MAGs, two contain an RNAP gene that are 95.5% and 97.8% identical to the RNAP sequences of two AG-7 phages (IMGVR_UViG_3300020411_000001 and AFVG_25M217), respectively. These two MAGs also contain sequences highly identical to a Chloroflexota bacterium (93.6 and 99.1% identity), suggesting that these two AG-7 phages infect Chloroflexota bacteria. Chloroflexota is a widespread bacterial phylum abundant in deep ocean83,84.
The host information for eight Autographiviridae UViGs is provided in IMG/VR 4.0. Five are prophages originating from bacterial genomes, including one AG-1.1 phage identified from the genomes of a Synechococcus strain and four AG-9 members identified from the genomes of three Gammaproteobacteria strains (Spartinivicinus, Vibrio, and Marinobacter). Using the CRISPR spacer match method, it was inferred that one member in AG-5 might infect phylum Bacillota. Additionally, using the k-mer match method, it was speculated that one member each in AG-7 and the unclassified group potentially infect Rhizobiales within Alphaproteobacteria and Desulfobacteraceae within Desulfobacteria, respectively.
The potential hosts of Autographiviridae UViGs were then predicted using the RaFAH tool based on protein content. This analysis revealed the broad taxonomic diversity of the hosts (Supplementary Data 4). Most AG-1 members were predicted to infect cyanobacteria, whereas most AG-2 and AG-8 members were predicted to infect SAR11. We found some members in AG-9 were also predicted to infect SAR11, and these members have significantly lower genomic G + C content (30.4–32.2%) than other AG-9 members. Most AG-4 and AG-6 members were predicted to infect Roseobacter. Several AG-7, AG-10, AG-12, and AG-13 members were also predicted to infect Roseobacter; however, the prediction scores were low. The hosts of most AG-5 members were not determined, whereas some were predicted to infect Burkholderiaceae with low prediction scores. Some AG-7 members were predicted to infect Chloroflexota. Some members in AG-9 were predicted to infect Enterobacteriaceae with high prediction scores. Some AG-14 members are predicted to infect Vibrionaceae, Alteromonadaceae, and Oceanospirillaceae with high prediction scores.
Biogeography of Autographiviridae phages in global oceans
In this study, 220 marine viromic datasets were used to elucidate and compare the distribution and relative abundance of different Autographiviridae populations (Supplementary Data 5).
We first examined the distribution of the Autographiviridae populations in global ocean. Of the 1143 Autographiviridae populations, 1083 were detected in marine viromes. These 1083 populations were detected in 93.6% (206 of 220) of the analyzed viromes, covering various marine environments (Fig. 4A, Supplementary Fig. S4 and Data 5). Each of these 206 viromes contained at least 20 Autographiviridae populations. Collectively, Autographiviridae phages were detected from tropical to polar stations, and from coastal to open ocean stations (Fig. 4A, B). They were also detected in distinct water layers, from the surface (SRF) (0–10 m) to the bathypelagic (BATHY) zones (1000–4000 m) (Fig. 4B and Supplementary Fig. S4). These results suggest that Autographiviridae phages are widespread in oceans worldwide. More Autographiviridae populations were detected at the trade (0°–30° latitude) and westerlies (30°–60° latitude) stations than at the polar stations (60°–90° latitude) (p < 0.01, Mann–Whitney U tests) (Fig. 4A and Supplementary Fig. S4). The number of detected Autographiviridae populations was highest at open ocean stations (p < 0.05, Mann–Whitney U tests) (Fig. 4B). Vertically, the overall population richness was highest at the deep chlorophyll maximum (DCM) stations followed by the MES and SRF stations (p < 0.01, Mann–Whitney U tests) (Fig. 4B). This pattern could be linked to the richness of their hosts, as some studies have indicated that bacterial richness peaked at the DCM and its adjacent depths85,86. The DCM and SRF zones contain populations from various Autographiviridae groups, with AG-1 that infect cyanobacteria contributing to the majority of the population richness (Supplementary Fig. S5).
