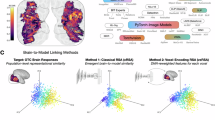
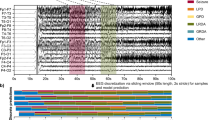
Abstract
Despite advancements in artificial intelligence, object recognition models still lag behind in emulating visual information processing in human brains. Recent studies have highlighted the potential of using neural data to mimic brain processing; however, these often rely on invasive neural recordings from non-human subjects, leaving a critical gap in understanding human visual perception. Addressing this gap, we present, ‘Re(presentational)Al(ignment)net’, a vision model aligned with human brain activity based on non-invasive EEG, demonstrating a significantly higher similarity to human brain representations. Our innovative image-to-brain multi-layer encoding framework advances human neural alignment by optimizing multiple model layers and enabling the model to efficiently learn and mimic the human brain’s visual representational patterns across object categories and different modalities. Our findings demonstrate that ReAlnets exhibit stronger alignment with human brain representations than traditional computer vision models, achieving an average similarity improvement of approximately 3% and a maximum relative improvement ratio reaching up to 40%. This alignment framework takes an important step toward bridging the gap between artificial and human vision and achieving more brain-like artificial intelligence systems.
Similar content being viewed by others
Data availability
The EEG data (THINGS EEG2 dataset) used are available as open data via the Open Science Framework (OSF) repository: https://osf.io/3jk45/35,70, and the fMRI data (Shen fMRI dataset) used are available as open data via the figshare repository: https://figshare.com/articles/Deep_Image_Reconstruction/703357736,71. The numerical Source data for all graphs in this paper can be found in Supplementary Dataset 1 and Dataset 2.
Code availability
The models and the analysis code can be assessed at https://github.com/ZitongLu1996/ReAlnet.
References
Lecun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Cichy, R. M., Khosla, A., Pantazis, D., Torralba, A. & Oliva, A. Comparison of deep neural networks to spatio-temporal cortical dynamics of human visual object recognition reveals hierarchical correspondence. Sci. Rep. 6, 1–13 (2016).
Güçlü, U. & van Gerven, M. A. Deep neural networks reveal a gradient in the complexity of neural representations across the ventral stream. J. Neurosci. 35, 10005–10014 (2015).
Kietzmann, T. C. et al. Recurrence is required to capture the representational dynamics of the human visual system. Proc. Natl. Acad. Sci. USA 116, 21854–21863 (2019).
Yamins, D. L. et al. Performance-optimized hierarchical models predict neural responses in higher visual cortex. Proc. Natl. Acad. Sci. USA 111, 8619–8624 (2014).
Lu, Z. & Golomb, J. Generate your neural signals from mine: individual-to-individual EEG converters. In Proc. Annual Meeting of the Cognitive Science Society (CogSci, 2023).
Rajalingham, R. et al. Large-scale, high-resolution comparison of the core visual object recognition behavior of humans, monkeys, and state-of-the-art deep artificial neural networks. J. Neurosci. 38, 7255–7269 (2018).
Kar, K., Kubilius, J., Schmidt, K., Issa, E. B. & DiCarlo, J. J. Evidence that recurrent circuits are critical to the ventral stream’s execution of core object recognition behavior. Nat. Neurosci. 22, 974–983 (2019).
Kubilius, J. et al. Brain-like object recognition with high-performing shallow recurrent ANNs. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2019).
Spoerer, C. J., McClure, P. & Kriegeskorte, N. Recurrent convolutional neural networks: a better model of biological object recognition. Front. Psychol. 8, 1551 (2017).
Tang, H. et al. Recurrent computations for visual pattern completion. Proc. Natl. Acad. Sci. USA 115, 8835–8840 (2018).
Bai, S., Li, Z. & Hou, J. Learning two-pathway convolutional neural networks for categorizing scene images. Multimed. Tools Appl. 76, 16145–16162 (2017).
