Abstract
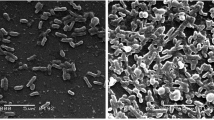
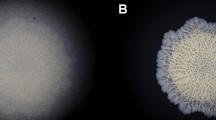
Biofilms are microbial communities of aggregated cells encased in extracellular matrix that are a pressing healthcare concern. Since biofilms have complex metabolic dynamics, in this study a new approach for studying biofilm metabolism is developed that employs optical-photothermal infrared (O-PTIR) spectroscopy imaging combined with 13C stable isotope probing and cryosectioning to track the carbon metabolism of cells at different depths of the biofilm. This approach demonstrated that metabolic gradients can be visualised using O-PTIR imaging, revealing a core of cells with low metabolic activity at the centre of the biofilm, with outer regions showing significantly higher metabolic activity. By incorporating the heavy stable isotope of carbon into bacterial biomass, we monitored the metabolic activity of gentamicin-resistant Salmonella Typhimurium within the biofilm structure upon exposure to various antibiotics. O-PTIR imaging revealed altered metabolic responses at various depths of the biofilm, with variations that depend on the bacterial antibiotic susceptibility profile.
Similar content being viewed by others
Data availability
Data presented in this study can be obtained in the supplementary data, with the underlying data for graphs in Figs. 1–2 found in supplementary data 1, the underlying data for graphs in Figs. 3–4 found in supplementary data 2, and the underlying data for graphs in Figs. 5–6 found in supplementary data 3. All other data are available from the corresponding author on reasonable request.
References
Hamilton, S. et al. The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics 10, 1–21 (2009).
Harrell, J. E. et al. Salmonella biofilm formation, chronic infection, and immunity within the intestine and hepatobiliary tract. Front. Cell. Infect. Microbiol. 10, 624622 https://doi.org/10.3389/FCIMB.2020.624622 (2021).
Jain, S. & Chen, J. Attachment and biofilm formation by various serotypes of salmonella as influenced by cellulose production and thin aggregative fimbriae biosynthesis. J. Food Prot. 70, 2473–2479 (2007).
Milho, C. et al. Escherichia coli and Salmonella Enteritidis dual-species biofilms: interspecies interactions and antibiofilm efficacy of phages. Sci. Rep. 9, 1–15 (2019).
Wang, H., Ding, S., Wang, G., Xu, X. & Zhou, G. In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. Int. J. Food Microbiol. 167, 293–302 https://doi.org/10.1016/j.ijfoodmicro.2013.10.005 (2013).
Wei, K. et al. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8, 894–907 (2013).
Werner, E. et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196 (2004).
Rani Suriani, A. et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 189, 4223–4233 (2007).
Dayton, H. et al. Cellular arrangement impacts metabolic activity and antibiotic tolerance in Pseudomonas aeruginosa biofilms. PLoS Biol. 22, e3002205 (2024).
Walters, M. C., Roe, F., Bugnicourt, A., Franklin, M. J. & Stewart, P. S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrobial Agents Chemother. 47, 317–323 (2003). https://doi.org/10.1128/AAC.47.1.317-323.2003
Chua, S. L. et al. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat. Commun. 7, 10750 (2016).
Jo, J., Cortez, K. L., Cornell, W. C., Price-Whelan, A. & Dietrich, L. E. An orphan cbb 3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. Elife 6, e30205 (2017).
Flemming, H.-C. & Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019).
Sanchez, C. J. et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 13, 47 (2013).
Ding, L. et al. Targeted treatment for biofilm-based infections using PEGylated tobramycin. J. Controlled Rel. 372, 43–58 (2024).
Krüger, G. I. et al. Mobile genetic elements drive the multidrug resistance and spread of Salmonella serotypes along a poultry meat production line. Front. Microbiol. 14, 1072793 https://doi.org/10.3389/FMICB.2023.1072793/FULL (2023).
Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005).
O’Neill, J., Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. Wellcome Trust and HM Government (2016). Accessed at: https://amr-review.org/sites/defaultfiles/160525_Final%20paper_with%20cover.pdf.
