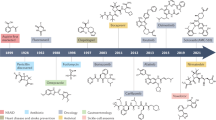
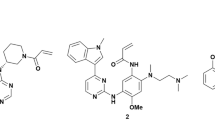
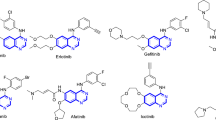
Abstract
Covalent drugs have long played an essential role in therapeutics, yet computational design approaches remain largely confined to virtual screening of existing libraries. Despite recent advances in deep generative models for drug discovery, methods specifically tailored to de novo covalent drug generation are still lacking. Here we introduce CovaGEN, a conditional latent diffusion framework for the de novo design of covalent inhibitors with enhanced drug-likeness and safety. CovaGEN generates ligands from a drug-like latent space while conditioning on target sequences and employing a classifier to guide the formation of desirable covalent warheads. A reinforcement learning strategy further optimizes the safety profiles of generated molecules. Experimental results demonstrate that CovaGEN effectively generates covalent drugs with the desired covalent warheads, exhibiting strong target protein affinity, favorable drug-likeness, and low toxicity. When applied to EGFR T790M and Mpro, the generated compounds exhibit higher probabilities of covalent binding. Overall, CovaGEN offers a pioneering approach for the de novo design of covalent inhibitors, advancing the discovery of covalent drugs with improved properties.
Similar content being viewed by others
Data availability
Source data for Figs. 2–5 are provided with this paper in Supplementary Data. All of the datasets used in this study are publicly available. The molecules used for the training of the molecular VAE and CovaGEN-cond are downloaded from the ZINC database (https://zinc.docking.org/). The raw data of the CrossDocked 2020 dataset were obtained from https://github.com/gnina/models/tree/master/data/CrossDocked2020. The small mouse intraperitoneal LD50 subdataset was obtained from TOXRIC.
Code availability
The source codes are available on GitHub: https://github.com/BioChemAI/CovaGENand deposited on Zenodo at https://doi.org/10.5281/zenodo.18374022(ref. 40).
References
Boike, L., Henning, N. J. & Nomura, D. K. Advances in covalent drug discovery. Nat. Rev. Drug Discov. 21, 881–898 (2022).
Lu, W. et al. Fragment-based covalent ligand discovery. RSC Chem. Biol. 2, 354–367 (2021).
Rachman, M. et al. Duckcov: a dynamic undocking-based virtual screening protocol for covalent binders. ChemMedChem 14, 1011–1021 (2019).
Soulère, L., Barbier, T. & Queneau, Y. Docking-based virtual screening studies aiming at the covalent inhibition of SARS-CoV-2 Mpro by targeting the cysteine 145. Comput. Biol. Chem. 92, 107463 (2021).
Zeng, X. et al. Deep generative molecular design reshapes drug discovery. Cell Rep. Med. 3, 100794 (2022).
Segler, M. H., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Gupta, A. et al. Generative recurrent networks for de novo drug design. Mol. Inform. 37, 1700111 (2018).
Jin, W., Barzilay, R., Jaakkola, T., Dy, J. & Krause, A. Junction tree variational autoencoder for molecular graph generation. In Proc. 35th International Conference on Machine Learning, Vol. 80 (eds. Dy, J. & Krause, A.) 2323–2332 (PMLR, 2018). https://proceedings.mlr.press/v80/jin18a.html.
Liu, Q., Allamanis, M., Brockschmidt, M. & Gaunt, A. Constrained graph variational autoencoders for molecule design. Adv. Neural Inf. Process. Syst. 31, 7806–7815 (2018).
De Cao, N. & Kipf, T. Molgan: An implicit generative model for small molecular graphs. In Proc. ICML 2018 Workshop on Theoretical Foundations and Applications of Deep Generative Models, Vol. 80, (PMLR, 2018)
Maziarka, Ł et al. Mol-cyclegan: a generative model for molecular optimization. J. Cheminform. 12, 2 (2020).
Schneuing, A. et al. Structure-based drug design with equivariant diffusion models. Nat. Comput. Sci. 4, 899–909 (2024).
Huang, L. et al. A dual diffusion model enables 3d molecule generation and lead optimization based on target pockets. Nat. Commun. 15, 2657 (2024).
Liebler, D. C. & Guengerich, F. P. Elucidating mechanisms of drug-induced toxicity. Nat. Rev. Drug Discov. 4, 410–420 (2005).