Choi, M., Han, K., Wang, X., Zhang, Y. & Liu, Z. A dual-stream neural network explains the functional segregation of dorsal and ventral visual pathways in human brains. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2023).
Han, Z. & Sereno, A. Modeling the ventral and dorsal cortical visual pathways using artificial neural networks. Neural Comput. 34, 138–171 (2022).
Han, Z. & Sereno, A. Identifying and localizing multiple objects using artificial ventral and dorsal cortical visual pathways. Neural Comput. 35, 249–275 (2023).
Sun, T., Wang, Y., Yang, J. & Hu, X. Convolution neural networks with two pathways for image style recognition. IEEE Trans. Image Process. 26, 4102–4113 (2017).
Finzi, D., Margalit, E., Kay, K., Yamins, D. L. K. & Grill-Spector, K. Topographic DCNNs trained on a single self-supervised task capture the functional organization of cortex into visual processing streams. In Proc. NeurIPS 2022 Workshop SVRHM (NeurIPS, 2022).
Lee, H. et al. Topographic deep artificial neural networks reproduce the hallmarks of the primate inferior temporal cortex face processing network. Preprint at bioRxiv https://doi.org/10.1101/2020.07.09.185116 (2020).
Lu, Z. et al. End-to-end topographic networks as models of cortical map formation and human visual behaviour. Nat. Hum. Behav. 9, 1975–1991 (2025).
Margalit, E. et al. A unifying framework for functional organization in early and higher ventral visual cortex. Neuron. 112, 2435–2451 (2024).
Konkle, T. & Alvarez, G. Cognitive steering in deep neural networks via long-range modulatory feedback connections. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2023).
Konkle, T. & Alvarez, G. A. A self-supervised domain-general learning framework for human ventral stream representation. Nat. Commun. 13, 1–12 (2022).
Prince, J. S., Alvarez, G. A. & Konkle, T. Contrastive learning explains the emergence and function of visual category-selective regions. Sci. Adv. 10, eadl1776 (2024).
O’Connell, T. P. et al. Approximating Human-Level 3D visual inferences with deep neural networks. Open Mind. 9, 305–324 (2025).
Dapello, J. et al. Aligning model and macaque inferior temporal cortex representations improves model-to-human behavioral alignment and adversarial robustness. In Proc. International Conference on Learning Representations (ICLR, 2023).
Federer, C., Xu, H., Fyshe, A. & Zylberberg, J. Improved object recognition using neural networks trained to mimic the brain’s statistical properties. Neural Netw. 131, 103–114 (2020).
Li, Z. et al. Learning from brains how to regularize machines. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2019).
Pirlot, C., Gerum, R. C., Efird, C., Zylberberg, J. & Fyshe, A. Improving the accuracy and robustness of CNNs using a deep CCA neural data regularizer. Preprint at https://doi.org/10.48550/arXiv.2209.02582 (2022).
Safarani, S. et al. Towards robust vision by multi-task learning on monkey visual cortex. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2021).
Fong, R. C., Scheirer, W. J. & Cox, D. D. Using human brain activity to guide machine learning. Sci. Rep. 8, 1–10 (2018).
Spampinato, C. et al. Deep learning human mind for automated visual classification. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 6809–6817 (IEEE, 2017).
Palazzo, S. et al. Decoding brain representations by multimodal learning of neural activity and visual features. IEEE Trans. Pattern Anal. Mach. Intell. 43, 3833–3849 (2021).
Fu, K., Du, C., Wang, S. & He, H. Improved video emotion recognition with alignment of CNN and human brain representations. IEEE Trans. Affect. Comput. 14, 1–15 (2023).
Kubilius, J. et al. CORnet: modeling the neural mechanisms of core object recognition.Preprint at bioRxiv https://doi.org/10.1101/408385 (2018).