A Map of the number of Autographiviridae populations detected in each virome. The size of each dot represents the number of Autographiviridae populations detected in that virome. Right: area plot showing the relationship between the population size (x-axis) and latitude (y-axis). Boxplot showing the numbers of Autographiviridae populations (y-axis) in different climate zones. B Box plots of the number of Autographiviridae populations (y-axis) detected in viromes from different ecological zones. The significance of pairwise comparisons calculated using t-test was shown with the asterisk corresponding to the p-value (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). C Box plots showing the number of stations (y-axis) where each Autographiviridae group was detected. The significance of pairwise comparisons was calculated using t-test and is indicated by the different letters above the boxes (p < 0.05). SRF surface, DCM deep chlorophyll maximum; MES Mesopelagic, BATHY bathypelagic.
Among distinct groups, AG-2 and AG-8, whose members are primarily predicted to infect SAR11, were the most prevalent globally (Figs. 4C, 5A and Supplementary Fig. S6). This wide distribution of AG-2 and AG-8 phages can be readily explained by the ubiquity of their putative host, the SAR11 clade, which is the most abundant bacterioplankton group in the ocean. This pattern also aligns with that of previously analyzed pelagiphages26,27,28. AG-1 members, particularly AG-1.1 and AG-1.4, were widely distributed globally except in the polar regions, and they were mostly distributed in SRF and DCM waters (Fig. 5A, Supplementary Fig. S5 and Fig. S6). This biogeographic pattern closely mirrors that of their putative hosts, the cyanobacteria Synechococcus and Prochlorococcus, which dominate the upper, warmer oceans87,88. Prochlorococcus is predominantly distributed between 40°S and 40°N latitude, whereas Synechococcus is distributed from the equator to subpolar regions, with a marked decline in abundance at high-latitude regions87,88. Some AG-1 populations were also detected in the mesopelagic (MES) (200–1000 m) and BATHY zones (Supplementary Figs. S5 and S7), possibly due to the sinking of cyanophages. This is consistent with findings that cyanophages can be exported from the photic to abyssal ocean via associating with sinking particles89. AG-10.1 members with large genome sizes were also ubiquitous globally (Figs. 4C, 5 and Supplementary Fig. S6). In addition, several other groups were also detected worldwide, such as AG-7 and AG-5 (Figs. 4C, 5A and Supplementary Fig. S6). Some AG-7 members were predicted to infect Chloroflexota and Roseobacter, which are also ubiquitously distributed in the ocean. Although the hosts of most AG-5 members remain unclear, their cosmopolitan distribution suggests that these phages may infect dominant and ubiquitous marine bacterial groups. The other groups displayed relatively narrow distributions (Figs. 4C, 5A and Supplementary Fig. S6). For example, AG-13 members were mostly detected at coastal, estuarine and polar stations; however, they were rarely detected in the open ocean, which may be related to the low abundance of their putative Roseobacter hosts in the open ocean90 (Fig. 5A and Supplementary Fig. S6). AG-3 members infecting unknown hosts were mostly detected at trade and westerlies stations, and were rarely detected in polar regions (Fig. 5A, Supplementary Fig. S6 and Fig. S7).
A Heatmap showing the relative abundance of each Autographiviridae phage in different marine viromic datasets. The relative abundance was normalized as reads per kilobase of genome per million mapped reads (RPKM). Environmental metadata associated with each station are shown above the heatmap using color bars. Box plots showing the relative abundance of Autographiviridae phages across different oceanic zones (B), biome types (C), and depth layers (D). The significance of pairwise comparisons calculated using the two-tailed Mann–Whitney U test is indicated with asterisks corresponding to the p-value (***p < 0.001, ****p < 0.0001).
Next, we examined the relative abundance of Autographiviridae phages in the global viromes (Fig. 5). Overall, the RPKM values of Autographiviridae phages were highest at the trade stations followed by westerlies stations (p < 0.01, Mann–Whitney U tests) (Fig. 5B). Autographiviridae phages had a higher RPKM at the coastal stations followed by open ocean stations (p < 0.01, Mann–Whitney U tests) (Fig. 5C). Vertically, the RPKM values of Autographiviridae phages in the DCM and SRF zones were significantly higher than those in the MES and BATHY zones, indicating that Autographiviridae phage were predominant in the upper ocean (Fig. 5D). Among the distinct groups, AG-1.1, AG-5, and AG-8 members showed the highest RPKM followed by AG-1.4, AG-7 and AG-1.3 (Fig. 6A). AG-1.1 and AG-1.4 members exhibited the highest RPKM at most trade stations (Fig. 6B, C and Supplementary Fig. S7). Linear regression analysis revealed that the relative abundance of AG-1.1 and AG-1.4 members was positively correlated with temperature (AG-1.1: p < 0.001, R2 = 0.66; AG-1.4: p < 0.001, R2 = 0.56) and negatively correlated with absolute latitude (AG-1.1: p < 0.001, R2 = 0.32; AG-1.4: p < 0.001, R2 = 0.42) (Supplementary Fig. S8). Similar correlations were observed in AG-1.2 and 1.3 (Supplementary Fig. S8). This is consistent with the distribution of their predicted hosts, Synechococcus and Prochlorococcus.
