Abstract
The Tibetan sheep is an ideal model animal for investigating adaptations to hypoxia and an important livestock species; however, the reproductive mechanisms that characterize its adaptive responses in extreme environments remain poorly understood. In this study, we employ single-nucleus RNA sequencing to characterize the transcriptomic landscape of the Tibetan sheep testis at four key developmental stages: newborn, pre-sexual maturity, and post-sexual maturity to adulthood. We constructed a single-nucleus transcriptomic atlas and identified two distinct subtypes of spermatogonial stem cells (SSCs): quiescent and active. Notably, we observed that pre-sexually mature Sertoli cells exhibit three distinct transcriptional states. Furthermore, we also identified a transitional state Sertoli cell that links immature and mature Sertoli cells. Analysis of testicular intercellular communication networks provides evidence for understanding somatic-germ cell interactions during spermatogenesis. Our study provides a comprehensive atlas of Tibetan sheep testicular development, revealing key insights into the dynamic changes and regulatory mechanisms of spermatogenesis and somatic cell maturation from birth to adulthood. These findings contribute to a detailed understanding of the genetic mechanisms underlying testicular development in Tibetan sheep and offer valuable insights and references for the study and comprehension of reproductive adaptations in other highland male domestic animals.
Similar content being viewed by others
Data availability
Single-nucleus transcriptomic datasets generated in this study are available in the Genome Sequence Archive (GSA) under accession number PRJCA035469. Source data underlying all graphs are provided in Supplementary Data 7. All additional data supporting the findings of this study are provided in Supplementary Data 1–6.
Code availability
Details of publicly available software used in the study are given in the Methods section. No custom code was used in the study.
References
Yang, J. et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations toextreme environments. Mol. Biol. Evol. 33, 2576–2592 (2016).
Yan, Z. et al. A time-resolved multi-omics atlas of transcriptional regulation in response to high-altitudehypoxia across whole-body tissues. Nat. Commun. 15, 3970 (2024).
Han, B. et al. Multiomics analyses provide new insight into genetic variation of reproductiveadaptability in Tibetan sheep. Mol. Biol. Evol. 41, https://doi.org/10.1093/molbev/msae058 (2024).
Morimoto, H. et al. ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell 12, 774–786 (2013).
Nishimura, H. & L’Hernault, S. W. Spermatogenesis. Curr. Biol. 27, R988–R994 (2017).
Guo, J. et al. Single-cell analysis of the developing human testis reveals somatic niche cell specificationand fetal germline stem cell establishment. Cell Stem Cell 28, 764–778 (2021).
Lord, T., Oatley, M. J. & Oatley, J. M. Testicular architecture is critical for mediation of retinoic acidresponsiveness by undifferentiated spermatogonial subtypes in the mouse. Stem Cell Rep 10, 538–552 (2018).
Tan, J. H. L. et al. Infertility-causing haploinsufficiency reveals TRIM28/KAP1 requirement inspermatogonia. Stem. Cell Rep. 14, 818–827 (2020).
De Rooij, D. G. Proliferation and differentiation of spermatogonial stem cells. Reproduction 121, 347–354 (2001).
Gassei, K., Valli, H. & Orwig, K. E. Whole-mount immunohistochemistry to study spermatogonial stemcells and spermatogenic lineage development in mice, monkeys, and humans. Methods Mol. Biol. 1210, 193–202 (2014).
Guo, J. et al. The dynamic transcriptional cell atlas of testis development during human puberty. CellStem Cell 26, 262–276 (2020).
Green, C. D. et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46, 651–667 (2018).
Zhao, L. et al. Single-cell analysis of developing and azoospermia human testicles reveals central roleof Sertoli cells. Nat. Commun. 11, 5683 (2020).
Wang, X. et al. Single-cell transcriptomic and cross-species comparison analyses reveal distinctmolecular changes of porcine testes during puberty. Commun. Biol. 7, 1478 (2024).
Tian, Y. et al. Single-cell RNA sequencing of the Mongolia sheep testis reveals a conserved anddivergent transcriptome landscape of mammalian spermatogenesis. FASEB J. 36, e22348 (2022).
Yang, H. et al. Characterization of sheep spermatogenesis through single-cell RNA sequencing. FASEB J. 35, e21187 (2021).
Su, J. et al. A dynamic transcriptional cell atlas of testes development after birth in Hu sheep. BMC Biol. 23, 78 (2025).