Balasubramanian, R. et al. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunotherapeutics 15, 1421–1426 (2019).
Kim, S. H., Jyung, S. & Kang, D. H. Comparative study of Salmonella Typhimurium biofilms and their resistance depending on cellulose secretion and maturation temperatures. LWT 154, 112700–112700 (2022).
Dong, N. et al. The phenotypic and molecular characteristics of antimicrobial resistance of Salmonella enterica subsp. enterica serovar Typhimurium in Henan Province, China. BMC Infect. Dis. 20, 511 https://doi.org/10.1186/s12879-020-05203-3 (2020).
Greeshma, S. S., Pillai, D. & Joseph, T. C. Multidrug resistance in salmonella serotypes across the globe: alarming rate of spread. 1-17 (Springer, 2023).
Wang, X. et al. Antibiotic resistance in salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 10, 985–985 (2019).
Vasicek, E. M. & Gunn, J. S. Invasive non-typhoidal Salmonella Lineage biofilm formation and gallbladder colonization vary but do not correlate directly with known biofilm-related mutations. Infect. Immun. 91, e00135-23 https://doi.org/10.1128/iai.00135-23 (2023).
Albicoro Francisco, J. et al. Lactate promotes the biofilm-to-invasive-planktonic transition in Salmonella enterica serovar Typhimurium via the de novo purine pathway. Infect. Immun. 92, e00266–00224 (2024).
Vlamakis, H., Aguilar, C., Losick, R. & Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–945 (2008).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021).
Aleksandrowicz, A. et al. Better together–Salmonella biofilm-associated antibiotic resistance. Gut Microbes 15, 2229937 (2023).
Namazoğlu, B., Aksoy, M., Memiş-Özgül, B. & Tulga-Öz, F. Anti-microbial efficiency of gaseous ozone’s combined use with fluoride and chlorhexidine on time-related oral biofilm: an: in situ: study on pediatric patients. Med. Gas. Res. 13, 192–197 (2023).
Mountcastle, S. E. et al. Biofilm viability checker: an open-source tool for automated biofilm viability analysis from confocal microscopy images. npj Biofilms Microbiomes 7, 44 (2021).
Gu, Y.-Q., Li, T.-T. & Li, H.-Q. Biofilm formation monitored by confocal laser scanning microscopy during startup of MBBR operated under different intermittent aeration modes. Process Biochem. 74, 132–140 (2018).
Schlafer, S. & Meyer, R. L. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods 138, 50–59 (2017).
Panwar, J. & Merten, C. A. Fluorescence crosstalk reduction by modulated excitation-synchronous acquisition for multispectral analysis in high-throughput droplet microfluidics. Lab a Chip 23, 2514–2520 (2023).
Hollmann, B., Perkins, M., Chauhan, V. M., Aylott, J. W. & Hardie, K. R. Fluorescent nanosensors reveal dynamic pH gradients during biofilm formation. npj Biofilms Microbiomes 7, 50 https://doi.org/10.1038/s41522-021-00221-8 (2021).
Prater, C. B., Kansiz, M. & Cheng, J.-X. A tutorial on optical photothermal infrared (O-PTIR) microscopy. APL Photonics 9, 091101 (2024).
Muhamadali, H. et al. Chicken, beams, and Campylobacter: rapid differentiation of foodborne bacteria via vibrational spectroscopy and MALDI-mass spectrometry. Analyst 141, 111–122 (2016).
Muhamadali, H. et al. Rapid, accurate, and comparative differentiation of clinically and industrially relevant microorganisms via multiple vibrational spectroscopic fingerprinting. Analyst 141, 5127–5136 (2016).
Helm, D. et al. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. Microbiology 137, 69 (1991).
Sayqal, A. et al. Metabolic analysis of the response of Pseudomonas putida DOT-T1E strains to toluene using Fourier transform infrared spectroscopy and gas chromatography mass spectrometry. Metabolomics 12, 112 https://doi.org/10.1007/s11306-016-1054-1 (2016).