Dollar, O., Joshi, N., Beck, D. A. & Pfaendtner, J. Attention-based generative models for de novo molecular design. Chem. Sci. 12, 8362–8372 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Polykovskiy, D. et al. Molecular sets (moses): a benchmarking platform for molecular generation models. Front. Pharmacol. 11, 565644 (2020).
Prykhodko, O. et al. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform. 11, 1–13 (2019).
Hoogeboom, E., Satorras, V. G., Vignac, C. & Welling, M. Chaudhuri, K. et al. Equivariant diffusion for molecule generation in 3D. In Proc. 39th International Conference on Machine Learning (eds Chaudhuri, K. et al.) 8867–8887 (PMLR, 2022). https://proceedings.mlr.press/v162/hoogeboom22a.html.
Durant, J. L., Leland, B. A., Henry, D. R. & Nourse, J. G. Reoptimization of mdl keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 42, 1273–1280 (2002).
Francoeur, P. G. et al. Three-dimensional convolutional neural networks and a cross-docked data set for structure-based drug design. J. Chem. Inf. Model. 60, 4200–4215 (2020).
Alhossary, A., Handoko, S. D., Mu, Y. & Kwoh, C.-K. Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics 31, 2214–2216 (2015).
Luo, S., Guan, J., Ma, J. & Peng, J. A 3d generative model for structure-based drug design. Adv. Neural Inf. Process. Syst. 34, 6229–6239 (2021).
Peng, X. et al. Chaudhuri, K. et al. Pocket2Mol: efficient molecular sampling based on 3D protein pockets. InProc. 39th International Conference on Machine Learning (eds Chaudhuri, K. et al.) Vol. 162, 17644–17655 (PMLR, 2022). https://proceedings.mlr.press/v162/peng22b.html.
Dhariwal, P. & Nichol, A. Diffusion models beat gans on image synthesis. Adv. Neural Inf. Process. Syst. 34, 8780–8794 (2021).
Singh, J., Petter, R. C., Baillie, T. A. & Whitty, A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011).
Fan, T., Sun, G., Zhao, L., Cui, X. & Zhong, R. Qsar and classification study on prediction of acute oral toxicity of n-nitroso compounds. Int. J. Mol. Sci. 19, 3015 (2018).
Uribe, M. L., Marrocco, I. & Yarden, Y. Egfr in cancer: signaling mechanisms, drugs, and acquired resistance. Cancers 13, 2748 (2021).
Jin, Z. et al. Structure of mpro from sars-cov-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).
Hongyu, H. et al. The binding mechanism of failed, in processing and succeed inhibitors target sars-cov-2 main protease. J. Biomol. Struct. Dyn. 42, 10565–10576 (2024).
Irwin, J. J. & Shoichet, B. K. Zinc- a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Lipinski, C. A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today. Technol. 1, 337–341 (2004).
Rombach, R., Blattmann, A., Lorenz, D., Esser, P. & Ommer, B. High-resolution image synthesis with latent diffusion models. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 10684–10695 (IEEE, 2022).
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 6000–6010 (2017).
Luo, S. et al. Antigen-specific antibody design and optimization with diffusion-based generative models for protein structures. Adv. Neural Inf. Process. Syst. 35, 9754–9767 (2022).
Steinegger, M. & Söding, J. Mmseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Black, K., Janner, M., Du, Y., Kostrikov, I. & Levine, S. Training diffusion models with reinforcement learning. In Proc. International Conference on Learning Representations https://openreview.net/forum?id=YCWjhGrJFD (2024).
Wu, L. et al. Toxric: a comprehensive database of toxicological data and benchmarks. Nucleic Acids Res. 51, D1432–D1445 (2023).
Zhang, W. & Li, P. CovaGen: De novo covalent drug generation with enhanced drug-likeness and safety. Zenodo https://doi.org/10.5281/zenodo.18374022 (2026).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grants 62572374, U22A2037, 62202353 and 62132015), and the Natural Science Basic Research Program of Shaanxi Province (2023-JC-QN-0707).
Author information
Authors and Affiliations
Contributions
P.L. and L.G. conceived and supervised the research project. W.Z. and P.L. jointly designed and implemented the overall framework. W.Z. and T.L. conducted the experiments and led the manuscript writing. T.L., X.D., and S.S. contributed to model training, and evaluation. X.Y. provided expert guidance on covalent drug design and revised the manuscript. All authors discussed the results and contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Feixiong Cheng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Laura Rodríguez Pérez. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, W., Liu, T., Dong, X. et al. De novo covalent drug generation with enhanced drug-likeness and safety. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09725-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09725-5