Mipam, T. et al. Single-cell transcriptome analysis and in vitro differentiation of testicular cells revealnovel insights into male sterility of the interspecific hybrid cattle-yak. BMC Genomics 24, 149 (2023).
Zhang, Y. W. et al. Single-cell analysis identifies critical regulators of spermatogonial development anddifferentiation in cattle-yak bulls. J. Dairy Sci. 107, 7317–7336 (2024).
Huang, D. et al. A single-nucleus transcriptomic atlas of primate testicular aging reveals exhaustion ofthe spermatogonial stem cell reservoir and loss of Sertoli cell homeostasis. Protein Cell 14, 888–907 (2023).
Murat, F. et al. The molecular evolution of spermatogenesis across mammals. Nature 613, 308–316 (2023).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Rotta, R., Noack, A. Multilevel local search algorithms for modularity clustering. ACM J. Exp.Algorithmics 16, https://doi.org/10.1145/1963190.1970376 (2011).
van der Maaten, L. J. P. & Hinton, G. E. Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605 (2008).
Zhang, G. et al. Single-cell and spatial transcriptomic investigation reveals the spatiotemporalspecificity of the beta-defensin gene family during mouse sperm maturation. Cell Commun. Signal 22, 267 (2024).
Chen, Q. et al. CNOT3 is required for male germ cell development and spermatogonial stem cellmaintenance. Development 152, https://doi.org/10.1242/dev.204557 (2025).
Griswold, M. D. Spermatogenesis: the commitment to meiosis. Physiol. Rev. 96, 1–17 (2016).
Ribeiro, J. & Crossan, G. P. GCNA is a histone binding protein required for spermatogonial stem cellmaintenance. Nucleic Acids Res. 51, 4791–4813 (2023).
Sohni, A. et al. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 26, 1501–1517 (2019).
Papetti, M. & Augenlicht, L. H. MYBL2, a link between proliferation and differentiation in maturingcolon epithelial cells. J. Cell Physiol. 226, 785–791 (2011).
Boheler, K. R. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatinstate and a checkpoint deficient cell cycle. J. Cell Physiol. 221, 10–17 (2009).
Shami, A. N. et al. Single-cell RNA sequencing of human, macaque, and mouse testes uncoversconserved and divergent features of mammalian spermatogenesis. Dev. Cell 54, 529–547 (2020).
Hermann, B. P. et al. The Mammalian spermatogenesis single-cell transcriptome, from spermatogonialstem cells to spermatids. Cell Rep. 25, 1650–1667 (2018).
Guo, J. et al. The adult human testis transcriptional cell atlas. Cell Res. 28, 1141–1157 (2018).
Wang, M. et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition duringhuman spermatogenesis. Cell Stem Cell 23, 599–614 (2018).
Arnold, K. et al. Sox2+ adult stem and progenitor cells are important for tissue regeneration andsurvival of mice. Cell Stem Cell 9, 317–329 (2011).
La, H. M. et al. Identification of dynamic undifferentiated cell states within the male germline. Nat.Commun. 9, 2819 (2018).
Adikusuma, F., Pederick, D., McAninch, D., Hughes, J. & Thomas, P. Functional equivalence of theSOX2 and SOX3 transcription factors in the developing mouse brain and testes. Genetics 206, 1495–1503 (2017).
Givelet, M. et al. Transcriptional profiling of β-2M(-)SPα-6(+)THY1(+) spermatogonial stem cells inhuman spermatogenesis. Stem. Cell Rep 17, 936–952 (2022).
Wang, S. et al. scRNA-seq and scATAC-seq reveal that Sertoli cell mediates spermatogenesis disordersthrough stage-specific communications in non-obstructive azoospermia. Elife 13, https://doi.org/10.7554/eLife.97958 (2025).
Lin, W. et al. Isoform-resolution single-cell RNA sequencing reveals the transcriptional panorama ofadult Baoshan pig testis cells. BMC Genomics 26, 459 (2025).
Litchfield, K. et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ celltumours. Nat. Commun. 6, 5973 (2015).
Ernst, C., Odom, D. T. & Kutter, C. The emergence of piRNAs against transposon invasion to preservemammalian genome integrity. Nat. Commun. 8, 1411 (2017).
Nie, X. et al. Single-cell analysis of human testis aging and correlation with elevated body mass index. Dev. Cell 57, 1160–1176 (2022).
Kubota, H. et al. Glial cell line-derived neurotrophic factor and endothelial cells promote self-renewalof rabbit germ cells with spermatogonial stem cell properties. FASEB J. 25, 2604–2614 (2011).
