Abstract
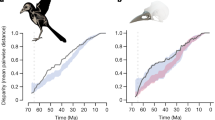
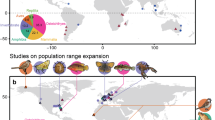
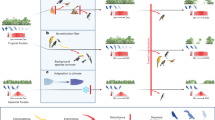
Dispersal is a fundamental ecological and evolutionary process, but identifying its determinants and predicting it across species remains a major challenge. Dispersal syndromes, which describe patterns of covariation among traits related to dispersal, are thought to capture general rules of dispersal evolution and its ecological consequences. Based on the most comprehensive empirical dispersal dataset available for European birds, we test how dispersal syndromes form and how well they predict dispersal across species. We found that distinct dispersal processes were governed by different trait combinations, with body mass consistently predicting overall dispersal, whereas flight efficiency was key for long-distance dispersal events. However, multi-trait dispersal syndromes performed poorly for phylogenetically distant species and were outperformed by models based on single mechanistic traits, especially body mass, life history, and, to a lesser extent, flight efficiency. Thus, single traits with clear mechanistic meaning predict avian dispersal ability better than complex syndromes. These findings highlight the complexity of avian dispersal and emphasize the need for refined mechanistic approaches to understand the constraints shaping dispersal evolution. Together, our study calls for broader empirical efforts and more mechanistic frameworks to uncover the evolutionary and ecological drivers of dispersal.
Similar content being viewed by others
Data availability
All data used in this study are derived from previously published sources. Dispersal kernel estimates were obtained from Fandos et al.8. Species trait data were compiled from Sheard et al.16, Reif et al.34, and Storchová and Hořák35. Phylogenetic data were obtained from Jetz et al.33. No new data were generated in this study.
Code availability
The code used to analyse the data and create the figures in this paper is available on Zenodo81 under the identifier: Fandos, G., Robinson, R., & Zurell, D. (2026). Code and data for: Simple mechanistic traits outperform complex syndromes in predicting avian dispersal distances (all versions). Zenodo https://doi.org/10.5281/zenodo.10713957.
References
Clobert, J., Baguette, M., Benton, T. G. & Bullock, J. M. Dispersal Ecology and Evolution (Oxford University Press, 2012).
Ronce, O. How does it feel to be like a rolling stone? Ten questions about dispersal evolution. Annu. Rev. Ecol. Evol. Syst. 38, 231–253 (2007).
Travis, J. M. J. et al. Dispersal and species’ responses to climate change. Oikos 122, 1532–1540 (2013).
Clobert, J., Le Galliard, J.-F., Cote, J., Meylan, S. & Massot, M. Informed dispersal, heterogeneity in animal dispersal syndromes and the dynamics of spatially structured populations. Ecol. Lett. 12, 197–209 (2009).
Goossens, S., Wybouw, N., Van Leeuwen, T. & Bonte, D. The physiology of movement. Mov. Ecol. 8, 5 (2020).
Ronce, O. & Clobert, J. Dispersal syndromes. Dispersal Ecol. Evol. 155, 119–138 (2012).
Bullock, J. M. et al. A synthesis of empirical plant dispersal kernels. J. Ecol. 105, 6–19 (2017).
Fandos, G. et al. Standardised empirical dispersal kernels emphasise the pervasiveness of long-distance dispersal in European birds. J. Animal Ecol. 92, 158–170 (2023).
Nathan, R. The challenges of studying dispersal. Trends Ecol. Evol. 16, 481–483 (2001).
Barbet-Massin, M., Thuiller, W. & Jiguet, F. The fate of European breeding birds under climate, land-use and dispersal scenarios. Glob. Change Biol. 18, 881–890 (2012).
Urban, M. C. et al. Improving the forecast for biodiversity under climate change. Science 353, aad8466–aad8466 (2016).
Paradis, E., Baillie, S. R., Sutherland, W. J. & Gregory, R. D. Patterns of natal and breeding dispersal in birds. J. Animal Ecol. 67, 518–536 (1998).