A Box plot showing the relative abundance of Autographiviridae groups in marine viromes. The relative abundance of each group at each virome was calculated by summing the RPKM values of all phages in each group. Autographiviridae groups were sorted according to their median RPKM value in marine viromes. The significance of pairwise comparisons was calculated using the two-tailed Mann–Whitney U test and is indicated by the different letters above the boxes (p < 0.05). B Map of the most abundant Autographiviridae groups detected in each virome. C Barchart showing the number of stations where each group displays the highest abundance. The number of stations with the highest abundance in different environments for each group is shown.
Further, we found that the top 10 Autographiviridae cyanophages with the highest RPKM were exclusively UViGs, suggesting that the most abundant Autographiviridae cyanophages have not yet been isolated (Supplementary Data 5). In the polar regions, AG-8, AG-7 and AG-5 were among the most abundant groups, exhibiting the highest relative abundance at most polar stations (38/42) (Fig. 6B, C). Host prediction analysis indicated that the potential hosts of AG-8 members were SAR11, and some AG-7 members might infect Roseobacter. Cold-adapted ecotypes of SAR11 and Roseobacter are metabolically active and constitute a significant portion of the microbial community in polar waters91,92,93,94,95, explaining the dominance of AG-8 and AG-7 clusters in polar environments. The hosts of AG-5 members remain unidentified, but it was speculated that they are highly abundant in polar environments due to their phages being prevalent in these regions. In AG-14, eight phages showed remarkably high relative abundance in Delaware Bay stations (Fig. 5A and Supplementary Data 5). They were predicted to infect members of the family Moraxellaceae (Predicted host score > 0.99). Comparative genomic analysis revealed that these eight phages exhibited high homology to Autographiviridae phage vB_AbaP_Indie (46.35–47.42% average amino acid identity), which infects Acinetobacter within Moraxellaceae. Phage vB_AbaP_Indie was originally isolated from influent wastewater at a treatment plant 108. This suggests that these eight phages may also originate from terrestrial environments.
Conclusions
Autographiviridae phages are important and dominant components of the marine virosphere and are notable for their wide distribution, high abundance, and diverse potential hosts. Previous studies addressing the genomic diversity of marine Autographiviridae phages were mostly limited to cyanophages, pelagiphages, and roseophages. In this study, we performed metagenomics-based analyses to assess the diversity and biogeography of marine Autographiviridae phages. Our analysis revealed an unprecedented diversity of marine phages in this family, as it comprises at least 14 subgroups that possess substantial genomic variation. The significant difference in the number of AMGs carried by phages may reflect their divergent survival strategies. The Autographiviridae UViGs discovered in this study substantially increase the known phylogenetic diversity of marine Autographiviridae phages and highlight how their infectivity and metabolic capabilities have influenced marine ecosystems. Furthermore, read-mapping analysis revealed that marine Autographiviridae groups were enriched in the upper ocean and that several groups infecting cosmopolitan marine bacteria were more prevalent. These distribution patterns mirror host ecology, emphasizing top-down control on microbial communities and biogeochemical cycles. Taken together, our findings reveal that marine Autographiviridae can infect various bacterial hosts and have a wide geographical distribution. Their ecological impacts warrant further investigation.
Limitations of the study
In this study, we report a comprehensive study of the genetic diversity and biogeography of marine Autographiviridae phages. Our analyses revealed that marine Autographiviridae phages harbor great genetic diversity and show considerable variations in genetic and ecological features. However, this study has several limitations. First, this analysis relies on Autographiviridae genomes retrieved from currently available marine metagenomic datasets. The investigation in this study may not include the Autographiviridae diversity from unexplored marine ecosystems. Second, reliable phage-host relationships were only predicted for limited UViGs. Hosts of marine Autographiviridae phages remain largely unknown. Finally, due to the lack of cultured representatives from different Autographiviridae groups, the understanding of biological characterization and ecological application of marine Autographiviridae phages is still limited. In conclusion, while our study provides extensive insights into the diversity and ecology of marine Autographiviridae phages and highlights their ecological importance, it also underscores the need for further study to address the above-mentioned limitations.