Lau, X., Munusamy, P., Ng, M. J. & Sangrithi, M. Single-cell RNA sequencing of the cynomolgusmacaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev.Cell 54, 548–566 (2020).
Tan, H. et al. Single-cell RNA-seq uncovers dynamic processes orchestrated by RNA-binding proteinDDX43 in chromatin remodeling during spermiogenesis. Nat. Commun. 14, 2499 (2023).
Fliegauf, M. et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primaryciliary dyskinesia. Am. J. Respir. Crit. Care Med. 171, 1343–1349 (2005).
Su, J. et al. Dosage compensation of the X chromosome during sheep testis development revealed bysingle-cell RNA sequencing. Animals 12, https://doi.org/10.3390/ani12172169 (2022).
Whelan, E. C. et al. Single-cell multiomic comparison of mouse and rat spermatogenesis reveals generegulatory networks conserved for over 20 million years. Stem Cell Rep. 20, 102449 (2025).
Yi, S. et al. Tcf12, A member of basic helix-loop-helix transcription factors, mediates bone marrowmesenchymal stem cell osteogenic differentiation in vitro and in vivo. Stem Cells 35, 386–397 (2017).
Takubo, K. et al. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNAdamage-induced cell-cycle arrest. Cell Stem Cell 2, 170–182 (2008).
Unhavaithaya, Y. et al. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cellself-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507–6519 (2009).
Wang, X. et al. A homozygous PIWIL2 frameshift variant affects the formation and maintenance ofhuman-induced pluripotent stem cell-derived spermatogonial stem cells and causes Sertoli cell-onlysyndrome. Stem Cell Res. Ther. 13, 480 (2022).
Lei, N. et al. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 77, 466–475 (2007).
Li, C. H. et al. Long-term propagation of tree shrew spermatogonial stem cells in culture andsuccessful generation of transgenic offspring. Cell Res. 27, 241–252 (2017).
Law, N. C., Oatley, M. J. & Oatley, J. M. Developmental kinetics and transcriptome dynamics of stemcell specification in the spermatogenic lineage. Nat. Commun. 10, 2787 (2019).
Dann, C. T. et al. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated duringretinoic acid-induced differentiation. Stem Cells 26, 2928–2937 (2008).
Buaas, F. W. et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 36, 647–652 (2004).
Zhang, T., Oatley, J., Bardwell, V. J. & Zarkower, D. DMRT1 is required for mouse spermatogonial stemcell maintenance and replenishment. PLoS Genet 12, e1006293 (2016).
Suzuki, H. et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 361, 301–312 (2012).
Li, Z. et al. Transcriptional priming as a conserved mechanism of lineage diversification in thedeveloping mouse and human neocortex. Sci. Adv. 6, eabd2068 (2020).
Delile, J. et al. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression inthe developing mouse spinal cord. Development 146, https://doi.org/10.1242/dev.173807 (2019).
Valli, H. et al. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrichhuman spermatogonial stem cells. Fertil. Steril. 102, 566–580 (2014).
Bardot, E. et al. Foxa2 identifies a cardiac progenitor population with ventricular differentiationpotential. Nat. Commun. 8, 14428 (2017).
Lord, T. & Nixon, B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev.Cell 52, 399–411 (2020).
Suzuki, S., McCarrey, J. R. & Hermann, B. P. An mTORC1-dependent switch orchestrates the transitionbetween mouse spermatogonial stem cells and clones of progenitor spermatogonia. Cell Rep 34, 108752 (2021).
Hyakutake, K. et al. Asymmetrical allocation of JAK1 mRNA during spermatogonial stem cell division inXenopus laevis. Dev. Growth Differ. 57, 389–399 (2015).
Wang, H. Q. et al. Meiotic transcriptional reprogramming mediated by cell-cell communications inhumans and mice revealed by scATAC-seq and scRNA-seq. Zool. Res. 45, 601–616 (2024).
Di Persio, S. & Neuhaus, N. Human spermatogonial stem cells and their niche in male (in) fertility:novel concepts from single-cell RNA-sequencing. Hum. Reprod. 38, 1–13 (2023).
Handel, M. A. & Schimenti, J. C. Genetics of mammalian meiosis: regulation, dynamics and impact onfertility. Nat. Rev. Genet. 11, 124–136 (2010).