Chu, J. J. & Claramunt, S. Determinants of natal dispersal distances in North American birds. Ecol. Evol. 13, e9789 (2023).
Weeks, B. C. et al. Morphological adaptations linked to flight efficiency and aerial lifestyle determine natal dispersal distance in birds. Funct. Ecol. 36, 1681–1689 (2022).
Claramunt, S. Flight efficiency explains differences in natal dispersal distances in birds. Ecology 102, e03442 (2021).
Sheard, C. et al. Ecological drivers of global gradients in avian dispersal inferred from wing morphology. Nat. Commun. 11, 2463 (2020).
Jenkins, D. G. et al. Does size matter for dispersal distance? Glob. Ecol. Biogeogr. 16, 415–425 (2007).
Cote, J. & Clobert, J. Risky dispersal: avoiding kin competition despite uncertainty. Ecology 91, 1485–1493 (2010).
Silverin, B. The stress response and autumn dispersal behaviour in willow tits. Animal Behav. 53, 451–459 (1997).
Peach, W. J., Hanmer, D. B. & Oatley, T. B. Do southern African songbirds live longer than their European counterparts? Oikos 93, 235–249 (2001).
Salewski, V. & Bruderer, B. The evolution of bird migration—a synthesis. Naturwissenschaften 94, 268–279 (2007).
Nur, N. The consequences of brood size for breeding Blue Tits. Iii. Measuring the cost of reproduction: survival, future fecundity, and differential dispersal. Evolution 42, 351–362 (1988).
Sutherland, G. D., Harestad, A. S., Price, K. & Lertzman, K. P. Scaling of natal dispersal distances in terrestrial birds and mammals. Ecol. Soc. 4, 16 (2000).
Stevens, V. M. et al. A comparative analysis of dispersal syndromes in terrestrial and semi-terrestrial animals. Ecol. Lett. 17, 1039–1052 (2014).
Bowman, J., Jaeger, J. A. & Fahrig, L. Dispersal distance of mammals is proportional to home range size. Ecology 83, 2049–2055 (2002).
Horswill, C. et al. Improving assessments of data-limited populations using life-history theory. J. Appl. Ecol. 58, 1225–1236 (2021).
Stevens, V. M. et al. Dispersal syndromes and the use of life-histories to predict dispersal. Evol. Appl. 6, 630–642 (2013).
Whitmee, S. & Orme, C. D. L. Predicting dispersal distance in mammals: a trait-based approach. J. Animal Ecol. 82, 211–221 (2013).
Ottaviani, D., Cairns, S. C., Oliverio, M. & Boitani, L. Body mass as a predictive variable of home-range size among Italian mammals and birds. J. Zool. 269, 317–330 (2006).
Dingle, H. Animal migration: is there a common migratory syndrome? J Ornithol. 147, 212–220 (2006).
Piersma, T., Pérez-Tris, J., Mouritsen, H., Bauchinger, U. & Bairlein, F. Is there a “migratory syndrome” common to all migrant birds? Annals N Y Acad. Sci. 1046, 282–293 (2005).
Winger, B. M., Auteri, G. G., Pegan, T. M. & Weeks, B. C. A long winter for the Red Queen: rethinking the evolution of seasonal migration. Biol. Rev. 94, 737–752 (2019).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Reif, J., Hořák, D., Krištín, A., Kopsová, L. & Devictor, V. Linking habitat specialization with species’ traits in European birds. Oikos 125, 405–413 (2016).
Storchová, L. & Hořák, D. Life-history characteristics of European birds. Glob Ecol. Biogeogr. 27, 400–406 (2018).
Stevens, V. M., Trochet, A., Van Dyck, H., Clobert, J. & Baguette, M. How is dispersal integrated in life histories: a quantitative analysis using butterflies: Dispersal life-history correlates. Ecol. Lett. 15, 74–86 (2012).
Sæther, B.-E. & Bakke, Ø Avian life history variation and contribution of demographic traits to the population growth rate. Ecology 81, 642–653 (2000).