Muhamadali, H. et al. Metabolic profiling of geobacter sulfurreducens during industrial bioprocess scale-up. Appl. Environ. Microbiol. 81, 3288–3298 (2015).
Bechtel, H. A., Muller, E. A., Olmon, R. L., Martin, M. C. & Raschke, M. B. Ultrabroadband infrared nanospectroscopic imaging. Proc. Natl. Acad. Sci. USA 111, 7191–7196 (2014).
Klementieva, O. et al. Super-resolution infrared imaging of polymorphic amyloid aggregates directly in neurons. Adv. Sci. 7, 1903004 (2020).
Lima, C., Muhamadali, H. & Goodacre, R. Simultaneous raman and infrared spectroscopy of stable isotope labelled escherichia coli. Sensors 22, 3928 https://doi.org/10.3390/S22103928 (2022).
Shams, S. et al. Optical photothermal infrared spectroscopy: a novel solution for rapid identification of antimicrobial resistance at the single-cell level via deuterium isotope labeling. Front. Microbiol. 14, 1077106 https://doi.org/10.3389/fmicb.2023.1077106 (2023).
Wang, Y., Huang, W. E., Cui, L. & Wagner, M. Single cell stable isotope probing in microbiology using Raman microspectroscopy. Curr. Opin. Biotechnol. 41, 34–42 (2016).
Muhamadali, H., Chisanga, M. & Subaihi, A. & Goodacre, R. Combining raman and FT-IR spectroscopy with quantitative isotopic labeling for differentiation of E. Coli cells at community and single cell levels. Anal. Chem. 87, 4578–4586 (2015).
Chisanga, M., Muhamadali, H., Kimber, R. & Goodacre, R. Quantitative detection of isotopically enriched E. coli cells by SERS. Faraday Discuss. 205, 331–343 (2017).
Shams, S. et al. Application of infrared spectroscopy to study carbon-deuterium kinetics and isotopic spectral shifts at the single-cell level. Spectrochimica Acta Part A Mol. Biomol. Spectrosc. 327, 125374 (2025).
Wang, Y. et al. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal. Chem. 88, 1602 https://doi.org/10.1021/acs.analchem.6b01602 (2016).
Chisanga, M. et al. Metabolism in action: stable isotope probing using vibrational spectroscopy and SIMS reveals kinetic and metabolic flux of key substrates. Analyst 146, 2319 https://doi.org/10.1039/d0an02319a (2021).
Goodacre, R., Lima, C., Muhamadali, H., Xu, Y. & Kansiz, M. Imaging isotopically labeled bacteria at the single-cell level using high-resolution optical infrared photothermal spectroscopy. Anal. Chem. 93, 3967 https://doi.org/10.1021/acs.analchem.0c03967 (2021).
Berry, D. et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. USA 110, 4720–4725 (2013).
Peteranderl, R. & Lechene, C. Measure of carbon and nitrogen stable isotope ratios in cultured cells. J. Am. Soc. Mass Spectrom. 15, 478–485 (2004).
Bae, K., Zheng, W., Ma, Y. & Huang, Z. Real-time monitoring of pharmacokinetics of antibiotics in biofilms with Raman-tagged hyperspectral stimulated Raman scattering microscopy. Theranostics 932043 https://doi.org/10.7150/thno.32043 (2019).
Shrestha, A., Shringi, S. & Shah, D. H. Rapid serotype-independent differential detection of biofilm-positive and biofilm-negative Salmonella using Fourier transform infrared biotyping. One Health 20, 101004 (2025).
Garg, A. et al. Microporous multiresonant plasmonic meshes by hierarchical micro–nanoimprinting for bio-interfaced SERS imaging and nonlinear nano-optics. Small 18, 2106887 (2022).
Crisp, A. R. et al. Investigating the chemical pathway to the formation of a single biofilm using infrared spectroscopy. Biofilm 6, 100141 (2023).