Data availability
The source data underlying main figures and Supplementary Figs. are provided as Supplementary Data files. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Details of publicly available software used in this study are provided in the Methods section. No custom code or mathematical algorithms that are central to the conclusions were generated during this study.
References
Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999).
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005).
Suttle, C. A. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007).
Breitbart, M. Marine viruses: truth or dare. Annu. Rev. Mar. Sci. 4, 425–448 (2012).
Mizuno, C. M., Rodriguez-Valera, F., Kimes, N. E. & Ghai, R. Expanding the marine virosphere using metagenomics. PLoS Genet. 9, e1003987 (2013).
Chow, C. E. et al. Combining genomic sequencing methods to explore viral diversity and reveal potential virus-host interactions. Front. Microbiol. 6, 265 (2015).
Roux, S. et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689–693 (2016).
Weynberg, K. D. et al. Coral-associated viral communities show high levels of diversity and host auxiliary functions. PeerJ 5, e4054 (2017).
Gregory, A. C. et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123.e14 (2019).
Liang, Y. et al. Metagenomic analysis of the diversity of DNA viruses in the surface and deep sea of the South China Sea. Front. Microbiol. 10, 1951 (2019).
Beaulaurier, J. et al. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 30, 437–446 (2020).
Wu, S. et al. Diverse and unique viruses discovered in the surface water of the East China Sea. BMC Genom. 21, 441 (2020).
Ignacio-Espinoza, J. C., Ahlgren, N. A. & Fuhrman, J. A. Long-term stability and Red Queen-like strain dynamics in marine viruses. Nat. Microbiol. 5, 265–271 (2020).
Luo, E., Eppley, J. M., Romano, A. E., Mende, D. R. & DeLong, E. F. Double-stranded DNA virioplankton dynamics and reproductive strategies in the oligotrophic open ocean water column. ISME J. 14, 1304–1315 (2020).
Hevroni, G., Flores-Uribe, J., Beja, O. & Philosof, A. Seasonal and diel patterns of abundance and activity of viruses in the Red Sea. Proc. Natl. Acad. Sci. USA 117, 29738–29747 (2020).
Jaiani, E. et al. Microbial diversity and phage-host interactions in the Georgian coastal area of the Black Sea revealed by whole genome metagenomic sequencing. Mar. Drugs 18, 558 (2020).
Sun, M. et al. Uncultivated viral populations dominate estuarine viromes on the spatiotemporal scale. mSystems 6, e01020-20 (2021).
Gao, C. et al. Virioplankton assemblages from challenger deep, the deepest place in the oceans. iScience 25, 104680 (2022).
Xu, B. et al. A holistic genome dataset of bacteria, archaea and viruses of the Pearl River estuary. Sci. Data 9, 49 (2022).
Jurgensen, S. K. et al. Viral community analysis in a marine oxygen minimum zone indicates increased potential for viral manipulation of microbial physiological state. ISME J. 16, 972–982 (2022).
Camargo, A. P. et al. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res. 51, D733–D743 (2023).
Zhao, Y. et al. Abundant SAR11 viruses in the ocean. Nature 494, 357–360 (2013).
Kang, I., Oh, H. M., Kang, D. & Cho, J. C. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc. Natl. Acad. Sci. USA 110, 12343–12348 (2013).
Zhang, Z. et al. Diverse, abundant, and novel viruses infecting the marine Roseobacter RCA lineage. mSystems 4, e00494–19 (2019).
Buchholz, H. H. et al. Efficient dilution-to-extinction isolation of novel virus-host model systems for fastidious heterotrophic bacteria. ISME J. 15, 1585–1598 (2021).
Du, S. et al. Genomic diversity, life strategies and ecology of marine HTVC010P-type pelagiphages. Microb. Genom.7, 000596 (2021).
Zhang, Z. et al. Culturing novel and abundant pelagiphages in the ocean. Environ. Microbiol. 23, 1145–1161 (2021).
Qin, F. et al. Newly identified HMO-2011-type phages reveal genomic diversity and biogeographic distributions of this marine viral group. ISME J. 16, 1363–1375 (2022).