Comte, L. & Olden, J. D. Evidence for dispersal syndromes in freshwater fishes. Proc. R. Soc. B: Biol. Sci. 285, 20172214 (2018).
Hämäläinen, A. M., Guenther, A., Patrick, S. C. & Schuett, W. Environmental effects on the covariation among pace-of-life traits. Ethology 127, 32–44 (2021).
Garrard, G. E., McCarthy, M. A., Vesk, P. A., Radford, J. Q. & Bennett, A. F. A predictive model of avian natal dispersal distance provides prior information for investigating response to landscape change. J. Animal Ecol. 81, 14–23 (2012).
Matthysen, E. Multicausality of dispersal. in Dispersal ecology and evolution/Clobert 3–18 (OUP Oxford, 2012).
Orme, C. D. L. et al. Global patterns of geographic range size in birds. PLOS Biol. 4, e208 (2006).
McNamara, J. M., Barta, Z., Wikelski, M. & Houston, A. I. A theoretical investigation of the effect of latitude on avian life histories. Am. Nat. 172, 331–345 (2008).
Bonte, D. et al. Costs of dispersal. Biol. Rev. 87, 290–312 (2012).
Kennedy, J. D. et al. The influence of wing morphology upon the dispersal, geographical distributions and diversification of the Corvides (Aves; Passeriformes). Proc. R. Soc. B: Biol. Sci. 283, 20161922 (2016).
Pigot, A. L., Jetz, W., Sheard, C. & Tobias, J. A. The macroecological dynamics of species coexistence in birds. Nat. Ecol. Evol. 2, 1112–1119 (2018).
Nathan, R. An emerging movement ecology paradigm. Proc. Natl. Acad. Sci. 105, 19050–19051 (2008).
Soriano-Redondo, A., Gutiérrez, J. S., Hodgson, D. & Bearhop, S. Migrant birds and mammals live faster than residents. Nat. Commun. 11, 5719 (2020).
Teitelbaum, C. S. & Mueller, T. Beyond migration: causes and consequences of nomadic animal movements. Trends Ecol. Evol. 34, 569–581 (2019).
Losos, J. B. Convergence, adaptation, and constraint. Evolution 65, 1827–1840 (2011).
Hadfield, J. D. & Nakagawa, S. General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J. Evol. Biol. 23, 494–508 (2010).
Böhning-Gaese, K. & Oberrath, R. Phylogenetic effects on morphological, life-history, behavioural and ecological traits of birds. Evol. Ecol. Res. 1, 347–364 (1999).
Crouch, N. M. & Tobias, J. A. The causes and ecological context of rapid morphological evolution in birds. Ecol. Lett. 25, 611–623 (2022).
Cote, J. et al. Evolution of dispersal strategies and dispersal syndromes in fragmented landscapes. Ecography 40, 56–73 (2017).
Cote, J. et al. Dispersal syndromes in challenging environments: a cross-species experiment. Ecol. Lett. 25, 2675–2687 (2022).
Nathan, R., Klein, E., Robledo-Arnuncio, J. J. & Revilla, E. Dispersal kernels: review. In Dispersal Ecology and Evolution. (eds J. Clobert, M. Baguette, T. G. Benton, & J. M. Bullock) 25, 187–210 (Oxford University Press, 2012).
Bonte, D. & Dahirel, M. Dispersal: a central and independent trait in life history. Oikos 126, 472–479 (2017).
Fan, H. et al. The influence of wing morphology upon intraspecific divergence in birds: a global study of subspecies richness. Avian Res. 15, 100188 (2024).
Delgado, M.D.M., Penteriani, V., Revilla, E. & Nams, V.O. The effect of phenotypic traits and external cues on natal dispersal movements. J. Animal Ecol. 79, 620–632 (2010).
Raffard, A. et al. Dispersal syndromes can link intraspecific trait variability and meta-ecosystem functioning. Trends Ecol Evol 37, 322–331 (2022).
Zurell, D. Integrating demography, dispersal and interspecific interactions into bird distribution models. J Avian Biol 48, 1505–1516 (2017).