Lanni, E. J. et al. Correlated imaging with C60-SIMS and confocal raman microscopy: visualization of cell-scale molecular distributions in bacterial biofilms. Anal. Chem. 86, 10885–10891 (2014).
van Hoogstraten, S. W. G., Kuik, C., Arts, J. J. C. & Cillero-Pastor, B. Molecular imaging of bacterial biofilms—a systematic review. Crit. Rev. Microbiol. 50, 971–992 (2024).
Molina, C. et al. Comparison of different vibrational spectroscopic probes (ATR-FTIR, O-PTIR, Micro-Raman, and AFM-IR) of lipids and other compounds found in environmental samples: case study of substrate-deposited sea spray aerosols. ACS Measurement Science Au https://doi.org/10.1021/acsmeasuresciau.4c00033 (2024).
Burr, D. J. et al. Stable isotope probing-nanoFTIR for quantitation of cellular metabolism and observation of growth-dependent spectral features. Small 20, e2400289 https://doi.org/10.1002/smll.202400289 (2024).
Schiessl, K. T. et al. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Commun. 10, 8733 https://doi.org/10.1038/s41467-019-08733-w (2019).
Xie, C. et al. Identification of single bacterial cells in aqueous solution using confocal laser tweezers raman spectroscopy. Anal. Chem. 77, 4390–4397 (2005).
Movasaghi, Z., Rehman, S. & Ur Rehman, D. I. Fourier Transform Infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 43, 134–179 (2008).
Maquelin, K. et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51, 255–271 (2002).
Mecozzi, M., Pietroletti, M., Scarpiniti, M., Acquistucci, R. & Conti, M. E. Monitoring of marine mucilage formation in Italian seas investigated by infrared spectroscopy and independent component analysis. Environ. Monit. Assess. 184, 6025–6036 (2011).
Stewart Philip, S. Diffusion in Biofilms. J. Bacteriol. 185, 1485–1491 (2003).
Kosztołowicz, T. & Metzler, R. Diffusion of antibiotics through a biofilm in the presence of diffusion and absorption barriers. Phys. Rev. E 102, 032408 (2020).
Nichols, W. W., Dorrington, S. M., Slack, M. P. & Walmsley, H. L. Inhibition of tobramycin diffusion by binding to alginate. Antimicrobial Agents Chemother. 32, 518–523 (1988).
Patra, S., Saha, S., Singh, R., Tomar, N. & Gulati, P. Biofilm battleground: unveiling the hidden challenges, current approaches and future perspectives in combating biofilm associated bacterial infections. Microb. Pathogenesis 198, 107155 (2025).
Azeredo, J. et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 43, 313–351 (2017).
Serra, D. O. & Hengge, R. Stress responses go three dimensional – the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 16, 1455–1471 (2014).
Klauck, G., Serra, D. O., Possling, A. & Hengge, R. Spatial organization of different sigma factor activities and c-di-GMP signalling within the three-dimensional landscape of a bacterial biofilm. Open Biol. 8, 180066 (2018).
Serra, D. O. & Hengge, R. A c-di-GMP-based switch controls local heterogeneity of extracellular matrix synthesis which is crucial for integrity and morphogenesis of escherichia coli macrocolony biofilms. J. Mol. Biol. 431, 4775–4793 (2019).
Kannan, H. et al. Spatiotemporal development of expanding bacterial colonies driven by emergent mechanical constraints and nutrient gradients. Nat. Commun. 16, 4878 (2025).
Stewart Philip, S. et al. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. J. Bacteriol. 201, 00307–00319 (2019).
Li, P., Seneviratne Chaminda, J., Alpi, E., Vizcaino Juan, A. & Jin, L. Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of candida albicans biofilm persisters. Antimicrobial Agents Chemother. 59, 6101–6112 (2015).
Davies, S. K. et al. Visualizing antimicrobials in bacterial biofilms: three-dimensional biochemical imaging using TOF-SIMS. mSphere 2, e00211–e00217 (2017).
