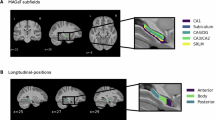
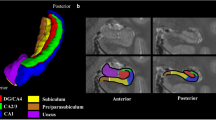
Abstract
The hippocampus plays a crucial role in cognition, yet its microstructural development during childhood and adolescence remains poorly understood. Here, we investigate age-related differences in hippocampal microstructure using diffusion MRI with ultra-strong gradients (300 mT/m) in a cohort of 88 participants aged 8–19 years. Surface-based hippocampal modelling was combined with established microstructural approaches, and a more advanced biophysical model (Soma and Neurite Density Imaging: SANDI) suited for studying cortical microstructure. Hippocampal volume, gyrification, and thickness remained stable across this developmental window, however we observed significant differences across age related to MR-derived neurite and soma parameters. Diffusion-derived changes across age were found to be correlated with adult microstructure maps related to myelin and iron content, synaptic density, and hippocampal interneurons derived from MRI, PET and histology. These findings highlight age-related differences of MR-derived neurite and soma parameters in the human hippocampus during late childhood and adolescence, offering insights into structural maturation during this critical period.
Similar content being viewed by others
Data availability
The histology, ex vivo MRI, and PET maps can be found in both the HippoMaps (osf.io/92p34/) and NeuroMaps (https://github.com/netneurolab/neuromaps) toolbox. Due to the inclusion of minors (under 18 participants), the availability of derived or identifiable data from the participant cohort is restricted due to privacy concerns. Derived data supporting the findings of the imaging analyses are available by contacting the authors in writing via email, allowing 4 weeks for access requests to be granted. Numerical Source data underlying all graphs in this article can be found in the Supplementary data file.
Code availability
The SANDI model was fit to the diffusion data using an open-source MATLAB toolbox (https://github.com/palombom/SANDI-Matlab-Toolbox-v1.0). Code to repeat the study analyses can be found at: https://doi.org/10.17605/OSF.IO/4AQXR.
References
Ding, S. & Van Hoesen, G. W. Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of cyto- and chemoarchitecture. J. Comp. Neurol. 523, 2233–2253 (2015).
Duvernoy, H. M., Cattin, F. & Risold, P. Y. The Human Hippocampus: Functional Anatomy, Vascularization, and Serial Sections with MRI (Springer, 2013).
Giaccio, R. G. The dual origin hypothesis: an evolutionary brain-behavior framework for analyzing psychiatric disorders. Neurosci. Biobehav. Rev. 30, 526–550 (2006).
Sanides, F. Functional architecture of motor and sensory cortices in primates in the light of a new concept of neocortex evolution. Adv. Primatol. 1, 137–208 (1970).
Sanides, F. The cyto-myeloarchitecture of the human frontal lobe and its relation to phylogenetic differentiation of the cerebral cortex. J. Fur Hirnforsch. 6, 269–282 (1964).
Buzsáki, G. & Moser, E. I. Memory, navigation and theta rhythm in the hippocampal-entorhinal system. Nat. Neurosci. 16, 130–138 (2013).
Squire, L. R. Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol. Rev. 99, 195–231 (1992).
Strange, B. A. et al. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 15, 655–669 (2014).
Sweatt, J. D. Hippocampal function in cognition. Psychopharmacol. 174, 99–110 (2004).
Callow, D. D., Canada, K. L. & Riggins, T. Microstructural integrity of the hippocampus during childhood: relations with age and source memory. Front. Psychol. 11, 620 (2020).
Giedd, J. N. et al. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: ages 4-18 years. J. Comp. Neurol. 366, 223–230 (1996).
Gogtay, N. et al. Dynamic mapping of normal human hippocampal development. Hippocampus 16, 664–672 (2006).
Krogsrud, S. K. et al. Development of hippocampal subfield volumes from 4 to 22 years. Hum. Brain Mapp. 35, 5646–5657 (2014).
Langnes, E. et al. Anterior and posterior hippocampus macro- and microstructure across the lifespan in relation to memory—a longitudinal study. Hippocampus 30, 678–692 (2020).
Lee, J. K., Ekstrom, A. D. & Ghetti, S. Volume of hippocampal subfields and episodic memory in childhood and adolescence. NeuroImage 94, 162–171 (2014).
Pfluger, T. et al. Normative volumetric data of the developing hippocampus in children based on magnetic resonance imaging. Epilepsia 40, 414–423 (1999).
Solar, K. G., Treit, S. & Beaulieu, C. High resolution diffusion tensor imaging of the hippocampus across the healthy lifespan. Hippocampus 31, 1271–1284 (2021).