Gifford, A. T., Dwivedi, K., Roig, G. & Cichy, R. M. A large and rich EEG dataset for modeling human visual object recognition. NeuroImage 264, 119754 (2022).
Shen, G., Horikawa, T., Majima, K. & Kamitani, Y. Deep image reconstruction from human brain activity. PLoS Comput. Biol. 15, e1006633 (2019).
Schrimpf, M. et al. Brain-score: which artificial neural network for object recognition is most brain-like? Preprint at bioRxiv https://doi.org/10.1101/407007 (2020).
Teichmann, L., Hebart, M. N. & Baker, C. I. Dynamic representation of multidimensional object properties in the human brain. J. Neurosci. e1057252026, https://doi.org/10.1523/JNEUROSCI.1057-25.2026 (2026).
Khaligh-Razavi, S.-M., Cichy, R. M., Pantazis, D. & Oliva, A. Tracking the spatiotemporal neural dynamics of real-world object size and animacy in the human brain. J. Cogn. Neurosci. 30, 1559–1576 (2018).
Wang, R., Janini, D. & Konkle, T. Mid-level feature differences support early animacy and object size distinctions: evidence from electroencephalography decoding. J. Cogn. Neurosci. 34, 1670–1680 (2022).
Lu, Z. & Golomb, J. D. Human EEG and artificial neural networks reveal disentangled representations and processing timelines of object real-world size and depth in natural images. eLife 13, RP98117 (2025).
Geirhos, R. et al. Partial success in closing the gap between human and machine vision. In Proc. Advances in Neural Information Processing Systems (NeurIPS) Vol. 34 (Neural Information Processing Systems Foundation, Inc., 2021).
Hebart, M. N., Zheng, C. Y., Pereira, F. & Baker, C. I. Revealing the multidimensional mental representations of natural objects underlying human similarity judgements. Nat. Hum. Behav. 4, 1173–1185 (2020).
Shao, Z. et al. Probing Human Visual Robustness with Neurally-Guided Deep Neural Networks. Preprint at https://doi.org/10.48550/arXiv.2405.02564 (2025).
McMahon, E., Bonner, M. F. & Isik, L. Hierarchical organization of social action features along the lateral visual pathway. Curr. Biol. 33, 5035–5047.e8 (2023).
Bao, P., She, L., McGill, M. & Tsao, D. Y. A map of object space in primate inferotemporal cortex. Nature 583, 103–108 (2020).
Jagadeesh, A. V. & Gardner, J. L. Texture-like representation of objects in human visual cortex. Proc. Natl. Acad. Sci. USA 119, e2115302119 (2022).
Cichy, R. M. & Oliva, A. A M/EEG-fMRI fusion primer: resolving human brain responses in space and time. Neuron 107, 772–781 (2020).
Lee Masson, H. & Isik, L. Rapid processing of observed touch through social perceptual brain regions: an EEG-fMRI fusion study. J. Neurosci. 43, 7700–7711 (2023).
Hu, Y. & Mohsenzadeh, Y. Neural processing of naturalistic audiovisual events in space and time. Commun. Biol. 8, 1–16 (2025).
Conwell, C., Prince, J. S., Kay, K. N., Alvarez, G. A. & Konkle, T. A large-scale examination of inductive biases shaping high-level visual representation in brains and machines. Nat. Commun. 15, 1–18 (2024).
Muukkonen, I. & Salmela, V. Entropy predicts early MEG, EEG and fMRI responses to natural images. Preprint at bioRxiv https://doi.org/10.1101/2023.06.21.545883 (2023).
Li, D., Wei, C., Li, S., Zou, J. & Liu, Q. Visual decoding and reconstruction via EEG embeddings with guided diffusion. In Proc. Advances in Neural Information Processing Systems (NeurIPS, 2024).
Du, C., Fu, K., Li, J. & He, H. Decoding visual neural representations by multimodal learning of brain-visual-linguistic features. IEEE Trans. Pattern Anal. Mach. Intell. 45, 10760–10777 (2023).