Han, Q. et al. Regrowth of microcosm biofilms on titanium surfaces after various antimicrobial treatments. Front. Microbiol. 10, 2693 (2019).
Chadwick, G. L., Jiménez Otero, F., Gralnick, J. A., Bond, D. R. & Orphan, V. J. NanoSIMS imaging reveals metabolic stratification within current-producing biofilms. Proc. Natl. Acad. Sci. 116, 20716–20724 (2019).
Schlechter, R. O. et al. Chromatic bacteria - a broad host-range plasmid and chromosomal insertion toolbox for fluorescent protein expression in bacteria. Front. Microbiol. 9, 3052 (2018).
McKenzie, G. J. & Craig, N. L. Fast, easy and efficient: site-specific insertion of transgenes into Enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 6, 39 (2006).
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. (EUCAST, 2025).
Lu, M., Liu, B., Xiong, W. & Liu, X. The combination of salmonella phage ST-3 and antibiotics to prevent salmonella typhimurium in vitro. Curr. Microbiol. 79, 371 (2022).
Mon Khin, K. Z., Si, Z., Chan-Park Mary, B. & Kenney Linda, J. Polyimidazolium protects against an invasive clinical isolate of salmonella typhimurium. Antimicrobial Agents Chemother. 66, e00597–00522 (2022).
Eckert, J. A., Rosenberg, M., Rhen, M., Choong, F. X. & Richter-Dahlfors, A. An optotracer-based antibiotic susceptibility test specifically targeting the biofilm lifestyle of Salmonella. Biofilm 4, 100083 (2022).
Patel, J., Singh, M., Macarisin, D., Sharma, M. & Shelton, D. Differences in biofilm formation of produce and poultry Salmonella enterica isolates and their persistence on spinach plants. Food Microbiol. 36, 388–394 (2013).
Prabhukhot, G. S., Eggleton, C. D., Kim, M. & Patel, J. Impact of surface topography and hydrodynamic flow conditions on single and multispecies biofilm formation by Escherichia coli O157:H7 and Listeria monocytogenes in presence of promotor bacteria. LWT 201, 116240 (2024).
Tan, Y. C. et al. EtOH-LN cryoembedding workflow to minimize freezing artifact in frozen tissues: a pilot study in preparing tissues compatible with mass spectrometry-based spatial proteomics application. Cryobiology 114, 104843 (2024).
Martens, H., Nielsen, J. P. & Engelsen, S. B. Light scattering and light absorbance separated by extended multiplicative signal correction. application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 75, 394–404 (2003).
Acknowledgements
We would like to express our gratitude to the University of Liverpool for providing funding (DS, HM and RG). HM would also like to thank the Analytical Chemistry Trust Fund (ACTF) and Community for Analytical Measurement Science (CAMS) for funding and support (600310/22/09). RR is grateful for funding from the BBSRC and Innovate UK via the UK National Biofilm Innovation Centre (grant numbers BB/R012415/1, BB/X002950/1). DS would also like to thank the University of Liverpool Shared Research Histology Core Facility, RRID:SCR_026606 for their help and support with developing the cryosectioning protocol.
Author information
Authors and Affiliations
Contributions
D.S.: Experimental design, sample collection and preparation, O-PTIR data collection, data analysis and interpretation, manuscript writing; X.Z.: preparation of Salmonella Typhimurium from strain collection, engineering resistance into strains and manuscript preparation; J.H.: provision of Salmonella Typhimurium from strain collection, experimental design and manuscript preparation; R.R.: supervision, experimental design and manuscript preparation; R.G. provision of O-PTIR & Bioscreen instruments for analysis and manuscript preparation; H.M.: principal investigator, experimental design, data interpretation and manuscript preparation. All authors contributed to the project and viewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Dr Sridhar Mani and Dr Ophelia Bu.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Smaje, D., Zhu, X., Hinton, J.C.D. et al. Investigating Salmonella biofilm responses to antibiotic treatment using optical photothermal infrared spectroscopy. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09655-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09655-2