Buchholz, H. H. et al. Novel pelagiphage isolate Polarivirus skadi is a polar specialist that dominates SAR11-associated bacteriophage communities at high latitudes. ISME J. 17, 1660–1670 (2023).
Yang, M. et al. Genomic characterization and distribution pattern of a novel marine OM43 phage. Front. Microbiol. 12, 651326 (2021).
Zaragoza-Solas, A., Rodriguez-Valera, F. & Lopez-Perez, M. Metagenome mining reveals hidden genomic diversity of pelagimyophages in aquatic environments. mSystems 5, e00905–19 (2020).
Zheng, K. et al. Identification of hidden N4-like viruses and their interactions with hosts. mSystems 8, e0019723 (2023).
Zucker, F. et al. New Microviridae isolated from Sulfitobacter reveals two cosmopolitan subfamilies of single-stranded DNA phages infecting marine and terrestrial Alphaproteobacteria. Virus Evol. 8, veac070 (2022).
Rihtman, B. et al. A new family of globally distributed lytic roseophages with unusual deoxythymidine to deoxyuridine substitution. Curr. Biol. 31, 3199–3206.E3194 (2021).
Molineux, I. J. in The Bacteriophages Vol. 2 (ed Calendar, R.) 277–301 (Oxford University Press, 2006).
Lavigne, R., Seto, D., Mahadevan, P., Ackermann, H. W. & Kropinski, A. M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159, 406–414 (2008).
Turner, D. et al. Create one new family (Autographiviridae) including nine subfamilies and one hundred and thirty-two genera in the order Caudovirales. (International Committee on Taxonomy of Viruses, 2019).
Chen, F. & Lu, J. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68, 2589–2594 (2002).
Sullivan, M. B., Coleman, M. L., Weigele, P., Rohwer, F. & Chisholm, S. W. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, e144 (2005).
Pope, W. H. et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine Synechococcus. J. Mol. Biol. 368, 966–981 (2007).
Labrie, S. J. et al. Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ. Microbiol. 15, 1356–1376 (2013).
Huang, S., Zhang, S., Jiao, N. & Chen, F. Comparative genomic and phylogenomic analyses reveal a conserved core genome shared by estuarine and oceanic cyanopodoviruses. PLoS One. 10, e0142962 (2015).
Zhao, Y. et al. Pelagiphages in the Podoviridae family integrate into host genomes. Environ. Microbiol. 21, 1989–2001 (2019).
Cai, L. et al. Abundant and cosmopolitan lineage of cyanopodoviruses lacking a DNA polymerase gene. ISME J. 17, 252–262 (2023).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 11, 119 (2010).
Katoh, K., Asimenos, G. & Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537, 39–64 (2009).
Finn, R. D., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011).
Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2021).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinform 28, 3150–3152 (2012).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157 (2015).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: a genome comparison visualizer. Bioinform 27, 1009–1010 (2011).
Klemetsen, T. et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 46, D692–D699 (2018).
Pachiadaki, M. G. et al. Charting the complexity of the marine microbiome through single-cell genomics. Cell 179, 1623–1635 e1611 (2019).
Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Paoli, L. et al. Biosynthetic potential of the global ocean microbiome. Nature 607, 111–118 (2022).
Shitrit, D. et al. Genetic engineering of marine cyanophages reveals integration but not lysogeny in T7-like cyanophages. ISME J. 16, 488–499 (2022).
Coutinho, F. H. et al. RaFAH: Host prediction for viruses of Bacteria and Archaea based on protein content. Patterns 2, 100274 (2021).
Nishimura, Y. et al. ViPTree: the viral proteomic tree server. Bioinform 33, 2379–2380 (2017).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinform 25, 1972–1973 (2009).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Grote, J. et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3, e00252-12 (2012).
Thrash, J. C. et al. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J. 8, 1440–1451 (2014).
Yang, M. et al. Genomic diversity and biogeographic distributions of a novel lineage of bacteriophages that infect marine OM43 bacteria. Microbiol. Spectr. 11, e0494222 (2023).
Liu, D., Wang, Y. S. & Wyss, D. F. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J. Biomol. NMR. 27, 389–392 (2003).
Clokie, M. R. et al. Transcription of a ‘photosynthetic’ T4-type phage during infection of a marine cyanobacterium. Environ. Microbiol. 8, 827–835 (2006).
Lindell, D. et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449, 83–86 (2007).