Gardner, E. et al. A family of process-based models to simulate landscape use by multiple taxa. Landsc. Ecol 39, 102 (2024).
Briscoe, N. J. et al. Forecasting species range dynamics with process-explicit models: matching methods to applications. Ecol. Lett. 22, 1940–1956 (2019).
Zurell, D. et al. Spatially explicit models for decision-making in animal conservation and restoration. Ecography 2022, https://doi.org/10.1111/ecog.05787 (2022).
Schaub, M., Looft, V., Plard, F. & Von Rönn, J. A. C. Dynamics of a goshawk population across half a century is driven by the variation of first-year survival. Ecol. Evol. 14, e70058 (2024).
Du Feu, C. R., Clark, J. A., Schaub, M., Fiedler, W. & Baillie, S. R. The EURING Data Bank–a critical tool for continental-scale studies of marked birds. Ringing Migr. 31, 1–18 (2016).
Koschová, M., Rivas-Salvador, J. & Reif, J. Continent-wide test of the efficiency of the European union’s conservation legislation in delivering population benefits for bird species. Ecol. Indic. 85, 563–569 (2018).
Hanzelka, J., Horká, P. & Reif, J. Spatial gradients in country-level population trends of European birds. Divers. Distrib. 25, 1527–1536 (2019).
Schielzeth, H. Simple means to improve the interpretability of regression coefficients. Methods Ecol. Evol. 1, 103–113 (2010).
Bürkner, P.-C. brms: an R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, 1–28 (2017).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2, 217–223 (2012).
Lüdecke, D., Ben-Shachar, M. S., Patil, I., Waggoner, P. & Makowski, D. Performance: an R package for assessment, comparison and testing of statistical models. J. Open Sour. Softw. 6, 3139 (2021).
Hartig, F. & Lohse, L. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level / Mixed) Regression Models. R package version 0.4.7. https://CRAN.R-project.org/package=DHARMa (2022).
Rodríguez-Sánchez, F. DHARMa.helpers: Helper functions to check models not (yet) directly supported by DHARMa (version 0.0.2) [R package]. GitHub. https://github.com/Pakillo/DHARMa.helpers (2024).
Piironen, J., Paasiniemi, M. & Vehtari, A. Projective inference in high-dimensional problems: prediction and feature selection. Electron. J. Stat. 14, 2155–2197 (2020).
Burnham, K. & Anderson, D. Model Selection and Multi-Model Inference (Springer, 2002).
Piironen, J., Paasiniemi, M., Catalina, A., Weber, F. & Vehtari, A. projpred: Projection Predictive Feature Selection (version 2.6.0) [R package]. https://github.com/Pakillo/DHARMa.helpers (2023).
Freckleton, R. P. On the misuse of residuals in ecology: regression of residuals vs. multiple regression. J. Animal Ecol. 542–545 (2002).
Arlot, S. & Celisse, A. A survey of cross-validation procedures for model selection. Stat. Surv. 4, 40–79 (2010).
Gelman, A., Goodrich, B., Gabry, J. & Vehtari, A. R-squared for Bayesian regression models. Am. Stat. 73, 307–309 (2019).
Fandos, G., Robinson, R. & Zurell, D. Code and data for: Simple mechanistic traits outperform complex syndromes in predicting avian dispersal distances. Zenodo https://doi.org/10.5281/zenodo.10713957 (2026).
Acknowledgements
D.Z. and G.F. received funding from the German Science Foundation DFG (grant no. ZU 361/1-1), G.F. also received funding from the Community of Madrid (Spain) and the Universidad Complutense de Madrid (Grant No. PR17/24-31914).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
G.F. and D.Z. designed research; G.F. performed research; G.F. analyzed data; G.F. and D.Z. conception and management of the project; G.F., R.R., and D.Z. wrote the paper. D.Z. acquired the funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fandos, G., Robinson, R.A. & Zurell, D. Simple mechanistic traits outperform complex syndromes in predicting avian dispersal distances. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09676-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09676-x