Tamnes, C. K. et al. Longitudinal development of hippocampal subregions from childhood to adulthood. Dev. Cogn. Neurosci. 30, 212–222 (2018).
Uematsu, A. et al. Developmental trajectories of amygdala and hippocampus from infancy to early adulthood in healthy individuals. PLoS ONE 7, e46970 (2012).
Vinci-Booher, S. et al. Development of human hippocampal subfield microstructure and relation to associative inference. Cereb. Cortex 33, 10207–10220 (2023).
Wierenga, L. et al. Typical development of basal ganglia, hippocampus, amygdala and cerebellum from age 7 to 24. NeuroImage 96, 67–72 (2014).
Huttenlocher, P. R. & Dabholkar, A. S. Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol. 387, 167–178 (1997).
Petanjek, Z. et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. USA 108, 13281–13286 (2011).
Goldman-Rakic, P. S. Development of cortical circuitry and cognitive function. Child Dev. 58, 601 (1987).
Le Bihan, D. Molecular diffusion, tissue microdynamics and microstructure. NMR Biomed. 8, 375–386 (1995).
Lebel, C. et al. Microstructural maturation of the human brain from childhood to adulthood. NeuroImage 40, 1044–1055 (2008).
Basser, P. J., Mattiello, J. & LeBihan, D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 66, 259–267 (1994).
Jelescu, I. O. & Budde, M. D. Design and validation of diffusion MRI models of white matter. Front. Phys. 5, 50 (2017).
Jensen, J. H. & Helpern, J. A. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed. 23, 698–710 (2010).
Karat, B. G. et al. Diffusion MRI of the hippocampus. J. Neurosci. 44, 11501–11515 (2024).
Jones, D. K. et al. Microstructural imaging of the human brain with a ‘super-scanner’: 10 key advantages of ultra-strong gradients for diffusion MRI. NeuroImage 182, 8–38 (2018).
McNab, J. A. et al. The human connectome project and beyond: initial applications of 300MT/m gradients. NeuroImage 80, 234–245 (2013).
Setsompop, K. et al. Pushing the limits of in vivo diffusion MRI for the human connectome project. NeuroImage 80, 220–233 (2013).
Palombo, M. et al. SANDI: a compartment-based model for non-invasive apparent soma and neurite imaging by diffusion MRI. NeuroImage 215, 116835 (2020).
Genc, S. et al. Impact of b-value on estimates of apparent fibre density. Hum. Brain Mapp. 41, 2583–2595 (2020).
Genc, S. et al. MRI signatures of cortical microstructure in human development align with oligodendrocyte cell-type expression. Nat. Commun. 16, 3317 (2025).
DeKraker, J. et al. Automated hippocampal unfolding for morphometry and subfield segmentation with HippUnfold. eLife 11, e80021 (2022).
Zhang, H. et al. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage 61, 1000–1016 (2012).
DeKraker, J. et al. HippoMaps: multiscale cartography of human hippocampal organization. Nat. Methods 22, 2211–2222 (2024).
Markello, R. D. et al. Neuromaps: structural and functional interpretation of brain maps. NeuroImage 245, 118665 (2022).
Alkemade, A. et al. A unified 3D map of microscopic architecture and MRI of the human brain. Sci. Adv. 8, eabm2589 (2022).
Finnema, S. J. et al. Kinetic evaluation and test–retest reproducibility of [11C]UCB-J, a novel radioligand for positron emission tomography imaging of synaptic vesicle glycoprotein 2a in humans. J. Cereb. Blood Flow. Metab. 38, 2041–2052 (2018).
Amunts, K. et al. BigBrain: an ultrahigh-resolution 3D human brain model. Science 340, 1472–1475 (2013).
Naganawa, M. et al. First-in-human evaluation of 18F-SynVesT-1, a radioligand for PET imaging of synaptic vesicle glycoprotein 2A. J. Nucl. Med. 62, 561–567 (2020).
Hansen, J. Y. et al. Local molecular and global connectomic contributions to cross-disorder cortical abnormalities. Nat. Commun. 13, 4682 (2022).
Rossi, R. et al. Synaptic vesicle glycoprotein 2a: features and functions. Front. Neurosci. 16, 864514 (2022).
Lee, J. K., Johnson, E. G. & Ghetti, S. Hippocampal development: structure, function and implications. in the Hippocampus from Cells to Systems (eds Hannula, D. & Duff, M.) 61–87 (Springer, 2017).
Zeineh, M. M. et al. Direct visualization and mapping of the spatial course of fiber tracts at microscopic resolution in the human hippocampus. Cereb. Cortex 27, 1779–1794 (2017).