Baier, K., Nicklisch, S., Grundner, C., Reinecke, J. & Lockau, W. J. F. M. L. Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC6803. FEMS Microbiol. Ecol. 195, 35–39 (2001).
Baier, K., Lehmann, H., Stephan, D. P. & Lockau, W. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 150, 2739–2749 (2004).
Nadel, O. et al. Oceanic photosynthesis is directly affected by cyanophage NblA proteins. bioRxiv, 2024.2011.2010.622831 (2024).
Thompson, L. R. et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 108, E757–E764 (2011).
Sprenger, G. A. Genetics of pentose-phosphate pathway enzymes of Escherichia coliK-12. Arch. Microbiol. 164, 324–330 (1995).
Pegg, A. E., Xiong, H., Feith, D. J. & Shantz, L. M. S-Adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochem. Soc. Trans. 26, 580–586 (1998).
Pegg, A. E. S-Adenosylmethionine decarboxylase. Essays Biochem. 46, 25–45 (2009).
Giovannoni, S. J. SAR11 bacteria: the most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9, 231–255 (2017).
Michael, A. J. Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 291, 14896–14903 (2016).
Smith, D. P. et al. Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique” describe the first PII-independent response to nitrogen limitation in a free-living alphaproteobacterium. mBio 4, e00133-12 (2013).
Sullivan, M. B. et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12, 3035–3056 (2010).
Groth, A. C. & Calos, M. P. Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678 (2004).
Fogg, P. C., Colloms, S., Rosser, S., Stark, M. & Smith, M. C. New applications for phage integrases. J. Mol. Biol. 426, 2703–2716 (2014).
Morris, R. M., Rappe, M. S., Urbach, E., Connon, S. A. & Giovannoni, S. J. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 70, 2836–2842 (2004).
Varela, M. M., van Aken, H. M. & Herndl, G. J. Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ. Microbiol. 10, 1903–1911 (2008).
Mende, D. R., Boeuf, D. & DeLong, E. F. Persistent core populations shape the microbiome throughout the water column in the North Pacific subtropical gyre. Front. Microbiol. 10, 2273 (2019).
Mende, D. R. et al. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat. Microbiol. 2, 1367–1373 (2017).
Flombaum, P. et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 110, 9824–9829 (2013).
Visintini, N., Martiny, A. & Flombaum, P. Prochlorococcus, Synechococcus, and picoeukaryotic phytoplankton abundances in the global ocean. Limnol. Oceanogr. Lett. 6, 207–215 (2021).
Luo, E., Leu, A. O., Eppley, J. M., Karl, D. M. & DeLong, E. F. Diversity and origins of bacterial and archaeal viruses on sinking particles reaching the abyssal ocean. ISME J. 16, 1627–1635 (2022).
Buchan, A., Gonzalez, J. M. & Moran, M. A. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677 (2005).
Kirchman, D. L., Cottrell, M. T. & Lovejoy, C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 12, 1132–1143 (2010).
Thiele, S., Storesund, J. E., Fernandez-Mendez, M., Assmy, P. & Ovreas, L. A Winter-to-summer transition of bacterial and archaeal communities in Arctic Sea Ice. Microorganisms 10, 1618 (2022).
Voget, S. et al. Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J. 9, 371–384 (2015).
Liu, Y. et al. Metagenome-assembled genomes reveal greatly expanded taxonomic and functional diversification of the abundant marine Roseobacter RCA cluster. Microbiome 11, 265 (2023).
Selje, N., Simon, M. & Brinkhoff, T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427, 445–448 (2004).
Acknowledgements
This research was funded by the National Natural Science Foundation of China (42276144, 42206096, and 42076105).
Author information
Authors and Affiliations
Contributions
YL.Z., Z.Z., and S.D. designed the research; S.D. retrieved the metagenomic viral genomes; S.D., Z.W., M.Y., YH.Z., and C.D. performed genome annotation and comparative genomic analysis and phylogenomic analysis; Z.W. and Y.W. performed host prediction analysis; Z.Z. and S.D. performed metagenomic read recruitment and statistical analysis. YL.Z., Z.Z., F.C., and S.D. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Mohammad Moniruzzaman and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Michele Repetto.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Du, S., Wu, Z., Wu, Y. et al. Marine Autographiviridae phages exhibit high genetic diversity and global-scale biogeographic patterns. Commun Biol 8, 1240 (2025). https://doi.org/10.1038/s42003-025-08611-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08611-w