Song, Y. et al. Decoding natural images from eeg for object recognition. In Proc. International Conference on Learning Representations (ICLR, 2024).
Ayzenberg, V., Blauch, N. & Behrmann, M. Using deep neural networks to address the how of object recognition. Preprint at https://doi.org/10.31234/osf.io/6gjvp (2023).
Cichy, R. M. & Kaiser, D. Deep neural networks as scientific models. Trends Cogn. Sci. 23, 305–317 (2019).
Doerig, A. et al. The neuroconnectionist research programme. Nat. Rev. Neurosci. 24, 431–450 (2023).
Kanwisher, N., Khosla, M. & Dobs, K. Using artificial neural networks to ask ‘why’ questions of minds and brains. Trends Neurosci. 46, 240–254 (2023).
Lu, Z. & Ku, Y. Bridging the gap between EEG and DCNNs reveals a fatigue mechanism of facial repetition suppression. iScience 26, 108501 (2023).
Hebart, M. N. et al. THINGS: a database of 1,854 object concepts and more than 26,000 naturalistic object images. PLoS ONE 14, 1–24 (2019).
Kriegeskorte, N., Mur, M. & Bandettini, P. Representational similarity analysis-connecting the branches of systems neuroscience. Front. Syst. Neurosci. 2, 249 (2008).
Nili, H. et al. A toolbox for representational similarity analysis. PLOS Comput. Biol. 10, e1003553 (2014).
Cichy, R. M., Pantazis, D. & Oliva, A. Resolving human object recognition in space and time. Nat. Neurosci. 17, 455–462 (2014).
Grootswagers, T., Wardle, S. G. & Carlson, T. A. Decoding dynamic brain patterns from evoked responses: a tutorial on multivariate pattern analysis applied to time series neuroimaging data. J. Cogn. Neurosci. 29, 677–697 (2017).
Xie, S., Kaiser, D. & Cichy, R. M. Visual imagery and perception share neural representations in the alpha frequency Band. Curr. Biol. 30, 2621–2627 (2020).
Kappenman, E. S. & Luck, S. J. The effects of electrode impedance on data quality and statistical significance in ERP recordings. Psychophysiology 47, 888–904 (2010).
Luck, S. J. An Introduction to the Event-Related Potential Technique 2nd edn (MIT Press, 2014).
Lu, Z. & Ku, Y. NeuroRA: a Python toolbox of representational analysis from multi-modal neural data. Front. Neuroinformatics 14, 61 (2020).
Gifford, A. T., Dwivedi, K., Roig, G. & Cichy, R. M. A large and rich EEG dataset for modeling human visual object recognition https://osf.io/3jk45/ (2022).
Shen, G. et al. Deep image reconstruction dataset https://figshare.com/articles/Deep_Image_Reconstruction/7033577 (2019).
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01-EY025648) and the National Science Foundation (NSF 1848939) to Julie D. Golomb. We thank the Ohio Supercomputer Center and Georgia Stuart for providing the essential computing resources and support. We thank Yuxuan Zeng for the “ReAlnet” name suggestion. We thank Tianyu Zhang, Shuai Chen, Jiaqi Li, and some other members in the Memory and Perception Reviews Reading Group (RRG) for helpful discussions about the methods and results. We thank Yuxin Wang for constructive feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: Z.L. Formal analysis: Z.L., and Y.W. Funding acquisition: J.D.G. Investigation: Z.L., and Y.W. Methodology: Z.L., and Y.W. Resources: J.D.G. Project administration: Z.L. Visualization: Z.L. Writing—original draft preparation: Z.L. Writing—review and editing: Z.L., Y.W., and J.D.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Ilya Kuzovkin, Bhavin Choksi and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Jasmine Pan. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lu, Z., Wang, Y. & Golomb, J.D. Achieving more human brain-like vision via human EEG representational alignment. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09685-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09685-w