Coupé, P. et al. Towards a unified analysis of brain maturation and aging across the entire lifespan: a MRI analysis. Hum. Brain Mapp. 38, 5501–5518 (2017).
Narvacan, K. et al. Evolution of deep gray matter volume across the human lifespan. Hum. Brain Mapp. 38, 3771–3790 (2017).
Hu, S. et al. Volumetric analysis of medial temporal lobe structures in brain development from childhood to adolescence. NeuroImage 74, 276–287 (2013).
Robillard, K. N. et al. Glial cell morphological and density changes through the lifespan of rhesus macaques. Brain Behav. Immun. 55, 60–69 (2016).
Jabès, A. et al. Postnatal development of the hippocampal formation: a stereological study in macaque monkeys. J. Comp. Neurol. 519, 1051–1070 (2010).
Lenroot, R. K. et al. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. NeuroImage 36, 1065–1073 (2007).
Sisk, C. L. & Foster, D. L. The neural basis of puberty and adolescence. Nat. Neurosci. 7, 1040–1047 (2004).
Peper, J. S. et al. Sex steroids and brain structure in pubertal boys and girls: a mini-review of neuroimaging studies. Neuroscience 191, 28–37 (2011).
Herting, M. M. et al. The impact of sex, puberty, and hormones on white matter microstructure in adolescents. Cereb. Cortex 22, 1979–1992 (2012).
Chase, H. W. et al. Evidence for an anterior-posterior differentiation in the human hippocampal formation revealed by meta-analytic parcellation of fMRI coordinate maps: focus on the subiculum. NeuroImage 113, 44–60 (2015).
Nichols, E. S. et al. Hippocampus long-axis specialization throughout development: a meta-analysis. Hum. Brain Mapp. 44, 4211–4224 (2023).
Poppenk, J. et al. Long-axis specialization of the human hippocampus. Trends Cogn. Sci. 17, 230–240 (2013).
Wolf, D. et al. Differential associations of age with volume and microstructure of hippocampal subfields in healthy older adults. Hum. Brain Mapp. 36, 3819–3831 (2015).
Ábrahám, H. et al. Myelination in the human hippocampal formation from midgestation to adulthood. Int. J. Dev. Neurosci. 28, 401–410 (2010).
Mellström, B. et al. Specific cytoarchitectural changes in hippocampal subareas in daDREAM mice. Mol. Brain 9, 22 (2016).
Canada, K. L. et al. A (sub)field guide to quality control in hippocampal subfield segmentation on high-resolution T2-weighted MRI. Hum. Brain Mapp. 45, e70004 (2024).
Yushkevich, P. A. et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. NeuroImage 111, 526–541 (2015).
Olsen, R. K. et al. Progress update from the hippocampal subfields group. Alzheimer’s. Dement. 11, 439–449 (2019).
Wisse, L. E. M. et al. A harmonized segmentation protocol for hippocampal and parahippocampal subregions: why do we need one and what are the key goals? Hippocampus 27, 3–11 (2016).
Ábrahám, H. et al. Ontogeny of calbindin immunoreactivity in the human hippocampal formation with a special emphasis on granule cells of the dentate gyrus. Int. J. Dev. Neurosci. 27, 115–127 (2008).
Ábrahám, H. et al. Development of parvalbumin-immunoreactive neurons in the postnatal human hippocampal formation. Front. Neuroanat. 17, 1058370 (2023).
Urbán, Z., Maglóczky, Z. & Freund, T. F. Calretinin-containing interneurons innervate both principal cells and interneurons in the CA1 region of the human hippocampus. Acta Biol. Hung. 53, 205–220 (2002).
Tóth, Z., Györimolnár, I., Ábrahám, H. & Hevesi, Á Cannulation and cardiopulmonary bypass produce selective brain lesions in pigs. Asian Cardiovasc. Thorac. Ann. 14, 273–278 (2006).
Petersen, A. C. et al. A self-report measure of pubertal status: reliability, validity, and initial norms. J. Youth Adolesc. 17, 117–133 (1988).
Caruyer, E. et al. Design of multishell sampling schemes with uniform coverage in diffusion MRI. Magn. Reson. Med. 69, 1534–1540 (2013).
Veraart, J., Fieremans, E. & Novikov, D. S. Diffusion MRI noise mapping using random matrix theory. Magn. Reson. Med. 76, 1582–1593 (2016).
Sairanen, V., Leemans, A. & Tax, C. M. W. Fast and accurate slicewise outlier detection (SOLID) with informed model estimation for diffusion MRI data. NeuroImage 181, 331–346 (2018).