Suzuki, S., Diaz, V. D. & Hermann, B. P. What has single-cell RNA-seq taught us about mammalianspermatogenesis? Biol. Reprod. 101, 617–634 (2019).
Li, L. et al. A novel frameshift mutation in ubiquitin-specific protease 26 gene in a patient with severeoligozoospermia. Biosci. Rep. 40, https://doi.org/10.1042/bsr20191902 (2020).
Zhao, Y. et al. Connexin-43 is a promising target for lycopene preventing phthalate-inducedspermatogenic disorders. J. Adv. Res. 49, 115–126 (2023).
Mays-Hoopes, L. L., Bolen, J., Riggs, A. D. & Singer-Sam, J. Preparation of spermatogonia,spermatocytes, and round spermatids for analysis of gene expression using fluorescence-activatedcell sorting. Biol. Reprod. 53, 1003–1011 (1995).
Sabari, B. R., Zhang, D., Allis, C. D. & Zhao, Y. Metabolic regulation of gene expression through histoneacylations. Nat. Rev. Mol. Cell Biol. 18, 90–101 (2017).
Ward, W. S. Function of sperm chromatin structural elements in fertilization and development. Mol.Hum. Reprod. 16, 30–36 (2010).
Liu, C. et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 12, 814–832 (2016).
Gewaily, M. S. et al. Comparative expression of cell adhesion molecule1 (CADM1) in the testes ofexperimental mice and some farm animals. Acta Histochem 122, 151456 (2020).
Cai, Z. et al. ALKBH5 in mouse testicular Sertoli cells regulates CDH2 mRNA translation to maintainblood-testis barrier integrity. Cell Mol. Biol. Lett. 27, 101 (2022).
Mruk, D. D. & Cheng, C. Y. The mammalian blood-testis barrier: its biology and regulation. Endocr. Rev. 36, 564–591 (2015).
Simorangkir, D. R. et al. Sertoli cell differentiation in rhesus monkey (Macaca mulatta) is an earlyevent in puberty and precedes attainment of the adult complement of undifferentiatedspermatogonia. Reproduction 143, 513–522 (2012).
Meroni, S. B. et al. Molecular mechanisms and signaling pathways involved in Sertoli cell proliferation. Front Endocrinol 10, 224 (2019).
Ou, Y. et al. Primary cilia in the developing pig testis. Cell Tissue Res 358, 597–605 (2014).
Li, X. et al. Genomic analyses of wild argali, domestic sheep, and their hybrids provide insights intochromosome evolution, phenotypic variation, and germplasm innovation. Genome Res. 32, 1669–1684 (2022).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNAsequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Camp, J. G. et al. Multilineage communication regulates human liver bud development frompluripotency. Nature 546, 533–538 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporalordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cellcommunication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Acknowledgements
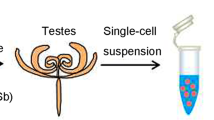
This work was supported by the National Natural Science Foundation of China (No. 32260833 & No. 32202675), and Discipline Team Project of Gansu Agricultural University (NO. GAU-XKTD-2022-20). We thank the Guangzhou Genedenovo Biotechnology Co., Ltd., for assisting in single-nucleus RNA sequencing. Schematic illustration elements in Figs. 1C, F, 3E, 4D, and 8F were created using Figdraw (www.figdraw.com), an online platform for scientific illustration. The use of these illustration elements complies with the platform’s Terms of Use for academic publishing. License information (Agreement No.) is as follows: Fig. 1C, SSUYUdd744; Fig. 1F, PRRSOc2e8d; Fig. 3E, OIPTOb8ffc; Fig. 4D, YUPUPae06c; Fig. 8F, YSRAO2ad8d.
Author information
Authors and Affiliations
Contributions
Conceptualization: H.H.W., Y.J.M. Methodology: H.H.W., D.P.L., Z.L.L., C.H.W. Investigation: X.X.Z., Y.J.Z., T.T.L., T.Z.S. Formal analysis: H.H.W., T.T.L. Visualization: H.H.W. Supervision: T.Z.S., Y.J.M. Writing—original draft: H.H.W. Writing—review & editing: H.H.W., T.T.L., Y.J.M. Funding acquisition: T.T.L., Y.J.M. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: David Favero.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Hh., Li, Tt., Li, Dp. et al. Single-nucleus RNA sequencing provides insights into the genetic mechanisms underlying reproductive adaptability in Tibetan sheep (Ovis aries). Commun Biol (2026). https://doi.org/10.1038/s42003-026-09729-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09729-1