Vos, S. B. et al. The importance of correcting for signal drift in diffusion MRI. Magn. Reson. Med. 77, 285–299 (2016).
Andersson, J. L. R. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage 125, 1063–1078 (2016).
Kellner, E. et al. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn. Reson. Med. 76, 1574–1581 (2016).
Tustison, N. J. et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010).
Rudrapatna, S. et al. Can we correct for interactions between subject motion and gradient-nonlinearity in diffusion MRI? Proc. Int. Soc. Mag. Reson. Med. 1206 (2018).
Karat, B. G. Developing Hippocampus (version 1.0) [Computer software]. OSF. https://doi.org/10.17605/OSF.IO/4AQXR (2025).
Isensee, F. et al. NNU-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Smith, S. M. et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23, S208–S219 (2004).
Mattiello, J., Basser, P. J. & Le Bihan, D. The B matrix in diffusion tensor echo-planar imaging. Magn. Reson. Med. 37, 292–300 (1997).
Jones, D. K. et al. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. NeuroImage 73, 239–254 (2013).
Harms, R. L. et al. Robust and fast nonlinear optimization of diffusion MRI microstructure models. NeuroImage 155, 82–96 (2017).
Ianuş, A. et al. Soma and neurite density MRI (SANDI) of the in-vivo mouse brain and comparison with the Allen brain Atlas. NeuroImage 254, 119135 (2022).
Karat, B. G. et al. Mapping the macrostructure and microstructure of the in vivo human hippocampus using diffusion MRI. Hum. Brain Mapp. 44, 5485–5503 (2023).
Nieuwenhuys, R., van Huijzen, C. & Voogd, J. The Human Central Nervous System (Springer, 2008).
Jeurissen, B. et al. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage 103, 411–426 (2014).
Tournier, J.-D. et al. MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. NeuroImage 202, 116137 (2019).
Alkemade, A. et al. A unified 3D map of microscopic architecture and MRI of the human brain [Data set]. Uvaauas https://doi.org/10.21942/uva.16834114 (2022).
Hansen, J. Y. et al. Local molecular and global connectomic contributions to cross-disorder cortical abnormalities [Data set]. Github https://github.com/netneurolab/hansen_crossdisorder_vulnerability (2022).
Lutti, A. et al. Using high-resolution quantitative mapping of R1 as an index of cortical myelination. NeuroImage 93, 176–188 (2014).
Stüber, C. et al. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. NeuroImage 93, 95–106 (2014).
Uchihara, T. Silver diagnosis in neuropathology: principles, practice and revised interpretation. Acta Neuropathol. 113, 483–499 (2007).
Amunts, K. et al. BigBrain: an ultrahigh-resolution 3D human brain model [Data set]. BigBrain Project https://bigbrainproject.org (2013).
Acknowledgements
The data were acquired at the UK National Facility for In Vivo MR Imaging of Human Tissue Microstructure, funded by the EPSRC (grant EP/M029778/1) and The Wolfson Foundation. B.G.K. is supported by a post-graduate scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). S.G. is supported by the Royal Children’s Hospital Foundation (RCHF 2022-1402). E.P.R. is supported by NICHD at NIH (F32HD103313). M.P. is funded by UKRI Future Leaders Fellowship (MR/T020296/2). A.R.K. is supported by the Canada Research Chairs program #950-231964, NSERC Discovery Grants RGPIN-2015-06639 and RGPIN-2023-05558, Canadian Institutes for Health Research Project grant #366062, Canada Foundation for Innovation (CFI) John R. Evans Leaders Fund project #37427, the Canada First Research Excellence Fund, and Brain Canada. D.K.J. is supported by a Wellcome Trust Investigator Award (096646/Z/11/Z) and a Wellcome Trust Strategic Award (104943/Z/14/Z). We thank the children/adolescents (and their carers) for generously donating their time to participate in this study.
Author information
Authors and Affiliations
Contributions
S.G. and E.P.R. acquired the data with input from M.P. and D.K.J.; B.G.K. devised the study methods and analysis; B.G.K. performed the analysis and wrote the code; S.G., E.P.R., M.P., A.R.K., and D.K.J. refined the analysis; B.G.K. wrote the manuscript; S.G., E.P.R., M.P., A.R.K., and D.K.J. edited the manuscript; All authors commented on and discussed the results.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editors: Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Karat, B.G., Genc, S., Raven, E.P. et al. Microstructural variation of hippocampal substructures across childhood and adolescence quantified with high-gradient diffusion MRI. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09622-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09622-x